A FRET Flow Cytometry Technique to Detect Tau-Seed Induced Reporter Protein Aggregation
Abstract
Source: Furman, J. L., et al. Sensitive Detection of Proteopathic Seeding Activity with FRET Flow Cytometry. J. Vis. Exp. (2015).
The video demonstrates a fluorescence resonance energy transfer (FRET) flow cytometry assay to detect the seeding activity of protein aggregates isolated from biological samples. Mammalian cells expressing tau reporter proteins are incubated with liposome transduction complexes containing tau seeds. These seeds mediate the aggregation of the reporter proteins, leading to generating a FRET positive signal in the flow cytometer.
Protocol
NOTE: This protocol emphasizes the use of FRET flow cytometry for detecting seeding activity from mouse biological samples. It is also compatible with recombinant fibrils and human biological samples. Mouse euthanasia and brain harvesting was performed in accordance with IACUC-approved procedures.
1. Brain Extraction
- Following deep anesthetization with isoflurane (2%), perfuse a mouse with ice-cold PBS containing 0.03% heparin, and extract the brain following the details described in Gage et al.
- Place extracted tissue in a cryo-vial, and snap freeze by placing in liquid nitrogen. Alternatively, freeze tissue on dry ice. Once frozen, transfer tissue to -80 °C and store for an extended period of time, if necessary.
NOTE: This protocol is compatible with whole brain homogenates or micro-dissected brain regions. See Hagihara et al. for a detailed micro-dissection protocol.
2. Preparation of Biological Seed Material
- Prepare homogenization buffer by dissolving protease inhibitors (ethylenediaminetetraacetic acid (EDTA) free) into 1x TBS (Tris-buffered saline, 1 tablet per 10 ml). Vortex vigorously, and store on ice. Protease inhibitors must be EDTA free in order to preserve biosensor cell viability.
- Weigh frozen brain tissue in a disposable weigh boat, and transfer to a 5 ml conical tube. Add ice-cold homogenization buffer such that the final solution is 10% weight/volume. Temporarily store on ice. Tissue mass and subsequent homogenization buffer volume will vary depending on the brain size and/or regions used. Here, an adult mouse hemibrain weighing 0.2 g is suspended in 2 ml for a 10% w/vol solution.
- Transfer samples into a cold room, and adjust a probe sonicator to the appropriate settings. For probe sonication with an Omni Ruptor (shown here), set the power to 20%, which corresponds to approximately 75 W. Use a pulse mode to ensure samples are not over-heated during sonication. Set the pulser to 30%, corresponding to approximately 500 msec.
- Clean the tip of the probe sonicator by rinsing with water, isopropanol, and water again, wiping off the probe in between each solution. Be certain to rinse and wipe both the sides and bottom of the probe with lab-wipes.
- Working with one sample at a time, submerge the probe tip into the homogenization buffer, and start the sonicator. Using the power and pulser settings described above, deliver 25 total pulses. Ensure that tissue is completely in suspension. Be careful to avoid foaming of the sample during this step.
NOTE: Samples are susceptible to foaming if a) the total volume is low or b) the homogenate warms up. With this specific instrument, do not sonicate with volumes less than 250 µl. To ensure samples stay chilled, tubes may be stored on ice during this step. - Clean the probe by wiping away residual lysate, then deliver 10 pulses into a beaker of clean water. Rinse the sides and bottom of the probe with water, isopropanol, and water again (as in step 2.4), wiping dry in between each step. To avoid contamination, thoroughly rinse the probe in between samples.
NOTE: With tau aggregates, we have not found that more stringent cleaning methods are necessary for preventing sample-to-sample contamination. Other amyloids, however, may require additional care which has been extensively described in literature. - Store homogenates on ice until all samples have been processed.
- Spin homogenates at 21,300 x g for 15 min at 4 °C.
- Transfer the supernatant to a clean tube, taking care not to disturb the pellet. Discard the pellet. Aliquot the supernatant, and store lysates at -80 °C for further use. Avoid freeze/thaw cycles of the homogenates, however, as this reduces seeding efficiency.
3. Replating Biosensor Cells
NOTE: Use four cell lines for this assay: HEK 293T (cell line #1), RD-P301S-CFP (cell line #2), RD-P301S-YFP (cell line #3), and RD-P301S-CFP/YFP (cell line #4). Please see reference Table 1 for each cell line's contribution to the assay.
- In a sterile environment, aspirate culture medium. Rinse cells with warm phosphate-buffered saline (PBS), and aspirate. Trypsinize cells (3 ml of trypsin-EDTA (0.05%)) for 3 min, quench with warm culture medium (9 ml of DMEM, 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, 1% GlutaMax), and immediately transfer cells to a conical tube.
- Centrifuge cells at 1,000 x g for 5 min at RT. Aspirate medium, and resuspend cell pellet in warm culture medium.
- Using a hemocytometer, determine the cell density for each cell line. Cell density will vary depending on confluency at time of harvesting and resuspension volume. Resuspend cells harvested from a 10 cm dish at 90% confluency in 10 ml media, for cell density to be approximately 1 million cells/ml.
- Make a master mix of cells + media such that each well of a 96 well plate will contain 35,000 cells in 130 µl media (e.g., to make a master mix for 100 wells, resuspend 3.5 million cells in 13 ml media). Modify the cell number to fit individual experimental designs and timelines. For cell treatment ~18 hr after plating, add 35,000 cells to each well of a 96-well plate.
- Using a multi-channel pipet, slowly pipette 130 µl of master mix cell suspension into each well of a flat bottom, tissue culture-treated 96 well plate. While plating, place the pipet tip in the center of the well, not touching the bottom of the plate.
NOTE: Though plating format can be modified, making n = 4 wells for each of cell lines 1-3 per plate is recommended. The remainder of the plate (n = 84) is reserved for cell line #4 (biosensor cells). - Allow the cells to settle by leaving the plate undisturbed for 10 min at RT. Incubate O/N at 37 °C, 5% CO2, and ≥80% relative humidity.
4. Treating Cells
NOTE: The following day, when tau biosensor cells are 60-65% confluent, prepare seed transduction complexes as follows:
- In one tube, combine transfection reagent, such as Lipofectamine, with reduced-serum media, such as Opti-MEM, to make a master mix. Per well, add 1.25 µl transfection reagent and 8.75 µl reduced-serum media. Flick tube gently to mix, briefly spin down, and incubate for 5 min at RT.
- In another tube, combine seed material (aliquot of lysate from step 2.9) with reduced-serum media.
NOTE: Volume of seed material will vary according to the individual experiment. When choosing seed volume, take into consideration: abundance of seeds in a sample and potential toxicity. Crude homogenates can be toxic to cells. As such, use the lowest volume possible to monitor your desired effect. Previously tested volumes of lysate/well range from 1-5 µl, corresponding to 5-20 µg total protein. Ensure that the total volume per well (seeds + reduced-serum media) is 10 µl. - Combine contents from the two tubes described in 4.1 and 4.2, flick gently to mix, briefly spin down (1,000 x g, 5 sec), and incubate at RT for at least 20 min and up to 2 hr.
- Gently pipette 20 µl of transduction complex on the side of individual biosensor wells. Use three technical replicates, if possible. Return treated cells to the incubator for 24-48 hr. Incubate under the same conditions as described in step 3.6.
NOTE: The appropriate negative control condition for this setup is biosensor cells treated with empty liposomes (i.e., liposome reagent + reduced-serum media) because application of phospholipids introduces a slight shift in the fluorescence profile relative to biosensor cells unexposed to liposome reagent.
5. Harvesting Cells for FRET Flow Cytometry
NOTE: Before harvesting cells—generally 24-48 hr post-treatment—it is possible to get a preliminary readout of seeding activity using the green fluorescent protein (GFP) filter on a standard inverted fluorescence microscope. Cells treated without seed material (i.e., empty liposomes) will show diffuse fluorescence, whereas cells treated with seed material will show intense punctate and reticular intracellular inclusions (Figure 1A–B).
- Using a multi-channel pipet, aspirate all cell medium (150 µl) . Trypsinize (50 µl) cells for 5 min, and quench with 150 µl chilled culture medium. Do not include a PBS rinse prior to trypsinization at this step, as it may cause cell lifting and subsequent cell loss. Exclude any contaminating dead cells and debris during flow cytometry via gating strategies.
- Immediately after quenching, transfer cells to a 96 well round bottom plate, and centrifuge at 1,000 x g for 5 min at RT.
- Aspirate and discard medium, taking care to avoid disturbing the cell pellet.
- Gently, but thoroughly, resuspend the cell pellet in 50 µl 2% paraformaldehyde, and incubate for 10 min. Alternatively, run cells live, although fixation provides cleaner and more consistent results.
- Centrifuge cells at 1,000 x g for 5 min at RT. Aspirate and discard paraformaldehyde, taking care to avoid disturbing the cell pellet.
- Gently, but thoroughly, resuspend the cell pellet in 200 µl chilled flow cytometry buffer (Hanks' Balanced Salt Solution (HBSS), 1% FBS, 1 mM EDTA) and run the plate as soon as possible to avoid cell clumping.
6. FRET Flow Cytometry
NOTE: Use a flow cytometer such as the MACSQuant VYB, which is equipped with FRET-compatible laser lines and filter sets (Table 2). For each step within this section, click the well of interest using the software's 96 well template, and click "play" to begin sample uptake and flow. Make plots or statistics tables by clicking the 'new analysis window' icon. Change axis parameters on individual bivariate plots by clicking the title on either the X or Y axis and selecting the appropriate filter. To shift cell populations or fluorescence signals, increase or decrease the voltages associated with the appropriate filters. With this instrument, run <1,000 events/sec to ensure accurate single-cell monitoring.
- Make a Side Scatter-Area (SSC-A) vs Forward Scatter-Area (FSC-A) bivariate plot, and with cell line #1 running, adjust the SSC and FSC voltages until the cell population is in the lower left quadrant. Click the polygon tool, and define the cell population (Gate P1). For all gating parameters see Figure 2.
- Make a FSC-H (height) vs FSC-A bivariate plot, and apply Gate P1 to this plot by clicking "live" above the plot, and selecting P1. With cell line #1 running, click the polygon tool, and define single cells (Gate P2).
- Make 3 histogram plots, one for each of the filters of interest: CFP, YFP, and FRET. Apply Gate P2 to all of these plots by clicking "live" above the plots, and selecting P2. With cell line #1 running, adjust voltages such that the median cell population in the CFP, YFP, and FRET histograms are all between 0 and 1. To measure CFP and FRET, excite cells with the 405 nm laser, and capture fluorescence with a 450/50 nm and 525/50 nm filter, respectively. To measure YFP, excite cells with a 488 nm laser and capture fluorescence with a 525/50 nm filter. Laser and filter settings are further described in Table 2.
- Perform compensation for CFP spillover into the FRET and YFP channels.
NOTE: Compensation is the process of fluorescence spillover correction. Fluorescence spillover occurs whenever the fluorescence emission of one fluorochrome is detected within a filter designed to measure signal from another fluorochrome. With proper compensation, the CFP spillover fluorescence can be removed from the FRET and YFP channels.
NOTE: Compensation requires the presence of both fluorescence -positive and -negative cells. Thus, to compensate on CFP-positive cells (cell line #2), negative cells (cell line #1) must be added to the sample prior to the run.- Spike in 30 µl of cell line #1 suspension from a single well into 200 µl of cell line #2 suspension immediately prior to compensating.
- Click the "Instrument Settings" icon, then 'compensation' tab, and check 'matrix' to open the compensation table.
- Make a FRET-A vs CFP-A bivariate plot, and apply gate P2, as described in step 6.3. With cell lines 1+2 running, click the 'quadrant' icon and draw quadrants such that the fluorescence -negative and -positive populations are separated by the lower two quadrants (Gates LL3 and LR3, respectively).
- Create a statistics table that displays the median fluorescence intensity (MFI) of FRET for the LL3 and LR3 gates. Adjust the FRET parameter in the compensation matrix until the MFI of the FRET signal is equivalent between LL3 and LR3. Following this step, the MFI of the FRET signal is equal between unstained cells and CFP-positive cells, suggesting the absence of CFP spillover into the FRET channel.
- Make a YFP-A vs CFP-A bivariate plot, and apply gate P2, as described in step 6.3. Draw quadrants (LL4 and LR4) similar to that done in step 6.4.3.
- Create a statistics table that displays the MFI of YFP for LL4 and LR4. Adjust the YFP parameter in the compensation matrix until the MFI of the YFP signal is equivalent between LL4 and LR4. Following this step, the MFI of a YFP signal is equal between unstained cells and CFP-positive cells, suggesting the absence of CFP spillover into the YFP channel.
- Following gate setup and compensation, highlight wells of interest and run the remainder of the plate.
- After all samples have been run, export data files by clicking 'file', then 'copy'. Select the 'data files' tab, highlight the experiment folder, and click 'copy'.
Representative Results
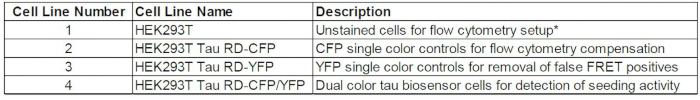
Table 1: Cell lines used with FRET flow cytometry. HEK 293T cells are used for flow cytometry setup. CFP single-positive cells are used for compensation (*). YFP single-positive cells are used to eliminate false FRET signal due to direct activation of YFP by 405 nm excitation. CFP/YFP dual-positive cells are FRET-compatible and serve as the biosensor cells.
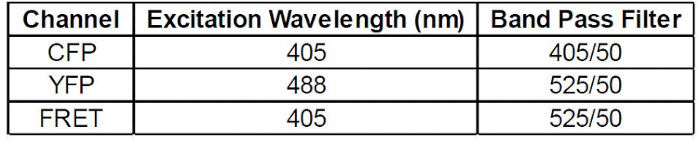
Table 2: Flow cytometer laser and filter settings.
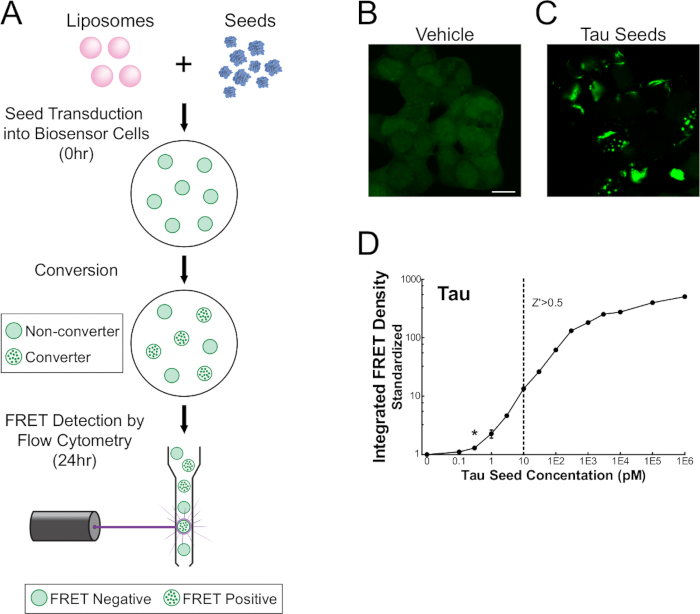
Figure 1. FRET flow cytometry sensitively detects tau seeding activity. Monoclonal HEK 293T cells expressing tau-RD-CFP/YFP were transduced with recombinant or biological samples, incubated for 24-48 hr, and analyzed on a single cell basis using flow cytometry (A). Unstimulated cells maintain tau-RD in a monomeric state (B), whereas cells treated with seed-containing material display prominent inclusions (C). Quantitative assessment (mean ± S.E.M.) of seeding activity shows that FRET flow cytometry is sensitive to femtomolar concentrations (monomer equivalent) of recombinant seed material and detection spans three orders of magnitude (D). Monomer equivalent represents the total amount of protein contained within the fibrillization reaction and does not correct for the incomplete incorporation of monomer into aggregated material. Thus, the concentration of actual aggregates (seeds) must be less than or equal to its 'monomer equivalent'. *Modified from Holmes and Furman et al.
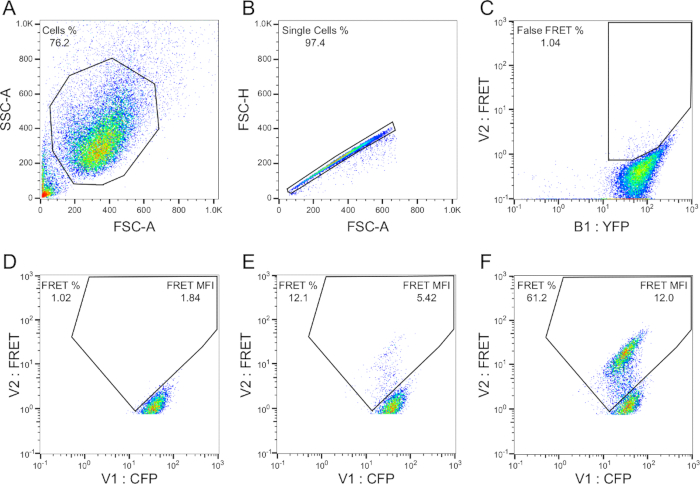
Figure 2. Gating strategy for FRET flow cytometry. Cell population (A) and singlet/doublet (B) gates are drawn with standard flow cytometry methodology. A false FRET gate (C) is drawn from YFP single-positive cells to eliminate YFP bleedthrough into the FRET filter. A FRET gate (D) is constructed from empty liposome-treated cells, such that background FRET is ≥1%. A population shift into the FRET gate appears following treatment with seed-positive material (E) and the shift becomes increasingly prominent with higher amounts of seed material (F). Final readouts include: percent FRET positivity, median fluorescence intensity (MFI) of FRET-positive events, and the integrated FRET density (Integrated FRET Density = Percent positive cells * MFI).
Disclosures
The authors have nothing to disclose.
Materials
| TBS | Sigma | T5912 | |
| Complete Protease Inhibitors (EDTA-free) | Roche | 4693159001 | |
| Cryo-vials | Sarstedt | 72.694.006 | |
| Analytical Balance | Mettler Toledo | XSE 105DU | |
| Weighing Boats | Fisher Scientific | 13-735-743 | |
| 15 mL conical tube | USA Scientific | 1475-0501 | |
| Omni Sonic Ruptor Ultrasonic Homogenizer | Omni International | 18-000-115 | |
| Micro-Tip for Ultrasonic Homogenizer | Omni International | OR-T-156 | |
| 2-Propanol | Fisher Scientific | A451 | |
| Noise Cancelling Ear Muffs | Fisher Scientific | 19-145-412 | |
| Kimwipes | Fisher Scientific | S47299 | |
| 1.5 mL tubes | USA Scientific | 1615-5510 | |
| Microcentrifuge | Eppendorf | 5424 000.215 | |
| DPBS | Life Technologies | 14190-136 | |
| DMEM | Life Technologies | 11965-084 | |
| Fetal Bovine Serum | HyClone | SH30071.03 | |
| Penicillin-Streptomycin | Life Technologies | 15140-122 | |
| GlutaMax | Life Technologies | 35050-061 | |
| Trypsin-EDTA | Life Technologies | 25300-054 | |
| 50 mL Conical Tubes | Phenix Research | SS-PH15 | |
| 25 mL reagent resevoirs | VWR | 41428-954 | |
| Multi channel pipet | Fisher Scientific | TI13-690-049 | |
| 96 well flat bottom plates | Corning | 3603 | |
| Opti-MEM | Life Technologies | 31985-070 | |
| Lipofectamine 2000 | Invitrogen | 11668019 | |
| 96 well round bottom plates | Corning | 3788 | |
| 16% Paraformaldehyde | Electron Microscopy Sciences | RT 15710 | |
| PBS | Sigma-Aldrich | P5493 | |
| EDTA | Sigma-Aldrich | ED2SS | |
| HBSS | Life Technologies | 14185-052 | |
| Sorvall ST 40 Centrifuge | Thermo Scientific | 75004509 | |
| BIOLiner Swinging Bucket Rotor | Thermo Scientific | 75003796 | |
| Hemacytometer | VWR | 15170-172 | |
| MACSQuant VYB Flow Cytomter | Miltenyi Biotec | 130-096-116 | |
| Chill 96 Rack | Miltenyi Biotec | 130-094-459 | |
| Flow Jo analysis software | Flow Jo | ||
| 20 uL pipet tips | Rainin | GPS-L10 | |
| 200 uL pipet tips | Rainin | GPS-250 | |
| 1 mL pipet tips | Rainin | GPS-1000 | |
| 200 uL pipet tips | USA Scientific | 1111-1800 | |
| 5 mL serological pipett | Phenix Research | SPG-606180 | |
| 10 mL serological pipett | Phenix Research | SPG-607180 | |
| 25 mL Serological pipett | Phenix Research | SPG-760180 |

