iCLIP – Transcriptome-wide Mapping of Protein-RNA Interactions with Individual Nucleotide Resolution
Summary
The spatial arrangement of RNA-binding proteins on a transcript is a key determinant of post-transcriptional regulation. Therefore, we developed individual-nucleotide resolution UV crosslinking and immunoprecipitation (iCLIP) that allows precise genome-wide mapping of the binding sites of an RNA-binding protein.
Abstract
The unique composition and spatial arrangement of RNA-binding proteins (RBPs) on a transcript guide the diverse aspects of post-transcriptional regulation1. Therefore, an essential step towards understanding transcript regulation at the molecular level is to gain positional information on the binding sites of RBPs2.
Protein-RNA interactions can be studied using biochemical methods, but these approaches do not address RNA binding in its native cellular context. Initial attempts to study protein-RNA complexes in their cellular environment employed affinity purification or immunoprecipitation combined with differential display or microarray analysis (RIP-CHIP)3-5. These approaches were prone to identifying indirect or non-physiological interactions6. In order to increase the specificity and positional resolution, a strategy referred to as CLIP (UV cross-linking and immunoprecipitation) was introduced7,8. CLIP combines UV cross-linking of proteins and RNA molecules with rigorous purification schemes including denaturing polyacrylamide gel electrophoresis. In combination with high-throughput sequencing technologies, CLIP has proven as a powerful tool to study protein-RNA interactions on a genome-wide scale (referred to as HITS-CLIP or CLIP-seq)9,10. Recently, PAR-CLIP was introduced that uses photoreactive ribonucleoside analogs for cross-linking11,12.
Despite the high specificity of the obtained data, CLIP experiments often generate cDNA libraries of limited sequence complexity. This is partly due to the restricted amount of co-purified RNA and the two inefficient RNA ligation reactions required for library preparation. In addition, primer extension assays indicated that many cDNAs truncate prematurely at the crosslinked nucleotide13. Such truncated cDNAs are lost during the standard CLIP library preparation protocol. We recently developed iCLIP (individual-nucleotide resolution CLIP), which captures the truncated cDNAs by replacing one of the inefficient intermolecular RNA ligation steps with a more efficient intramolecular cDNA circularization (Figure 1)14. Importantly, sequencing the truncated cDNAs provides insights into the position of the cross-link site at nucleotide resolution. We successfully applied iCLIP to study hnRNP C particle organization on a genome-wide scale and assess its role in splicing regulation14.
Protocol
1. UV cross-linking of tissue culture cells
- Remove the media and add 6 ml ice-cold PBS to cells grown in a 10 cm plate (enough for three experiments).
- Remove lid and place on ice. Irradiate once with 150 mJ/cm2 at 254 nm.
- Harvest the cells by scraping with a cell lifter.
- Transfer 2 ml cell suspension to each of three microtubes. Spin at top speed for 10 sec at 4°C to pellet cells, then remove supernatant.
- Snap-freeze the cell pellets on dry ice and store at -80°C until use.
2. Bead preparation
- Add 100 μl of protein A Dynabeads (Dynal, 100.02) per experiment to a fresh microtube (Use protein G Dynabeads for a mouse or goat antibodies).
- Wash beads 2x with lysis buffer (50 mM Tris-HCl, pH 7.4; 100 mM NaCl; 1% NP-40; 0.1% SDS; 0.5% sodium deoxycholate; 1/100 protease inhibitor cocktail III, Calbiochem).
- Resuspend beads in 100 μl lysis buffer with 2-10 μg antibody.
- Rotate tubes at room temperature for 30-60 min.
- Wash 3x with 900 μl lysis buffer and leave in the last wash until ready to proceed to step 4.1.
3. Cell lysis and partial RNA digestion
- Resuspend the cell pellet in 1 ml lysis buffer and transfer to 1.5 ml microtubes.
- Prepare a 1/500 dilution of RNase I (Ambion, AM2295). Add 10 μl RNase I dilution as well as 2 μl Turbo DNase to the cell lysate (1/500 RNase I dilutions [low RNase] are used for library preparation; 1/50 dilutions [high RNase] are necessary to control for antibody specificity).
- Incubate the samples for exactly 3 min at 37°C, shaking at 1,100 rpm. Immediately transfer to ice.
- Spin at 4°C and 22,000 g for 20 min to clear the lysate. Carefully collect the supernatant (leave about 50 μl lysate with the pellet).
4. Immunoprecipitation
- Remove the wash buffer from the beads (from step 2.5), then add the cell lysate (from step 3.4).
- Rotate the samples for 2 h at 4°C.
- Discard the supernatant and wash the beads 2x with 900 μl high-salt buffer (50 mM Tris-HCl, pH 7.4; 1 M NaCl; 1 mM EDTA; 1% NP-40; 0.1% SDS; 0.5% sodium deoxycholate).
- Wash 2x with 900 μl wash buffer (20 mM Tris-HCl, pH 7.4; 10 mM MgCl2; 0.2% Tween-20).
5. Dephosphorylation of RNA 3'ends
- Discard the supernatant and resuspend the beads in 20 μl PNK mix (15 μl water; 4 μl 5x PNK pH 6.5 buffer [350mMTris-HCl, pH 6.5; 50mMMgCl2 25mMdithiothreitol]; 0.5 μl PNK enzyme; 0.5 μl RNasin [Promega]).
- Incubate for 20 min at 37°C.
- Add 500 μl wash buffer and wash 1x with high-salt buffer.
- Wash 2x with wash buffer.
6. Linker ligation to RNA 3′ ends
- Carefully remove the supernatant and resuspend the beads in 20 μl ligation mix (9 μl water; 4 μl 4x ligation buffer [200 mMTris-HCl; 40m MM gCl2; 40 mM dithiothreitol]; 1 μl RNA ligase [NEB]; 0.5 μl RNasin [Promega]; 1.5 μl pre-adenylated linker L3 [20 μM]; 4 μl PEG400 [81170, Sigma]).
- Incubate overnight at 16°C.
- Add 500 μl wash buffer and then wash 2x with 1 ml high-salt buffer.
- Wash 2x with 1 ml wash buffer and leave in 1 ml of the second wash.
7. RNA 5' end labelling
- Remove the supernatant and resuspend the beads in 8 μl of hot PNK mix (0.4 μl PNK [NEB]; 0.8 μl 32P-γ-ATP; 0.8 μl 10x PNK buffer [NEB]; 6 μl water).
- Incubate for 5 min at 37°C.
- Remove the hot PNK mix and resuspend the beads in 20 μl 1x Nupage loading buffer (Invitrogen).
- Incubate on a thermomixer at 70°C for 10 min.
- Immediately place on a magnet to precipitate the empty beads and load the supernatant on the gel (see step 8).
8. SDS-PAGE and membrane transfer
- Load the samples on a 4-12% NuPAGE Bis-Tris gel (Invitrogen) according to the manufacturer's instructions. Use 0.5 l of 1x MOPS running buffer (Invitrogen). Also load 5 μl of a pre-stained protein size marker (for example PAGE ruler plus, Fermentas, SM1811).
- Run the gel for 50 min at 180 V.
- Remove the gel front and discard as solid waste (contains free radioactive ATP).
- Transfer the protein-RNA complexes from the gel to a nitrocellulose membrane using the Novex wet transfer apparatus according to the manufacturer's instructions (Invitrogen, transfer 1 h at 30 V).
- After the transfer, rinse the membrane in PBS buffer, then wrap it in saran wrap and expose it to a Fuji film at -80°C (place a fluorescent sticker next to the membrane to later align the film and the membrane; perform exposures for 30 min, 1h and over night).
9. RNA isolation
- Isolate the protein-RNA complexes from the low-RNase experiment using the autoradiograph from step 8.5 as a mask. Cut this piece of membrane into several small slices and place them into a 1.5 ml microtube.
- Add 200 μl PK buffer (100 mM Tris-HCl pH 7.4; 50 mM NaCl; 10 mM EDTA) and 10 μl proteinase K (Roche, 03115828001) to the membrane pieces. Incubate shaking at 1,100 rpm for 20 min at 37°C.
- Add 200 μl of PKurea buffer (100 mM Tris-HCl pH 7.4; 50 mM NaCl; 10 mM EDTA; 7 M urea) and incubate for 20 min at 37°C.
- Collect the solution and add it together with 400 μl of RNA phenol/chloroform (Ambion, 9722) to a 2 ml Phase Lock Gel Heavy tube (713-2536, VWR).
- Incubate for 5 min at 30°C, shaking at 1,100 rpm. Separate the phases by spinning for 5 min at 13,000 rpm at room temperature.
- Transfer the aqueous layer into a new tube (be careful not to touch the gel with the pipette). Add 0.5 μl glycoblue (Ambion, 9510) and 40 μl 3 M sodium acetate pH 5.5 and mix. Then add 1 ml 100% ethanol, mix again and precipitate over night at -20°C.
10. Reverse transcription
- Spin for 20 min at 15,000 rpm and 4°C. Remove the supernatant and wash the pellet with 0.5 ml 80% ethanol.
- Resuspend the pellet in 7.25 μl RNA/primer mix (6.25 μl water; 0.5 μl Rclip primer [0.5pmol/μl]; 0.5 μl dNTP mix [10mM]). For each experiment or replicate, use a different Rclip primer containing individual barcode sequences (see 14).
- Incubate for 5 min at 70°C before cooling to 25°C.
- Add 2.75 μl RT mix (2 μl 5x RT buffer; 0.5 μl 0.1M DTT; 0.25 μl Superscript III reverse transcriptase [Invitrogen]).
- Incubate 5 min at 25°C, 20 min at 42°C, 40 min at 50°C and 5 min at 80°C before cooling to 4°C.
- Add 90 μl TE buffer, 0.5 μl glycoblue and 10 μl sodium acetate pH 5.5 and mix. Then add 250 μl 100% ethanol, mix again and precipitate over night at -20°C.
11. Gel purification of cDNA
- Spin down and wash the samples (see 10.1), then resuspend the pellets in 6 μl of water.
- Add 6 μl 2x TBE-urea loading buffer (Invitrogen). Heat samples to 80°C for 3 min directly before loading.
- Load the samples on a precast 6% TBE-urea gel (Invitrogen) and run for 40 min at 180 V as described by the manufacturer. Also load a low molecular weight marker for subsequent cutting (see below).
- Cut three bands at 120-200 nt (high), 85-120 nt (medium) and 70-85 nt (low). Use theupper dye and the marks on the plastic gel support to guide excision (see Figure 3). Note that the Rclip primer and the L3 sequence together account for 52 nt of the CLIP sequence.
- Add 400 μl TE and crush the gel slice into small pieces using a 1 ml syringe plunger. Incubate shaking at 1,100 rpm for 2 h at 37°C.
- Place two 1 cm glass pre-filters (Whatman, 1823010) into a Costar SpinX column (Corning Incorporated, 8161). Transfer the liquid portion of the sample to the column. Spin for 1 min at 13,000 rpm into a 1.5 ml tube.
- Add 0.5 μl glycoblue and 40 μl sodium acetate pH 5.5, then mix the sample. Add 1 ml 100% ethanol, mix again and precipitate over night at -20°C.
12. Ligation of primer to the 5’end of the cDNA
- Spin down and wash the samples (see 10.1), then resuspend the pellets in 8 μl ligation mix (6.5 μl water; 0.8 μl 10x CircLigase Buffer II; 0.4 μl 50 mM MnCl2; 0.3 μl; Circligase II [Epicentre]) and incubate for 1 h at 60°C.
- Add 30 μl oligo annealing mix (26 μl water; 3 μl FastDigest Buffer [Fermentas]; 1 μl cut_oligo [10 μM]). Incubate for 1 min at 95°C. Then decrease the temperature every 20 sec by 1°C until 25°C are reached.
- Add 2 μl BamHI (Fast Fermentas) and incubate for 30 min at 37°C.
- Add 50 μl TE and 0.5 μl glycoblue and mix. Add 10 μl sodium acetate pH 5.5 and mix, then add 250 μl 100% ethanol. Mix again and precipitate over night at -20°C.
13. PCR amplification
- Spin down and wash the samples (see 10.1), then resuspend the pellet in 19 μl water.
- Prepare the PCR mix (19 μl cDNA; 1 μl primer mix P5/P3 solexa, 10 μM each; 20 μl Accuprime Supermix 1 enzyme [Invitrogen]).
- Run the following PCR programme: 94°C for 2 min, [94°C for 15 sec, 65°C for 30 sec, 68°C for 30 sec]25-35 cycles, 68°C for 3 min, 4°C for ever.
- Mix 8 μl PCR product with 2 μl of 5x TBE loading buffer and load on a precast 6% TBE gel (Invitrogen). Stain the gel with Sybrgreen I (Invitrogen) and analyse with a gel imager.
- The barcode in the Rclip primers allow to multiplex different samples before submitting for high throughput sequencing. Submit 15 μl of the library for sequencing and store the rest.
14. Linker and primer sequences
Pre-adenylated 3′ linker DNA:
[We order the DNA adapter from IDT and then make aliquots of 20μM.]

15. Representative Results:
Prior to sequencing of the iCLIP library, the success of the experiment can be monitored at two steps: the autoradiograph of the protein-RNA complex after membrane transfer (step 8.5) and the gel image of the PCR products (step 13.4). In the autoradiograph of the low-RNase samples, diffuse radioactivity should be seen above the molecular weight of the protein (Figure 2, sample 4). For high-RNase samples, this radioactivity is focused closer to the molecular weight of the protein (Figure 2, sample 3). When no antibody is used in the immunoprecipitation, no signal should be detected (Figure 2, samples 1 and 2). Further important controls for specificity of the immunoprecipitation either omit UV irradiation or use cells that do not express the protein of interest14.
The gel image of the PCR products (step 13.4) should show a size range that corresponds to the cDNA fraction (high, medium or low) purified in step 11.4 (Figure 4, lanes 4-6). Note that the PCR primers P3Solexa and P5Solexa introduce an additional 76 nt to the size of the cDNA. If no antibody is used during the immunoprecipitation, no corresponding PCR products should be detected (Figure 4, lanes 1-3). Primer dimer product can appear at about 140 nt.
For representative results of high-throughput sequencing and subsequent bioinformatic analyses see14.
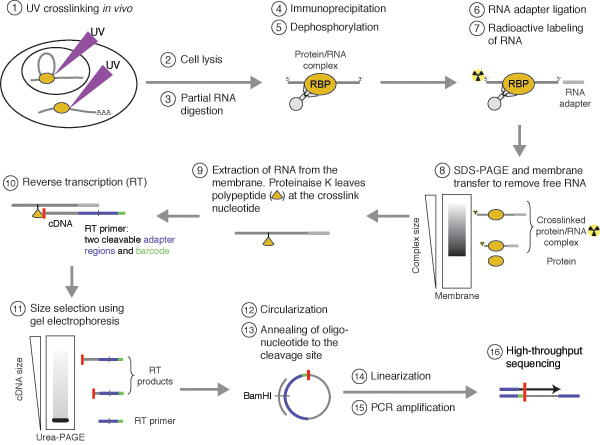
Figure 1. Schematic representation of the iCLIP protocol. Protein-RNA complexes are covalently cross-linked in vivo using UV irradiation (step 1). The protein of interest is purified together with the bound RNA (steps 2-5). To allow for sequence-specific priming of reverse transcription, an RNA adapter is ligated to the 3′ end of the RNA, whereas the 5′ end is radioactively labelled (steps 6 and 7). Cross-linked protein-RNA complexes are purified from free RNA using SDS-PAGE and membrane transfer (step 8). The RNA is recovered from the membrane by digesting the protein with proteinase K leaving a polypeptide remaining at the cross-link nucleotide (step 9). Reverse transcription (RT) truncates at the remaining polypeptide and introduces two cleavable adapter regions and barcode sequences (step 10). Size selection removes free RT primer before circularization. The following linearization generates suitable templates for PCR amplification (steps 11-15). Finally, high-throughput sequencing generates reads in which the barcode sequences are immediately followed by the last nucleotide of the cDNA (step 16). Since this nucleotide locates one position upstream of the cross-linked nucleotide, the binding site can be deduced with high resolution.
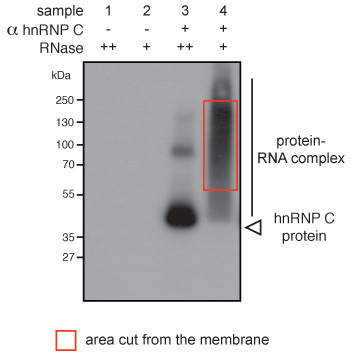
Figure 2. Autoradiograph of cross-linked hnRNP C-RNA complexes using denaturing gel electrophoresis and membrane transfer. hnRNP C-RNA complexes were immuno-purified from cell extracts using an antibody against hnRNP C (α hnRNP C, samples 3 and 4). RNA was partially digested using low (+) or high (++) concentration of RNase. Complexes shifting upwards from the size of the protein (40 kDa) can be observed (sample 4). The shift is less pronounced when high concentrations of RNase were used (sample 3). The radioactive signal disappears when no antibody was used in the immunoprecipitation (samples 1 and 2).
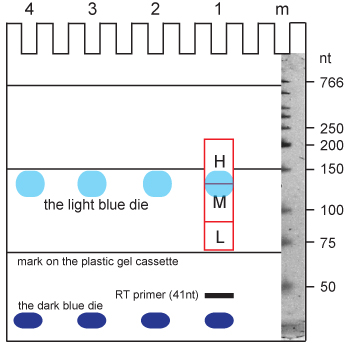
Figure 3. Schematic 6% TBE-urea gel (Invitrogen) to guide the excision of iCLIP cDNA products. The gel is run for 40 min at 180 V leading to a reproducible migration pattern of cDNAs and dyes (light and dark blue) in the gel. Use a razor blade to cut (red line) the high (H), medium (M), and low (L) cDNA fractions. Start by cutting in the middle of the light blue dye and immediately above the mark on the plastic gel cassette. Divide the medium and low fractions and trim the high fraction about 1 cm above the light blue dye. Use vertical cuts guided by the pockets and the dye to separate the different lanes (in this example 1-4). The marker lane (m) can be stained and imaged to control sizes after the cutting. Fragment sizes are indicated on the right.
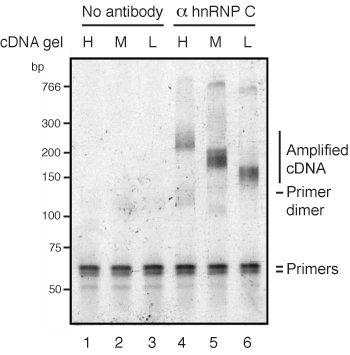
Figure 4. Analysis of PCR-amplified iCLIP cDNA libraries using gel electrophoresis. RNA recovered from the membrane (Figure 1) was reverse transcribed and size-purified using denaturing gel electrophoresis (Figure 2). Three size fractions of cDNA (high [H]: 120-200 nt, medium [M]: 85-120 nt and low [L]: 70-85 nt) were recovered, circularized, re-linearized and PCR-amplified. PCR products of different size distribution can be observed as a result of the different sizes of the input fractions. Since the PCR primer introduces 76 nt to the cDNA, sizes should range between 196-276 nt for high, 161-196 nt for medium and 146-161 nt for low size fractions. PCR products are absent when no antibody was used for the immunoprecipitation (lanes 1-3).
Discussion
Since the iCLIP protocol contains a diverse range of enzymatic reactions and purification steps, it is not always easy to identify a problem when an experiment fails. In order to control for the specificity of identified RNA cross-link sites, one or more negative controls should be maintained throughout the complete experiment and subsequent computational analyses. These controls can be the no-antibody sample, the non-cross-linked cells, or immunoprecipitation from knockout cells or tissue. Ideally, these control experiments should not purify any protein-RNA complexes, and therefore should give no signal on the SDS-PAGE gel, and no detectable products after PCR amplification. High-throughput sequencing of these control libraries should return very few unique sequences. Knockdown cells are not recommended as a sequencing control, since the resulting sequences still correspond to cross-link sites of the same protein, which is purified from knockdown cells in smaller quantities.
Precautions should also be taken to avoid contamination with PCR products from previous experiments. The best way to minimize this problem is to spatially separate pre- and post-PCR steps. Ideally, the analysis of the PCR products and all subsequent steps should be performed in a separate room. Moreover, each member of the laboratory should use their own set of buffers and other reagents. In this way, sources of contamination can be easier identified.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors thank all members of the Ule, Luscombe and Zupan laboratories for discussion and experimental assistance. We thank James Hadfield and Nik Matthews for high-throughput sequencing. We would like to point out that the iCLIP method described here shares several steps with the original CLIP protocol, developed by Kirk Jensen and J.U. in the laboratory of Robert Darnell. This work was supported by the European Research Council grant 206726-CLIP to J.U. and a Long-term Human Frontiers Science Program fellowship to J.K.
Materials
For gel electrophoresis and membrane transfer we recommend t he use of XCell SureLock® Mini-Cell and XCell II™ Blot Module Kit CE Mark (Invitrogen, EI0002), which is compatible with the use of the different precast minigels that are specified throughout the protocol. The brand and order number of all materials used is mentioned during the protocol. The list of enzymes used in the protocol is shown in the table below.
| Name of reagent | Company | Catalogue # | Comments |
| Protein A Dynabeads | Invitrogen | 10001D | use protein G for mouse or goat antibody |
| RNase I | Ambion | AM2295 | activity can change from batch to batch |
| T4 RNA ligase I | NEB | M0204S | |
| PNK | NEB | M0201S | |
| proteinase K | Roche | 03115828001 | |
| Superscript III reverse transcriptase | Invitrogen | 18080044 | |
| Circligase II | Epicentre | CL9021K | |
| FastDigest® BamHI | Fermentas | FD0054 | |
| AccuPrime™ SuperMix I | Invitrogen | 12342010 | this PCR mix gives the best results in our hands |
References
- Keene, J. D. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 8, 533-543 (2007).
- Wang, Z., Burge, C. B. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 14, 802-813 (2008).
- Trifillis, P., Day, N., Kiledjian, M. Finding the right RNA: identification of cellular mRNA substrates for RNA-binding proteins. RNA. 5, 1071-1082 (1999).
- Brooks, S. A., Rigby, W. F. Characterization of the mRNA ligands bound by the RNA binding protein hnRNP A2 utilizing a novel in vivo technique. Nucleic Acids Res. 28, E49-E49 (2000).
- Tenenbaum, S. A., Carson, C. C., Lager, P. J., Keene, J. D. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc Natl Acad Sci. 97, 14085-14090 (2000).
- Mili, S., Steitz, J. A. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 10, 1692-1694 (2004).
- Ule, J. CLIP identifies Nova-regulated RNA networks in the brain. Science. 302, 1212-1215 (2003).
- Ule, J., Jensen, K., Mele, A., Darnell, R. B. CLIP: A method for identifying protein-RNA interaction sites in living cells. Methods. 37, 376-386 (2005).
- Licatalosi, D. D. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 456, 464-469 (2008).
- Yeo, G. W. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 16, 130-137 (2009).
- Urlaub, H., Hartmuth, K., Lührmann, R. A two-tracked approach to analyze RNA-protein crosslinking sites in native, nonlabeled small nuclear ribonucleoprotein particles. Methods. 26, 170-181 (2002).
- König, J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 17, 909-915 (2010).

