Immunolocalization of Arabinogalactan Proteins and Pectins in Plant Tissue
Abstract
Source: Costa, M., et al. Fluorescent Immunolocalization of Arabinogalactan Proteins and Pectins in the Cell Wall of Plant Tissues. J. Vis. Exp. (2021).
This video demonstrates an immunostaining technique to localize arabinogalactan proteins (AGPs) and pectins in plant tissues. Fixed and resin-embedded plant tissue sections are stained with AGP- and pectin-specific primary and fluorescent secondary antibodies and a cellulose-binding fluorescent dye to reveal their tissue-specific colocalization in the cell wall.
Protocol
1. Immunolocalization
NOTE: The fluorescent immunolocalization procedure relies on sequential use of two antibodies. The primary antibody is raised against a specific target antigen. The secondary antibody is raised specifically against the primary antibody and for fluorescent techniques is conjugated to a fluorophore (FITC [fluorescein isothiocyanate] in this specific protocol). The primary antibody will be used to detect the target antigen in the sample, and the secondary antibody will be used to mark the location where the primary antibody connected to the sample after washing off the excess of primary antibody. Controls are an important part of this assay and must always be performed to insure the accuracy of the observations. One well on the slide should be reserved for use as a negative control, where the primary antibody treatment will be skipped, and therefore no signal should be observed at the end of the experiment. A positive control must be included in the experiment by treating one well with an antibody which labelling is already known and certain. The positive control is used to confirm the secondary antibody labelling effectiveness and reaction conditions while the negative control tests the secondary antibody specificity.
- Prepare an incubation chamber by placing some damped paper towels at the bottom of a pipette tips box and wrapping it with tin foil (Figure 1B-1E). Place the slides in the incubation chamber.
- Pipette a 50 μL drop per 8 mm well of blocking solution (5% nonfat dry milk (w/v) in 1 M phosphate-buffered saline [PBS]) and incubate for 10 min. Remove the blocking solution and wash all wells twice with PBS for 10 min.
- Prepare the primary antibody solutions (see Table 1), 1:5 (v/v) antibody in blocking solution; make an estimate of about 40 μL per 8 mm well.
NOTE: The concentration of the antibody must be adjusted according to the manufacture’s protocol. - Perform a final wash with double-distilled water (ddH2O) for 5 min, never let the wells dry completely.
- Pipette the primary antibody solution to the reaction wells. Pipette blocking solution to the control wells.
- Close the incubation chamber and let stand for 2 h at room temperature followed by overnight at 4 °C. Prepare the secondary antibody solution, 1% in blocking solution (v/v) about 40 μL per well. Keep it covered with tin foil.
- Wash all wells twice with PBS for 10 min, followed by 10 additional minutes with ddH2O. Make sure that no trace of the blocking solution or deposits is visible on the wells.
- Pipette the secondary antibody solution to all wells. From now on, protect the slides from light.
- Incubate for 3–4 h in the dark at room temperature.
- Wash all wells twice for 10 min with PBS followed by another wash of 10 min with ddH2O.
- Apply a drop of calcofluor (1:10,000 (w/v) fluorescent brighter 28 in PBS) to each well. Without washing, apply a drop of mounting medium (see Table of Materials) to each well and place a coverslip (Figure 2H).
- Observe with a fluorescence microscope, equipped with 10x/0.45, 20x/0.75, 40x/0.95 and 100x/1.40 lens, use UV (for calcofluor stain) and FITC filters, to detect cell wall and immunolocalization respectively (Figure 2I). Use the following wavelengths: excitation/emission (nm) 358/461 for UV and 485/530 for FITC.
- For a better visualization of the results overlap both images with ImageJ or similar.
Table 1: List of useful monoclonal antibodies. The above table represents an example of available antibodies, with information about their targets and where they can be purchased.
| Name | Affinity | Available at |
| LM2 | Arabinogalactan protein | Plant probes (Leeds, U.K) |
| LM5 | Pectins | |
| LM6 | Arabinan | |
| LM7 | Homogalacturonan | |
| LM8 | Xylogalacturonan | |
| LM13 | (1-5)-a-L-ARABINAN | |
| LM14 | Arabinogalactan protein | |
| LM16 | Processed Arabinan – RG-I | |
| LM18 | Homogalacturonan | |
| LM19 | Homogalacturonan | |
| LM20 | Homogalacturonan | |
| LM26 | BRANCHED-GALACTAN | |
| LM30 | Arabinogalactan protein | |
| JIM5 | Low methyl-esterified homogalacturonan | CCRC (Georgia, USA) |
| JIM7 | Highly methyl-esterified homogalacturonan | |
| JIM4 | Arabinogalactan protein | |
| JIM8 | Arabinogalactan protein | |
| JIM13 | Arabinogalactan protein | |
| JIM15 | Arabinogalactan protein | |
| JIM16 | Arabinogalactan protein | |
| JIM20 | Extensin | |
| MAC207 | Arabinogalactan protein |
Representative Results
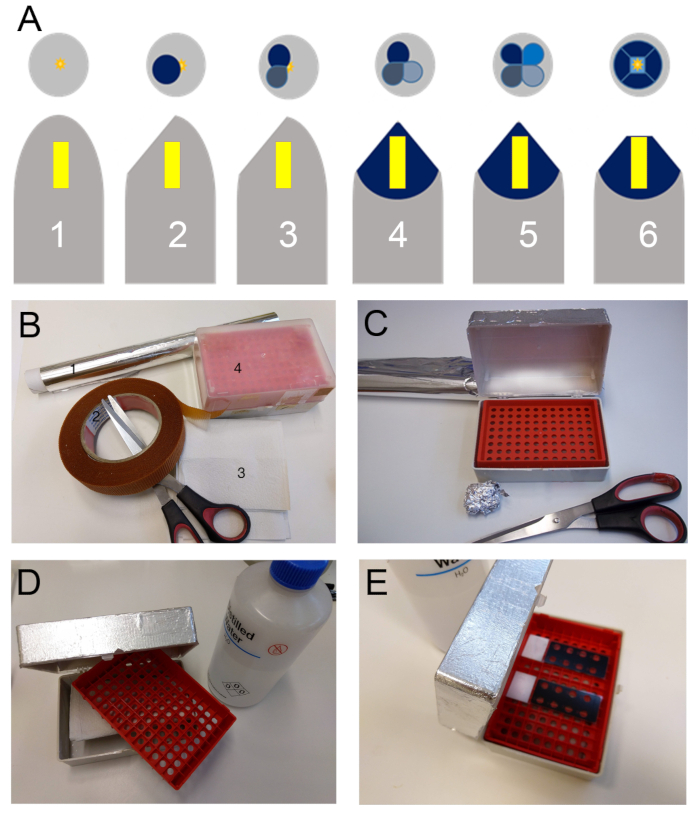
Figure 1. (A) Schematic representation of the block trimming sequence for preparing the sample (Yellow block) for sectioning with the ultramicrotome. Firstly, the gelatin capsule is removed (1). Then they are first trimmed at a 40° to 45° angle to the sides of the capsule tangentially to the sample (2), a second cut is made at 90° of the first (3) followed by a third (4) and fourth following the same rule (5). From this first phase of trimming results a pyramidal shaped structure which summit is located above the sample. Finally, with a sharp blade the summit is shaved off perpendicularly to the resin block major axis to reach the embedded sample. A square or trapezoid surface should be obtained at the end of the procedure (6). (B) Required supplies to make an incubation chamber; tin foil (1), double sided duct tape (2), paper towels (3) and a pipette tip box. (C) First step, the tin foil is fixed to the box with the double-sided duct tape. (D) Second step, the pipette tips holder is removed to place paper towels at the bottom of the box. (E) Third step, the paper towels are damped with water and the pipette tips holder is placed back to act has a tray for the slides.

Figure 2. Overview of the complete protocol.
Disclosures
The authors have nothing to disclose.
Materials
| 8 wells Glass reaction slides | Marinfeld | MARI1216750 | other brands may be used |
| Anti-Rat IgG (wole molecule)-FITC antibody produce in GOAT | Sigma-Aldrich | F6258 | |
| cover slips, 24 mm x 50 mm | Marinfeld | MARI0100222 | The cover slip should cover all the wells. Other brands may be used |
| ddH2O | na | na | |
| Ethanol absolute | na | na | |
| Fluorescent brightner 28 | Sigma-Aldrich | F-6259 | |
| Non fat dry milk | Nestlé | na | any non fat dry milk is adequate |
| Oven | na | na | generic laboratory oven |
| Rat generated Monoclonal Anti-Body | Plant probes | na | Several antibodies that recognize cell wall components are available at both the Complex Carbohydrate research center (CCRC, Georgia University USA) and Plant Probes (Paul Knox Cell Wall Lab, at Leeds University UK). A short list of some commonly used MABS and where they can be purchased is presented in Supplemental Table 1 |
| upright epifluorescence microscope with UV and FITC fluorescence filters | Leica Mycrosistems | DMLb | |
| vaccum chamber | na | na | |
| vaccum pump | na | na | |
| Vectashield | vecta Labs | T-1000 | Other anti-fade may be used. Please do check for compatibility with FITC and the Fluorescente brightner 28. (Note: for a non-commercial alternative, (see Jonhson et al 198218) An antifade medium can be made by mixing 25 mg/mL of 1,4-Diazobicyclo-(2,2,2)octane (DABCO) in 9:1 (v/v) glycerol to 1xPBS. Adjust pH to 8.6 with diluted HCl.) |

