Detection of Functional Matrix Metalloproteinases by Zymography
Summary
This protocol describes an activity-based assay for detecting matrix metalloproteinases in culture supernatants or body fluids.
Abstract
Matrix metalloproteinases (MMPs) are zinc-containing endopeptidases. They degrade proteins by cleavage of peptide bonds. More than twenty MMPs have been identified and are separated into six groups based on their structure and substrate specificity (collagenases, gelatinases, membrane type [MT-MMP], stromelysins, matrilysins, and others). MMPs play a critical role in cell invasion, cartilage degradation, tissue remodeling, wound healing, and embryogenesis. They therefore participate in both normal processes and in the pathogenesis of many diseases, such as rheumatoid arthritis, cancer, or chronic obstructive pulmonary disease1-6. Here, we will focus on MMP-2 (gelatinase A, type IV collagenase), a widely expressed MMP. We will demonstrate how to detect MMP-2 in cell culture supernatants by zymography, a commonly used, simple, and yet very sensitive technique first described in 1980 by C. Heussen and E.B. Dowdle7-10. This technique is semi-quantitative, it can therefore be used to determine MMP levels in test samples when known concentrations of recombinant MMP are loaded on the same gel11.
Solutions containing MMPs (e.g. cell culture supernatants, urine, or serum) are loaded onto a polyacrylamide gel containing sodium dodecyl sulfate (SDS; to linearize the proteins) and gelatin (substrate for MMP-2). The sample buffer is designed to increase sample viscosity (to facilitate gel loading), provide a tracking dye (bromophenol blue; to monitor sample migration), provide denaturing molecules (to linearize proteins), and control the pH of the sample. Proteins are then allowed to migrate under an electric current in a running buffer designed to provide a constant migration rate. The distance of migration is inversely correlated with the molecular weight of the protein (small proteins move faster through the gel than large proteins do and therefore migrate further down the gel). After migration, the gel is placed in a renaturing buffer to allow proteins to regain their tertiary structure, necessary for enzymatic activity. The gel is then placed in a developing buffer designed to allow the protease to digest its substrate. The developing buffer also contains p-aminophenylmercuric acetate (APMA) to activate the non-proteolytic pro-MMPs into active MMPs. The next step consists of staining the substrate (gelatin in our example). After washing the excess dye off the gel, areas of protease digestion appear as clear bands. The clearer the band, the more concentrated the protease it contains. Band staining intensity can then be determined by densitometry, using a software such as ImageJ, allowing for sample comparison.
Protocol
1. Loading and Running the Gel
- All samples must be prepared adequately to maintain the function of the enzymes and used immediately after collection or stored frozen at -80°C. The samples must not contain reducing agents (such a β-mercaptoethanol) or be boiled before gel loading.
- Open the pouch containing a gel inside a cassette, rinse the cassette with deionized water.
- Remove the protective tape from the bottom of the cassette and the comb from the top of the gel.
- Rinse the wells three times with running buffer (diluted to 1X in deionized water).
- Place the gel into the Mini-Cell, ensuring that the smaller side of the cassette faces inwards. Lock into place with the Gel Tension Wedge. The Mini-Cell allows to run one or two gels in parallel.
- Fill the top (inside) chamber with 1X running buffer above wells level, check for any leaks. In case of leaks, remove the buffer and reposition the gel.
- Fill the lower chamber with 1X running buffer.
- Load 10 μL of protein molecular marker in one well.
- Mix an equal amount of gel-loading buffer and of sample and load into the wells of the gel using gel-loading tips (changed between each sample). These wells can be loaded with up to 20 μL total.
- Place the lid on the Mini-Cell and connect the electrode cords to the power supply. Switch the power supply on and set it to run at 125 V constant for 90 minutes. Check the formation of small bubbles on the wire of the lower chamber, indicating current circulation.
- When running your first gel, monitor progress of the migration every 15 minutes, using the bromophenol blue included in the loading buffer as an indicator. Let the gel run until the indicator dye reaches the bottom of the gel.
2. Renaturing and Developing the Gel
- For each gel, prepare 100 mL of 1X renaturing buffer and 200 mL of denaturing buffer, both in deionized water.
- When the bromophenol blue tracking dye reaches the bottom of the gel, switch the power supply off, open the Mini-Cell, and remove the gel. Separate the two sides of the cassette using the gel knife (or a weighing spatula). Cut a corner to mark the gel’s direction.
- Carefully remove the gel from the cassette and place into a container with 100 mL renaturing buffer. Incubate for 30 minutes at room temperature with gentle agitation.
- Remove the renaturing buffer and add 100 mL of developing buffer to the gel. Incubate for 30 minutes at room temperature with gentle agitation.
- Remove the developing buffer and add 100 mL more of developing buffer to the gel. Incubate overnight (16-18 hours) at 37°C.
- Remove the developing buffer and rinse three times (5 minutes each) with deionized water under gentle agitation at room temperature.
- Scan the gel to save the exact positioning of the protein standard bands as they will become less or not visible after gel staining.
- Stain the gel by adding 20 mL of SimplyBlue Safestain to the gel. Incubate for 1 hour at room temperature under gentle agitation.
- Remove the SimplyBlue SafeStain and de-stain the gel in 100 mL or more deionized water for one hour at room temperature under gentle agitation.
- For better results, replace with fresh deionized water and incubate for another hour or more at room temperature under gentle agitation.
3. Data Analysis
- Carefully remove the gel from the water and place in a plastic sheet protector.
- Scan the gel with a resolution of 300 dpi or higher. Save the image in the TIFF format (Figure 1A).
- Measure band intensities with ImageJ (or another similar software).
- Open the TIFF file in ImageJ (Figure 1A).
- Visualize in black and white by selecting “Image>Type>8-bit” (Figure 1B).
- Use the rectangular selection tool to outline the first band, drawing a rectangle at least twice higher than it is wide (this is a software requirement).
- Press “1” or select “Analyze>Gels>Select first lane” and the band will be outlined.
- A new rectangle will appear, move it to the next band and select “Analyze>Gels>Select next lane”. Repeat until all bands are selected (Figure 1B).
- Press “3” or select “Analyze>Gels>Plot lanes” to generate the profile plot for each band (Figure 1C).
- Use the straight line selection (should already be automatically selected) to draw base lines so that the peak of interest is a completely enclosed area.
- Select the wand tool and click inside each peak to select it.
- Select “Analyze>Gels>Label peaks” to obtain a table with the area for each selected peak. These data can be plotted as such (Figure 1D) or can be normalized to the value of a chosen band. This normalization is especially important when pooling values from several replicate gels to determine statistical significance of the results.
4. Representative Results
We show a gel with 11 wells loaded with different cell culture supernatants containing different amounts of MMP-2 (Figure 1A). Direct observation of this gel shows obvious differences in MMP-2 concentrations between some of the wells. For example, it is clear that wells # 4 and 5 contain much more MMP-2 than well # 11 or even well # 10. For objective quantification of bands we have used densitometry with the ImageJ software (Figure 1B-D), confirming an approximately 4-fold difference in MMP-2 amounts between well # 11 and wells # 4 and 5 and an approximately 2-fold difference in MMP-2 amounts between well # 10 and wells # 4 and 5.
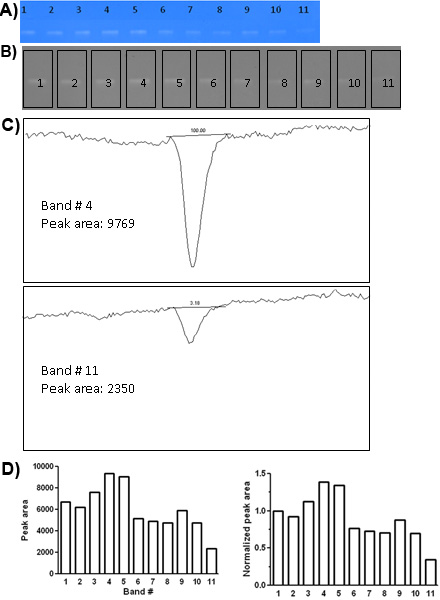
Figure 1. Detection of MMP-2 by gelatin zymography. A, Scanned image of a gelatin gel. Well numbers are indicated at the top of the gel. B, Same gel as in A shown in black and white for densitometry. Each band was selected with a rectangle in ImageJ. C, Two examples of densitometry profiles for the gel shown in A and B (bands # 4 and # 11). A straight line was drawn at the base of each peak to create a closed area. D, Plot of the peak areas for the gel shown in A and B before (left) and after (right) normalization to the density of band # 1.
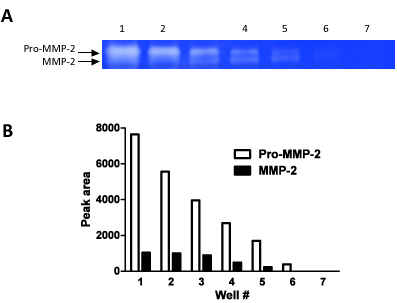
Figure 2. This figure demonstrates how zymography can be used as a semi-quantitative technique. We have loaded serial dilutions (steps of 1:1 dilutions) in 7 consecutive wells and measured the band densities for both pro-MMP-2 and MMP-2.
Discussion
We have demonstrated how to perform zymographic analysis of MMP-2 in cell culture supernatants.
The amount of sample to load on a gel must be determined empirically depending on the origin and MMP of interest. Loading too little will prevent detection while loading too much may lead to saturation as there is only so much substrate a protease can digest in the area of the band. If necessary, the sample can be diluted in deionized water prior to mixing with loading buffer or the developing time can de reduced from overnight to 4 hours. It is also possible to concentrate proteins in a solution using concentrators (e.g. Millipore catalog # UFC803024). If bands remain barely visible, it may be necessary to develop the gels for a longer period of time, even up to 48 hours. When preparing samples from tissue culture supernatants, it should be noted that fetal bovine serum (and other sera) contains MMPs that can affect the results. For this reason, we collect all of our supernatants after culture in serum-free medium.
The washes described in paragraphs 2.9 and 2.10 of the protocol are necessary to remove the background staining of digested bands. Longer wash times will remove more background, but will also dim the staining of the substrate throughout the gel. If longer wash times are necessary, we recommend scanning the gel every few hours to select the scan with the best contrast.
This technique can be used to detect other MMPs. For example, MMP-9 can be detected on gelatin gels, and also to a lower extent MMP-1, MMP-8, and MMP-13 as gelatin is not their preferred substrate. MMP-1 and MMP-13 are best detected on collagen zymography while casein is the preferred substrate for MMP-11 and also allows for the detection of MMP-1, MMP-3, MMP-7, MMP-12, and MMP-139. MMP-7 (matrilysin) and collagenases (MMP-1 and MMP-13) are difficult to detect when present at low levels in casein or gelatin gels. Addition of heparin to the sample at time of gel loading significantly improves the detection limit for MMP-7. For MMP-1 and MMP-13, the heparin needs to be added to the gel after the electrophoretic run is already under way12.
Since zymography measures enzymatic activity after denaturation and renaturation of the enzymes, it will measure the activity of all MMPs present in the sample. This include enzymes, pro-enzymes, and enzymes bound to endogenous inhibitors (e.g. TIMP-2). It is however possible to determine the level of MMP activation in the sample by comparing the band density of the active enzyme and of that of the pro-enzyme, which will have a slightly higher molecular weight.
Zymography is often not sufficient to identify an MMP. Comparison of the migration level of an MMP with known molecular weight standards does help in the identification but it should be noted that some of these standards contain a reducing agent and that when used under non-reducing conditions they may indicate different molecular weights9. An option is to load a soluble recombinant MMP on the same gel as the test samples. MMPs may however be associated with other proteins, inducing a change in apparent molecular weight. Selective MMP inhibitors can be added to the gel (or part of a gel cut in half) during the incubation in developing buffer as pharmacological tools to identity the MMP of interest.A second technique (Western Blot, immunocytochemistry, or ELISA) is recommended to help in the identification of the MMP of interest. It should be noted that detection limits for Western Blots are often much lower than those of zymographic gels, potentially leading to false-negative results when using this technique.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We would like to thank Dr. S. Ressler (Department of Molecular and Cellular Biology, Baylor College of Medicine) for suggesting the use of protein concentrators to increase the concentration of MMPs in cell culture supernatants.
This project was supported by grants from the Mrs. Clifford Elder White Graham Endowed Research Fund and the NIH/NIAMS (AR059838) to CB. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Materials
| Material Name | Type | Company | Catalogue Number | Comment |
|---|---|---|---|---|
| Xcell SureLock Mini-Cell CE mark electrophoresis apparatus | Invitrogen | EI0001 | ||
| Power supply (model 302) | VWR | 93000-744 | ||
| Novex 10% gelatin zymogram gels, 1.0 mm, 12 wells | Invitrogen | EC61752BOX | ||
| Blue Juice gel loading buffer | Invitrogen | 10816015 | ||
| Gel loading tips | VWR | 53509-015 | ||
| Protein molecular weight standard | Invitrogen | LC5800 | ||
| Novex Tris-Glycine-SDS running buffer | Invitrogen | LC2675 | ||
| Novex zymogram renaturing buffer | Invitrogen | LC2670 | ||
| Novex zymogram developing buffer | Invitrogen | LC2671 | ||
| SimplyBlue SafeStain | Invitrogen | LC6060 | ||
| Epson Perfection 4490 Photo Scanner | Amazon | n/a | ||
| ImageJ software | http://rsbweb.nih.gov/ij/ | n/a | Authored by W. Rasband, NIH/NIMH |
References
- Luo, J. The role of matrix metalloproteinases in the morphogenesis of the cerebellar cortex. Cerebellum. 4, 239-245 (2005).
- Pasternak, B., Aspenberg, P. Metalloproteinases and their inhibitors – diagnostic and therapeutic opportunities in orthopedics. Acta Orthop. 80, 693-703 (2009).
- Kessenbrock, K., Plaks, V., Werb, Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 141, 52-67 (2010).
- Lagente, V., Boichot, E. Role of matrix metallopreoteinases in the inflammatory process of respiratory diseases. J. Mol. Cell. Cardiol. 48, 440-444 (2010).
- Rodríguez, D., Morrison, C. J., Overall, C. M. Matrix metallopreoteinases: what do they not do? New substrates and biological roles identified by murine models and proteomics. Biochim. Biophys. Acta. 1803, 39-54 (2010).
- Bourboulia, D., Stetler-Stevenson, W. G. Matrix metalloproteinases (MMPs) and tissue inhibitors of metalloproteinases (TIMPs): positive and negative regulators in tumor cell adhesion. Semin. Cancer Biol. , (2010).
- Heussen, C., Dowdle, E. B. Electrophoretic analysis of plasminogen activators in polyacrylamide gels containing sodium dodecyl sulfate and copolymerized substrates. Analytical Biochem. , 102-196 (1980).
- Lombard, C., Saulnier, J., Wallach, J. Assays of matrix metalloproteinases (MMPs) activities: a review. Biochimie. 87, 265-272 (2005).
- Snoek-van Beurden, P. A., Von den Hoff, J. W. Zymographic techniques for the analysis of matrix metalloproteinases and their inhibitors. Biotechniques. 38, 73-83 (2005).
- Kupai, K., Szucs, G., Cseh, S., Hajdu, I., Csonka, C., Csont, T., Ferdinandy, P. Matrix metalloproteinase activity assays: importance of zymography. J. Pharmacol. Toxicol. Methods. 61, 205-209 (2010).
- Kleiner, D. E., Stetler-Stevenson, W. G. Quantitative zymography: detection of pictogram quantities of gelatinases. Anal. Biochem. 218, 325-329 (1994).
- Yu, W. H., Woessner, J. F. Heparin-enhanced zymographic detection of matrilysin and collagenases. Anal. Biochem. 293, 38-42 (2001).

