Mass Spectrometric Analysis of Glycosphingolipid Antigens
Summary
A specific and sensitive method to gain insight into the expression profile of glycosphingolipid antigens in immune organs and cells is described. The method takes advantage of the ion trap mass spectrometry allowing step-wise fragmentation of glycosphingolipid molecules for structural analysis in comparison to chemically synthesized standards.
Abstract
Glycosphingolipids (GSL’s) belong to the glycoconjugate class of biomacromolecules, which bear structural information for significant biological processes such as embryonic development, signal transduction, and immune receptor recognition1-2. They contain complex sugar moieties in the form of isomers, and lipid moieties with variations including fatty acyl chain length, unsaturation, and hydroxylation. Both carbohydrate and ceramide portions may be basis of biological significance. For example, tri-hexosylceramides include globotriaosylceramide (Galα4Galβ4Glcβ1Cer) and isoglobotriaosylceramide (Galα3Galβ4Glcβ1Cer), which have identical molecular masses but distinct sugar linkages of carbohydrate moiety, responsible for completely different biological functions3-4. In another example, it has been demonstrated that modification of the ceramide part of alpha-galactosylceramide, a potent agonist ligand for invariant NKT cells, changes their cytokine secretion profiles and function in animal models of cancer and auto-immune diseases5. The difficulty in performing a structural analysis of isomers in immune organs and cells serve as a barrier for determining many biological functions6.
Here, we present a visualized version of a method for relatively simple, rapid, and sensitive analysis of glycosphingolipid profiles in immune cells7-9. This method is based on extraction and chemical modification (permethylation, see below Figure 5A, all OH groups of hexose were replaced by MeO after permethylation reaction) of glycosphingolipids10-15, followed by subsequent analysis using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS) and ion trap mass spectrometry. This method requires 50 million immune cells for a complete analysis. The experiments can be completed within a week. The relative abundance of the various glycosphingolipids can be delineated by comparison to synthetic standards. This method has a sensitivity of measuring 1% iGb3 among Gb3 isomers, when 2 fmol of total iGb3/Gb3 mixture is present9.
Ion trap mass spectrometry can be used to analyze isomers. For example, to analyze the presence of globotriaosylceramide and isoglobtriaosylceramide in the same sample, one can use the fragmentation of glycosphingolipid molecules to structurally discriminate between the two (see below Figure 5). Furthermore, chemical modification of the sugar moieties (through a permethylation reaction) improves the ionization and fragmentation efficiencies for higher sensitivity and specificity, and increases the stability of sialic acid residues. The extraction and chemical modification of glycosphingolipids can be performed in a classic certified chemical hood, and the mass spectrometry can be performed by core facilities with ion trap MS instruments.
Protocol
1. Lab Safety Concerns
- All organic solvents must be stored in specific areas with clear labeling. See labels by manufactures for types of fire extinguishers required.
- Several reagents used are potentially carcinogenic, and may cause mental depression. These reagents must be handled in a chemical hood with ventilation (See table of specific reagents and equipment for specifics).
- It is recommended that when working with organic solvents and cells, personal protective equipment such as gloves, lab coat, and eye protection be utilized at all times.
2. Extraction of Glycosphingolipids from Rat Leukemia Cells
- Store RBL rat leukemia cells8 (10 million to 100 million) in 16×100 mm glass tubes under -80 °C conditions. Cells are counted by a hemocytometer after trypan blue staining. Viability of cells should be higher than 95%.
- Extract the lipids using extensive sonication four times for 1-2 hr each with mixed polarity solvents. Use 6 ml of chloroform-methanol 1:1 (v/v) as the first and last solvent. Six milliliter of isopropanol:hexane:H2O 55:25:20 (v/v/v, remove upper phase by aspiration before use) can then be used as the second and third solvent. This solvent is prepared by mixing isopropanol:hexane:H2O in a 55:25:20 ratio, shaking it vigorously, and allowing an upper phase to appear, which will be removed. Keep the lower phase for use.
- Follow sonication with centrifugation to pellet the insoluble material. Pool the supernatants. Dry them in a Speedvac for 4 hr. Nitrogen stream or rotary evaporator can be used if Speedvac is unavailable. Use a good cold trap to hold evaporated organic solvents. Contamination of organic solvent into vacuum pump oil will degrade the efficiency of the vacuum pump.
- Perform a preliminary analysis using high performance thin layer chromatography (HPTLC). To quantify glycolipids, prepare a 0.2% Orcinol in 50% sulfuric acid solution (100 mg in 50 ml) and a Galactose standard (40 mg in 100 ml stock solution). Prepare in 30 μl reaction volume a series of dilutions to use as standard curve (from 0 g/L to 20 g/L). Add 20 μl of Methanol to each dilution series sample. Dissolve each glycolipid sample in 200 μl of Methanol, sonicate, and use 20 μl for glycolipid quantity measurement. In 1.5 ml Eppendorf tubes with screwed caps (Sarstedt, 72.692.005), add 0.4 ml of 0.2% Orcinol in 50% sulfuric acid solution to each sample, with a series of Galactose-H2O-Methanol solutions which serve for standard curve. Boil for 5 min in heating block at 100 °C. Read optical density of color reaction at 440 nm using Sunrise Tecan Microplate Reader. To perform HPTLC, use chloroform:methanol:H2O 60:35:8 (v/v/v) as the solvent for the neutral lipids treated with chloroform-methanol 1:1 (v/v). Use isopropanol:hexane:H2O 55:25:20 (v/v/v) as the solvent for the acid lipids (gangliosides) . The isolated neutral GSLs was dissolved in 200 μl solvent chloroform:methanol 1:1(v/v) and loaded on Merck silica gel thin layer chromatography plate by Drummond Microcaps. The plates were run in solvent chloroform:methanol:H2O 60:35:8 (v/v/v) and dried. The glycosphingolipids were visualized by Orcinol-sulphuric acid at 300 °C.
- In order to separate out the neutral lipids from the acidic lipids, fraction out the neutral and acidic lipids by anion exchange chromatography on a small column of DEAE Sephadex A-25 [the DEAE Sephadex A25 resin can be prepared by soaking Sephadex A-25 resin powder in chloroform:methanol:H2O 30:60:8 (v/v/v) overnight. Aspirate the supernatant until the Sephadex A25 resin is all that is left. Add fresh chloroform:methanol:H2O 30:60:8 (v/v/v) to wash the resin. Repeat three times to remove the negatively charged residues from Sephadex A25 resin].
- Dissolve, sonicate (for no more than 10 min), and resuspend the lipids from step 2.1 in chloroform:methanol:H2O 30:60:8 (v/v/v) when needed.
- Prepare a small DEAE Sephadex A25 (Cl Form) column by using glass wool and a 9″ Pasteur pipette. Pack the glass wool carefully so that resin powder does not pass through the column. Add DEAE Sephadex A25(Cl Form) resin to the “neck” of the 9″ Pasteur pipette without letting the column dry. Make sure that you do not see any resin in eluent. The samples dissolved in chloroform:methanol:H2O 30:60:8 (v/v/v) were dissolved. The neutral lipid fraction is the flow through fraction.
- Elute the acidic lipid fraction (bound to the resins) with 8 ml of sodium acetate in methanol. Dry both the neutral and acidic fractions, desalt them by dialysis, dry them by Speedvac and analyze them by HPTLC.
The acidic fraction contains gangliosides, which can be dialyzed for the Step 3 reaction directly. The neutral fraction contains not only neutral glycosphingolipids, but also phospholipids (phosphatidylcholine and sphingomyelin), which must be removed by an acetylation reaction.
In our experience, the method of a dialysis cassette, is more efficient and convenient than a dialysis tube or reverse phase C18 column chromatography.
- The next step is the removal of phospholipids from neutral glycosphingolipids by an acetylation and peracetylation reaction. Dry the DEAE Sephadex A-25 pass-through fraction in the Speedvac (with cooling) for 4 hr. After the samples are dry, put them back into the Speedvac and dry for another 4 hr. Ensure that these samples are completely dry, as any residual water will prevent the peracetylation reaction.
- Peracetylate the samples with 1 ml of pyridine and 0.5 ml of acetic anhydride in the dark at room temperature or 37 °C overnight. This amount of pyridine and acetic anhydride is proper for up to 200 μg of glycosphingolipids. This reaction can be carried out at 37 °C, as the GSLs have better solubility.
- Dry the peracetylated material by Speedvac for 3 hr, with the addition of 1 ml of toluene 3x (coevaporation) to ensure complete evaporation. If the samples are still not completely dry, Speedvac until dry.
- Prepare a Florisil (Sigma-Aldrich) column (30-60 mesh, 10×80 mm) in a Pasteur pipette with glass wool, using Florisil beads. Florisil beads should be completely dried before use, by incubation in a 110 °C oven. Fill the beads into the Pasteur pipette up to the neck, and equilibrate in 1,2-dichloroethane-hexane 4:1 (v/v).
Elution from the Florisil column is very fast. Rapid volatility of the solvents can lead to column drying. Thus all solvent mixtures should be prepared before running the column since it is not possible to stop the column during the elution.
- Apply the peracetylated sample in 1 ml of 1,2-dichloroethane-hexane 4:1 (v/v), and wash the column with 6 ml of the same solvent, followed by 10 ml of 1,2-dichloroethane. Elute neutral peracetylated GSLs with 6 ml of 1,2-dichloroethane-acetone 1:1 (v/v). This step removes the peracetylated phospholipids from glycosphingolipids, because the peracetylated glycosphingolipids bind to the Florisil column, while peracetylated phospholipids do not.
- Dry the fractions by Speedvac for 2 hr and deacetylate with 2 ml 0.5 M sodium methoxide [Sigma-Aldrich, 403067] in 2 ml of methanol for 3 hr at room temperature.
- Neutralize the mixture with 3 ml of methanolic acetic acid [acetic acid/methanol 15/85 v/v], dry by Speedvac, desalt by dialysis [Dissolve the dried products in 3 ml of water, and inject into the Slide-A-Lyzer Dialysis Cassette, dialyze against water for 24 hr. Change the water at least twice]. Dry the dialyzed GSLs (in water solution) by Speedvac until the samples are completely dry.
3. Chemical Modification (Permethylation) of Glycosphingolipids13
- Introduce GSLs (1-20 μg) to a glass tube with a screw cap and Teflon liner, and add dimethyl sulfoxide (150 μl) without using special drying conditions or inert gas atmosphere.
- Prepare powdered sodium hydroxide (40-60 mg) from sodium hydroxide particles by grinding them in a chemical mortar with a pestle. Make sure to store the powders in a very dry environment and be careful to do the grinding in the chemical hood to avoid breathing the powders.
- Next add the sodium hydroxide powder to the sample solution with a spatula. Add iodomethane (80 μl) with a 100 microliter Halmilton syringe, [or a 100 microliter VWR calibrated pipette linked to a syringe through rubber tubing can be used], and shake the mixture at room temperature for 1 hr.
- Quench the methylation reaction with water (2 ml). Extract the permethylated products 3 times by the addition of dichloromethane (2 ml) [glycolipids are in the dichloromethane phase, which is the lower phase, so the upper phase can be transferred to a new glass tube, and extracted again with dichloromethane].
- Wash the combined dichloromethane extracts 3 times with 2 ml water [ the upper phase, which is the water, can then be disposed of]. Following the final wash, transfer the samples to a new tube, and dry using the Speedvac.
- Samples can then be dissolved in 100 to 200 μl of methanol for ESI-MS analysis.
4. MALDI-TOF Analysis of the Permethylated Gycosphingolipids
- Perform MALDI-TOF-MS and MALDI-TOF/TOF-MS/MS experiments on a MALDI TOF-TOF Mass Spectrometer (Applied Biosystems 4700, Foster City, CA). Prepare the matrix by mixing 50% volume of 10 mg/ml dihydroxybenzoic acid (DHB) in acetonitrile and 50% volume of 0.1% trifluoroacetic acid (TFA) in water.
- The DHB matrix can be spotted on the MALDI target (1 μl), followed by a 1 μl (containing 100 ng GSLs) spot of sample dissolved in methanol. Apply slowly and allow it to dry before analysis.
5. Ion Trap MS Analysis of Permethylated Glycosphingolipids
- Carry out mass spectrometry on a linear ion trap mass spectrometer (LTQ, ThermoScientific, San Jose, CA) using a nanoelectrospray source, 0.30 μl/min flow rate at 230 °C capillary temperature in positive ion mode with 30-50% collision energy. Set collision energies to leave a minimal residual abundance of precursor ion. Detect all ions as sodium adducts.
- Identify the iGb3 (Galα3Galβ4Glcβ1Cer) in an isobaric mixture of Gb3 (Galα4Galβ4Glcβ1Cer) and iGb3 (Galα3Galβ4Glcβ1Cer) standards by comparing the different patterns of MS4 product ions from the sodiated molecular ion via the glycan fragment m/z 667 and the terminal disaccharide 1-ene ion m/z 445 (i.e. X→667→445→, X means the molecular ion detected at MS1) of pure permethylated iGb3 (Galα3Galβ4Glcβ1Cer) standards via ESI-LIT-MS with those of permethylated Gb3 (Galα3Galβ4Glcβ1Cer).
Representative Results
An example of a MALDI MS analysis of the glycosphingolipids from rat leukemia cell line RBL (an antigen presenting cell type that presents glycosphingolipid antigens to NKT cells) is shown in Figure 3. The ions can be assigned to specific glycosphingolipid structures (Figure 3A). In RBL cells treated by NB-DGJ16, a drug which inhibits glycosphingolipid synthesis, significant reduction of all glycosphingolipids was observed (Figure 3B). Structural isomers cannot be distinguished. The MS/MS capacity of the Applied Biosystems 4700 allows fragmentation of MS1 ions to MS2 fragments, allowing limited structural information to be generated. However, MS2 analysis is often not sufficient to discriminate oligosaccharide isomers.
The Ion-Trap MS analysis of glycosphingolipids enables the study of isomers, such as iGb3 (Galα3Galβ4Glcβ1Cer) and Gb3 (Galα4Galβ4Glcβ1Cer) which may exist as the trihexosylceramide ions detected in Figure 3A. To detect such isomers, standard iGb3 and Gb3 were analyzed by multiple rounds of fragmentation, until differences were found (Figure 4A and 4B). In the example presented here (Figure 5C) iGb3 and Gb3 could be discriminated by comparing to standard iGb3 and Gb3.
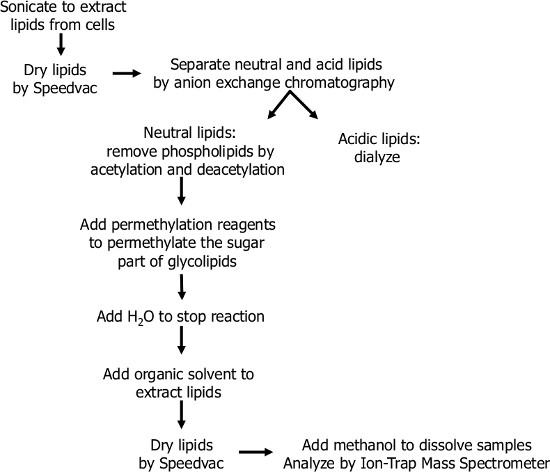
Figure 1. Flow chart of the glycosphingolipid extraction procedure, and chemical modification of glycosphingolipids (permethylation). Glycosphingolipids were extracted from cells by organic solvents. To remove non-glycosphingolipids, lipids can be peracetylated and deacetylated. Second, glycosphingolipids were chemically modified by a permethylation reaction, which improves sensitivity and specificity of MS detection. Finally, the glycosphingolipid profiles were determined using MALDI-TOF mass spectrometry and ion trap mass spectrometry.
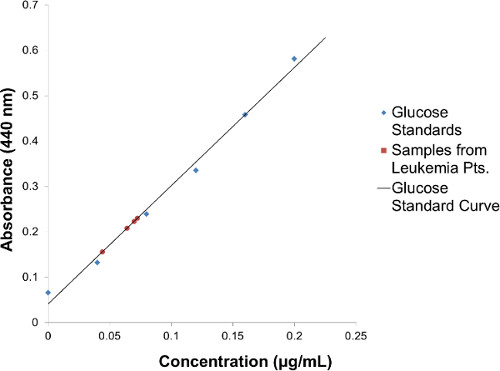
Figure 2. Glycosphingolipid Quantification by Orcinol Sulfuric Acid Method: Quantification of glycosphingolipids using a 0.2% Orcinol in 50% sulfuric acid solution and a Glucose Standard Curve.
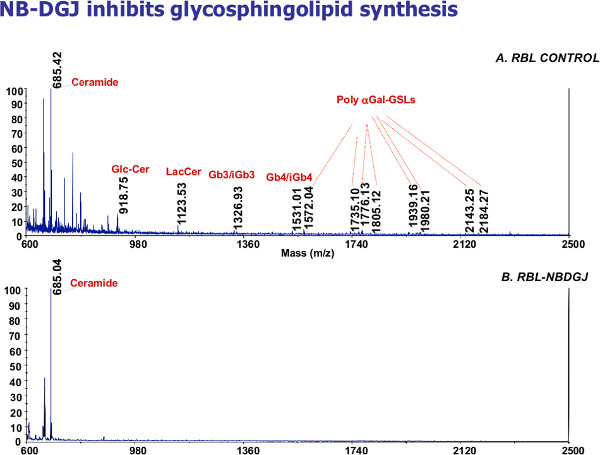
Figure 3. Glycosphingolipidomics of rat leukemia cells. A. Representative MALDI mass spectrum; each ion represents a specific glycosphingolipid structure, structural isomers cannot be distinguished. B. NB-DGJ, a drug that inhibits glycosphingolipid synthesis, significantly reduces all glycosphingolipids produced in RBL cells.
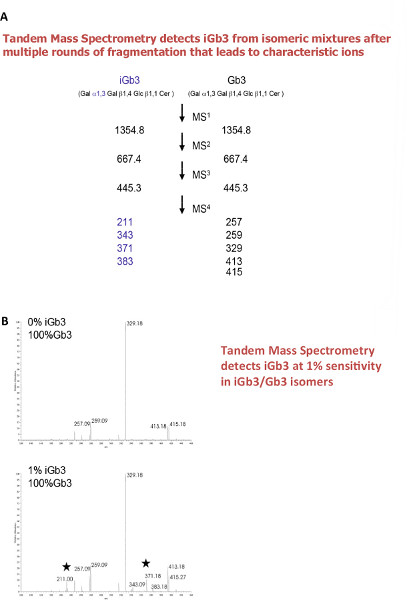
Figure 4. Tandem Mass Spectrometry of isomers of Glycosphingolipids. A. Characteristic decomposition pathways for positive mode ion trap mass spectrometry of permethylated GSLs Gb3Cer and iGb3Cer. Gb3 and iGb3 are clearly separated by their fragmentation patterns. The differences in linkage are reflected in the MS4 product ions consistent with the isobaric m/z 445 precursors. B. Representative results showing iGB3 can be measured in a mixture of iGb3/Gb3 isomers. Stars indicate diagnostic ions for iGb3 only. Click here to view larger figure.
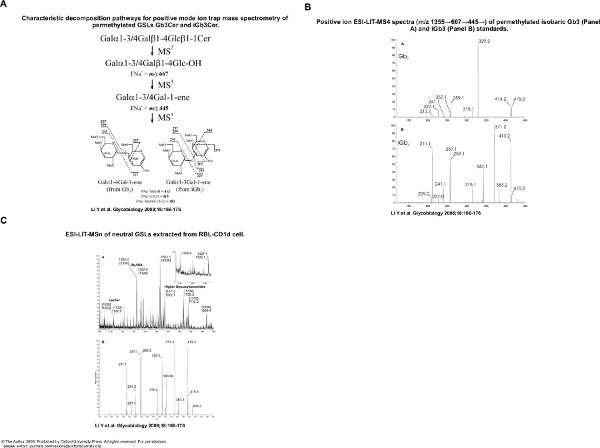
Figure 5. Ion trap MS allows analysis of isomers of glycosphingolipids. A. Standard iGb3 and Gb3 show difference of fragmentation profile after 4 rounds of fragmentation in an ion trap LTQ mass spectrometer. B. Characteristic MS4 profile of ions derived from iGb3 or Gb3. C. Trihexosyleramides are mixtures of iGb3 and Gb3 isomers which can be identified by ion trap MS method (Figure by Dapeng Zhou). Click here to view larger figure.
Discussion
The glycome, a term coined in analogy to the genome and proteome, refers to all saccharide structures of an organism. To fully comprehend the manifold functions of glycosylation will require an integrated approach including both functional and structural glycomics studies. Both are complicated by the non-template driven nature of glycan biosynthesis, the resulting complexity and diversity of glycan structures, the frequent involvement of aglycone structure in modulating glycan ligand-receptor interactions at the molecular level, and the functional importance of low abundance ligands.
It is generally accepted that mass spectrometry (MS) is an indispensable method for structural glycomics studies, especially for identifying and characterizing low abundance ligands. To observe glycans or glycoconjugates as molecular species, we often use a highly efficient, low energy ionization method, called electrospray ionization (ESI). For more detailed characterization of glycan structure, it is essential to select individual molecular species and break the glycans into smaller pieces. This is normally done by collision-induced dissociation, which involves activating the glycans through collision with inert gas molecules. The increased energy induces bond breakage, and systematic analysis of the resulting fragments provides information about the molecular structure of the glycan. Often, “signature” fragments can be generated that are diagnostic for particular glycan structural features. Together with the molecular mass, these fragments can sometimes be sufficient for identifying glycans, but in the past much more information has been required to fully characterize them, particularly if the structure is novel. These methods include sugar composition analysis, gas chromatography mass spectrometry analysis of partially methylated alditol acetates for linkage analysis, and specific glycosidase digestion17.
As demonstrated over the past decade18-19, multiple rounds of low energy fragmentation, which can be effectively carried out in an ion-trap mass spectrometer (IT-MS), greatly improves the information yield from glycan mass spectral analysis. With four or five rounds of fragmentation (which cannot be done with other MS instruments), it is possible to distinguish isomeric glycans that contain the same sugar components arranged in different sugar linkages, even when these are present together in mixtures of components with the same molecular mass (isobars). For this analysis, the glycans were derivatized, replacing all free hydroxyl groups with methyl groups using permethylation. While structural assignment of the glycans is possible without permethylation, the permethylation increases sensitivity17.
Levery and Zhou, have combined all of the potential advantages of IT-MS methodology, including the detection of isomeric structures using signature diagnostic ions, observable only in MS4 and MS5 spectra, for the highly sensitive identification and quantitation of glycosphingolipids present in the form of multiple isobaric mixtures7-9. In theory, we may be able to identify every existing glycolipid structure, pending the availability of standard glycolipids, which can be chemically synthesized.
The critical steps of this analysis are the recovery rate of GSLs from biological samples. Typically 80 μg of GSL can be recovered from 100 million tumor cells such as RBL. To generate sufficient molecular ions for multiple rounds of fragmentation in ion-trap mass spectrometers, at least 10 microgram of tumor GSLs are required. Low yield of GSLs during purification will lead to low abundance of ions, which could not be subjected to further fragmentation. The success of analysis is dependent on the amount of GSLs which are recovered. Typically, final concentration of permethylated GSLs should reach 1 mg/ml, dissolved in methanol, before being analyzed on a nano-electrospray source. The limit for a thorough MSn analysis is dependent on the abundance of specific ion in MS1 profile. A typical e7 ion abundance is required for a thorough MS5 analysis. The low yield of GSLs during purification can be caused by loss of GSLs during peracetylation step (which removes the phospholipids), when the per-acetylated GSLs must be bound to Florisil resin chromatography column, washed, and subsequently eluted. To ensure the binding of per-acetylated GSLs to silica gel, samples should be reloaded twice to the chromatography column after flowing through.
Disclosures
The authors have nothing to disclose.
Acknowledgements
DZ is supported by MD Anderson Cancer Center and NIH grants AI079232. MD Anderson Cancer Center is supported in part by NIH grant CA16672.
Materials
| Name of the reagent | Company | Catalogue number | Comments (optional) |
| 1,2 Dichloroethane | Sigma-Aldrich | 34872 | Carcinogenic |
| Acetic acid | VMR | JT9524-0 | |
| Acetic anhydride | Fluka | 45830 | |
| Acetone | Fischer | A18P-4 | |
| Chloroform | Fischer | C606-1 | Carcinogenic |
| DEAE Sephadex A25 (Cl Form) | GE Healthcare Biosciences | AB 17—170-01 | 100 gram |
| Decane (anhydrous): | Sigma-Aldrich | 457116 | 100 ml |
| Dichloroacetic acid | Sigma | D54702 | Carcinogenic |
| Dialysis cassette | Fischer(Pierce) | 66110 | Slide-A-Lyzer, 3500 MWCO, 3-12 ml |
| DMSO | Thermo Scientific | 20684 | |
| Ethyl acetate | Fischer | UN1173 | |
| Florisil | Fluka | 46384 | for packing chromatography column |
| Hexanes | EMD | HX0290-6 | |
| Iodomethane | Riedel de Haen, Germany | 03810 | stabilized with silver foil Carcinogenic |
| Methanol | VWR/EMD | MXO475-1 | |
| Neutral glycosphingolipid Qualmix | Matreya | 1505 | |
| Monosialganglioside mixture | Matreya | 1508 | |
| Disialoganglioside mixture | Matreya | 1509 | |
| Lactosyl ceramide and sialosylderivatives | Matreya | 1510 | |
| Gangliotetraosyl ceramide and sialosyl derivatives | Matreya | 1511 | |
| Pasteur pipettes | Fischer | 13-678-6A | 9″” |
| Pyridine: | Sigma | 270407 | |
| Sodium Hydroxide: | BDH | 0292 | 500 gram |
| Sodium methoxide | Sigma | 404367 | 0.5 M solution in methanol |
| Toluene | J.T. Baker | 9351-03 | 4 L |
| Name of the equipment | Company | Catalogue number | Comments (optional) |
| Pasteur pipettes | Fischer | 13-678-6A | 9″” |
| Fiber glass (glass wool) | Corning Incorporated | 3950 | Pyrex 9989 glass |
| 12×75 mm glass tubes | Fischer | 14-962-10B | |
| Calibrated pipettes (100 microlitter) | VMR | 53432-921 | |
| 16×100 mm disposible borosilicate tubes x1,000 | Fischer | 14-961-29 | With screw caps |
| Caps for glass tubes | Kimble | 40566C | size/13-415 |
| Merck TLC plates | Sigma | Z 29, 301-6 | |
| Glass tank for TLC | Sigma | Z 12,619-5 | |
| Drummond Microcaps | Drummond scientific company | 1-000-0100 | 10 μl, for loading of TLC samples |
| Sunrise Microplate Absorbance Reader | Tecan | A-5082 | |
| Whatman Chromatography Paper | Fischer | 05-716-3E | 18 cm x 34 cm For TLC Tank |
| Orcinol ferric chloride spray reagent | Sigma | O7875 | For detecting glycosphingolipids on TLC plates |
| Prevel Spray Unit | Sigma | Z 36,555-6 | |
| Sonicator | Fischer | FS20 | |
| Centrifuge | Sorvall | Legend RT | |
| Speedvac | Savant | AS160 | With chemical trap for organic solvants |
| MALDI TOF-TOF Mass spectrometer | Applied Biosystems | Proteomics 4700 | |
| Ion Trap Mass spectrometer | Thermo | LTQ |
References
- Schnaar, R. L., Suzuki, A., Stanley, P., Varki, A., Cummings, R. D., Esko, J. D., Freeze, H. H., Stanley, P., Bertozzi, C. R., Hart, G. W., Etzler, M. E. Chapter 10 Glycosphingolipids. Essentials of Glycobiology. , (2009).
- Bendelac, A., Paul, W. E. Chapter 17 NKT cells and other Innate-like T and B lineages. Fundamental Immunology. , (2011).
- Jacewicz, M., Clausen, H., Nudelman, E., Donohue-Rolfe, A., Keusch, G. T. Pathogenesis of shigella diarrhea. XI. Isolation of a shigella toxin-binding glycolipid from rabbit jejunum and HeLa cells and its identification as globotriaosylceramide. J. Exp. Med. 163 (6), 1391-1404 (1986).
- Zhou, D. The immunological function of iGb3. Curr. Protein Pept. Sci. 7 (4), 325-333 (2006).
- Miyamoto, K., Miyake, S., Yamamura, T. A synthetic glycolipid prevents autoimmune encephalomyelitis by inducing TH2 bias of natural killer T cells. Nature. 413 (6855), 531-534 (2001).
- Zhou, D., Levery, S. B., Hsu, F. F., Wang, P. G., Teneberg, S., Almeida, I. C., Li, Y., Xu, H., Wang, L. X., Xia, C., Ibrahim, N. K., Michael, K. Immunologic mapping of glycomes: implications for cancer diagnosis and therapy. Frontiers in Bioscience. S3, 1520-1532 (2011).
- Li, Y., Thapa, P., Hawke, D., Kondo, Y., Furukawa, K., Furukawa, K., Hsu, F. F., Adlercreutz, D., Weadge, J., Palcic, M. M., Wang, P. G., Levery, S. B., Zhou, D. Immunologic glycosphingolipidomics and NKT cell development in mouse thymus. J. Proteome Res. 8 (6), 2740-2751 (2009).
- Li, Y., Zhou, D., Xia, C., Wang, P. G., Levery, S. B. Sensitive quantitation of isoglobotriaosylceramide in the presence of isobaric components using electrospray ionization-ion trap mass spectrometry. Glycobiology. 18 (2), 166-176 (2008).
- Li, Y., Teneberg, S., Thapa, P., Bendelac, A., Levery, S. B., Zhou, D. Sensitive detection of isoglobo and globo series tetraglycosylceramides in human thymus by ion trap mass spectrometry. Glycobiology. 18 (2), 158-165 (2008).
- Hakomori, S. I. A rapid permethylation of glycolipid, and polysaccharide catalyzed by methylsulfonyl carbanion in dimethyl sulfoxide. J. Biochem. (Tokyo). 55, 205 (1964).
- Ciucanu, I., Kerek, F. Rapid and simultaneous methylation of fatty and hydroxy fatty acids for gas-liquid chromatographic analysis. Carbohydr. Res. 131, 209 (1984).
- Kang, P., Mechref, Y., Klouckova, I., Novotny, M. V. . Rapid Commun Mass Spectrom. 19 (23), 3421-3428 (2005).
- Levery, S. B., Hakomori, S. Microscale methylation analysis of glycolipids using capillary gas chromatography-chemical ionization mass fragmentography with selected ion monitoring. Methods Enzymol. 138, 13-25 (1987).
- Spooncer, E., Fukuda, M., Klock, J. C., Oates, J. E., Dell, A. Isolation and characterization of polyfucosylated lactosaminoglycan from human granulocytes. J. Biol. Chem. 259 (8), 4792-4801 (1984).
- Ciucanu, I., Costello, C. E. Elimination of oxidative degradation during the per-O-methylation of carbohydrates. J. Am. Chem. Soc. 125, 16213 (2003).
- Platt, F. M., Neises, G. R., Karlsson, G. B., Dwek, R. A., Butters, T. D. N-butyldeoxygalactonojirimycin inhibits glycolipid biosynthesis but does not affect N-linked oligosaccharide processing. J. Biol. Chem. 269 (43), 27108-27114 (1994).
- Mulloy, B., Hart, G. W., Stanley, P., Varki, A., Cummings, R. D., Esko, J. D., Freeze, H. H., Stanley, P., Bertozzi, C. R., Hart, G. W., Etzler, M. E. Chapter 47 Structural Analysis of Glycans. Essentials of Glycobiology. , (2009).
- Ashline, D., Singh, S., Hanneman, A., Reinhold, V. Congruent strategies for carbohydrate sequencing. 1. Mining structural details by MSn. Anal. Chem. 77 (19), 6250-6262 (2005).
- Zhang, H., Singh, S., Reinhold, V. N. Congruent strategies for carbohydrate sequencing. 2. FragLib: an MSn spectral library. Anal. Chem. 77 (19), 6263-6270 (2005).

