Genetic Manipulation in Δku80 Strains for Functional Genomic Analysis of Toxoplasma gondii
Summary
Here we report a method for using type I and type II Δku80 strains of Toxoplasma gondii to efficiently generate targeted gene deletions and gene replacements for functional genomic analysis.
Abstract
Targeted genetic manipulation using homologous recombination is the method of choice for functional genomic analysis to obtain a detailed view of gene function and phenotype(s). The development of mutant strains with targeted gene deletions, targeted mutations, complemented gene function, and/or tagged genes provides powerful strategies to address gene function, particularly if these genetic manipulations can be efficiently targeted to the gene locus of interest using integration mediated by double cross over homologous recombination.
Due to very high rates of nonhomologous recombination, functional genomic analysis of Toxoplasma gondii has been previously limited by the absence of efficient methods for targeting gene deletions and gene replacements to specific genetic loci. Recently, we abolished the major pathway of nonhomologous recombination in type I and type II strains of T. gondii by deleting the gene encoding the KU80 protein1,2. The Δku80 strains behave normally during tachyzoite (acute) and bradyzoite (chronic) stages in vitro and in vivo and exhibit essentially a 100% frequency of homologous recombination. The Δku80 strains make functional genomic studies feasible on the single gene as well as on the genome scale1-4.
Here, we report methods for using type I and type II Δku80Δhxgprt strains to advance gene targeting approaches in T. gondii. We outline efficient methods for generating gene deletions, gene replacements, and tagged genes by targeted insertion or deletion of the hypoxanthine-xanthine-guanine phosphoribosyltransferase (HXGPRT) selectable marker. The described gene targeting protocol can be used in a variety of ways in Δku80 strains to advance functional analysis of the parasite genome and to develop single strains that carry multiple targeted genetic manipulations. The application of this genetic method and subsequent phenotypic assays will reveal fundamental and unique aspects of the biology of T. gondii and related significant human pathogens that cause malaria (Plasmodium sp.) and cryptosporidiosis (Cryptosporidium).
Introduction
Toxoplasma gondii is a common obligate intracellular protozoan parasite that frequently and chronically infects a wide range of animals and humans5. It is estimated that more than 1 billion humans are currently and chronically infected by this pathogen. In addition to the importance of disease caused by T. gondii infection, the increasing availability of experimental tools, powerful genomics resources6, ease of in vitro growth and excellent mouse models have made T. gondii a leading model system for the broader study of intracellular eukaryotic pathogens and other significant apicomplexan parasites that cause devastating diseases such as malaria (Plasmodium sp.) and Cryptosporidiosis (Cryptosporidium)5,7. A significant limitation of Toxoplasma gondii as a model organism has been the inefficient recovery of progeny that carry targeted genetic manipulations. This problem in gene targeting is due to a low frequency of homologous recombination relative to the very high frequency of nonhomologous recombination in wild-type strains of T. gondii even when extensive DNA homology is provided in DNA target molecules used in genetic studies2.
We recently genetically blocked the major pathway of nonhomologous recombination in type I and type II strains of Toxoplasma gondii by deleting the gene encoding the KU80 protein1,2. The resulting type I and type II Δku80 strains exhibit normal growth rates, size and behavior both in vitro and in vivo during tachyzoite and bradyzoite stages in vitro and in vivo, however, these strains exhibit essentially a 100% frequency of homologous recombination and this phenotype increases the likelihood of rapidly isolating desired progeny of targeted genetic manipulations by several hundred to several thousand-fold1,2,4. Recent community-wide use of type I and type II Δku80 strains has significantly ramped up the pace, the variety, as well as the success rate of targeted genetic approaches in T. gondii1-4,8-13. Here we describe a comprehensive protocol for targeted gene deletion, gene replacement, and gene tagging in Δku80 strains of T. gondii. We demonstrate how to design and reliably target genetic manipulations to specific genes or genetic loci in Δku80 strains of Toxoplasma gondii.
Protocol
1. Gene Targeting to Delete a Gene of Interest
This protocol is designed for the efficient generation of a mutant strain that has a targeted gene deletion at a defined genetic locus. The previously described T. gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase (HXGPRT) selectable marker is used in this protocol for safe and reliable marker insertion following mycophenolic acid and xanthine (MPA + X) selection14-16. This protocol uses a validated and cost-effective method for constructing DNA targeting molecules based on yeast recombinational cloning17,18. Several alternative protocols are commercially available for constructing recombinant targeting molecules by using bacterial recombinational cloning methods, in vitro recombinational cloning methods, or fusion PCR. The protocol described below provides the highest overall efficiency and reliability of recovering cloned parasites possessing a targeted gene deletion in Δku80 strains of T. gondii.
1.1 Preparation of targeting DNA
- Access ToxoDB6 (http://www.toxodb.org) to obtain genomic sequences for the gene of interest (GOI).
Note: Genomic sequences should be obtained from the Type I, II or III genome sequence in the ToxoDB database that is closest to the strain being manipulated. For example: GT1 for RH and ME49 for Pru.
- Design specific overlap primers that amplify the 5' and 3' genomic targeting flanks of the gene of interest (GOI) incorporating a 33 bp overlap with the yeast shuttle vector pRS416 (or alternatively pRS426) and incorporate a 33 bp overlap with the selectable marker (HXGPRT) (Figures 1A-B). Add a unique restriction enzyme site in each primer at both ends of the 5' and 3' genomic targeting flanks, placed between the 33 bp overlap and the T. gondii-specific primer sequence(s) (Figures 1A-B).
Note: A greater than 30 bp overlap is required for efficient yeast recombination cloning. The 5' and 3' genomic targeting flanks are designed to amplify specific DNA fragments between 800 and 1,400 bp that define a targeted deletion. Be aware that shorter flanks will significantly reduce targeting efficiency in the Δku80Δhxgprt background2.
- PCR amplify the 5' target flank using primers F1 and R1, and PCR amplify the 3' target flank using primer F2 and R2 (Figures 1A-C) from genomic DNA3. Separately, PCR amplify the ~2 Kbp HXPGRT gene including the associated 5' and 3' dhfr flanking regions of the HXGPRT cDNA pminiHXGPRT cassette14 using primers HXF and HXR (Figures 1B-C).
Note: Amplify target flanks using genomic DNA from the parental strain to be transfected.
Note: Place the HXGPRT marker in the forward orientation relative to the 5' and 3' DNA targeting flanks to facilitate subsequent efficient genetic manipulations, such as the targeted removal of HXGPRT from the disrupted locus.
- Verify correct PCR product sizes by agarose gel electrophoresis and estimate the DNA fragment concentration using agarose gel standards or another method for determining DNA concentration.
- Generate competent yeast aliquots as described in Gietz and Schiestly17.
- Combine 50 ng of 5' genomic targeting flank, 50 ng of 3' genomic targeting flank, 100 ng of HXGPRT selection marker, and 50 ng of linearized shuttle vector with sterile H2O to achieve a final volume of 10 – 20 μl. Transform competent yeast with the three PCR products and yeast-E. coli shuttle vector for recombinational cloning using the protocol described by Gietz and Schiestly17. Plate transformed yeast on uracil-minus minimal medium agar plates and incubate at 30 °C for 2 – 3 days.
Note: The ordered assembly of the plasmid, the 5' target flank, the HXGPRT marker, and the 3' target flank is facilitated by the engineered 33 bp overlap between the DNA fragments (Figures 1A-B) and is mediated by homologous recombination in yeast.
- Harvest yeast by adding 2 ml of 2x YPAD to the uracil-minus plates and gently scraping to dislodge colonies without disrupting the agar. Centrifuge the yeast solution for 5 min at 5,000 x g and discard the supernatant.
- Resuspend the yeast pellet in 250 μl of a resuspension buffer containing RNaseA and add 50 – 100 μl of acid-washed glass beads to the solution. Vortex for 5 min at the highest speed to smash the yeast cells.
- Allow the glass beads to settle to the bottom of the tube and transfer the supernatant to a 2 ml Eppendorf tube.
- Isolate yeast DNA using a miniprep DNA isolation kit, resuspending in a final volume of ~100 μl.
- Mix 2 μl yeast DNA (~50 ng) with 40 μl of DH10B electroporation competent E. coli kept on ice.
- Transfer the solution to a chilled 2 mm gap electroporation cuvette and electroporate the bacterial cells at 2.4 kV and 129 Ω.
- Rescue cells by adding 0.8 ml SOC broth to each cuvette and transfer solution to a 14 ml snap-cap Falcon tube. Incubate cells at 37 °C on a roller drum for 40 – 60 min.
- Plate E. coli on 2XYT + ampicillin (AMP) plates and incubate the plates at 37 °C overnight.
- Select ~6 – 8 single colonies and grow in 3 ml 2XYT + AMP for ~16 hr at 37 °C.
- Prepare a 30% glycerol freezer stock of each E. coli clone.
- Isolate plasmid pΔGOI from E. coli clones using a miniprep kit, resuspending in a final volume of ~100 μl.
- Validate the pΔGOI using restriction enzyme digests to measure the DNA plasmid size and verify the expected pΔGOI DNA banding patterns.
- Finalize validation of pΔGOI by DNA sequencing of the 5' and 3' genomic targeting flanks to verify 100% DNA sequence based on genome sequence data in http://www.ToxoDB.org.
Note: Near perfect sequence homology is essential for maximizing gene targeting efficiency3 since each base pair difference constitutes a corresponding cut in the length of perfect homology.
Note: The genome sequence of the type II Δku80 strain based on the Pru parental strain is not available at this time. This work uses the type II ME49 genome sequence as the surrogate genome for the type II Δku80 strain. Based on sequence data at a number of genetic loci, it is estimated that the ME49 genome and the Pru genome exhibit single nucleotide polymorphisms at a frequency of ~1 per 10,000 nucleotides, or less.
- Generate >200 μg of pΔGOI stock by inoculating a ~250 ml 2XYT + AMP overnight culture with glycerol stocks from a validated E. coli miniprep clone.
- Isolate pΔGOI plasmid DNA from the large culture using a maxiprep DNA isolation kit, resuspending in a final volume of ~1,000 μl.
- Repeat restriction enzyme digests, or DNA sequencing of the pΔGOI maxiprep DNA to verify the correct targeting DNA molecule prior to transfection.
- Linearize ~15 μg pΔGOI at the 5' end using the unique restriction enzyme X (RE.X) digest site built into the 5' target flank (Figures 1B).
Note: Gene targeting in T. gondii requires a minimum of ~10 μg of targeting DNA to obtain an efficient frequency of targeting events at the gene locus of interest2.
- Validate linearization of pΔGOI and determine the DNA concentration by agarose gel electrophoresis or an alternative method.
Note: If restriction enzyme digestion at the 5' flank does not yield complete linearization, the designed 3' unique RE.Y digest site can be used as an alternative site for linearization of pΔGOI.
Note: A completely linearized targeting DNA molecule is essential for successful gene targeting by homologous recombination in the Δku80 strains.
- Neutralize the restriction enzyme by incubating at 68 °C for 15 – 20 min.
- Prepare at least 15 μg of linearized plasmid in 100 μl of sterile H2O. Store linearized pΔGOI at -20 °C until the time of transfection.
1.2 Preparation of T. gondii parasites, transfection, selection, subcloning, validation, archival and maintenance of genetically manipulated strains
General methods for the culture and manipulation of T. gondii in human foreskin fibroblast (HFF) cells are described19-21. The Δku80Δhxgprt strains replicate normally in parasite infection medium (Eagles Minimal Essential Medium (EMEM) growth medium supplemented with 1% fetal bovine serum (FBS) and 1% Antimycotic-Antibiotic diluted from a 100x stock)1,2. All work with Toxoplasma gondii must be performed using biosafety level 2 procedures. The T. gondii RHΔku802 parental strain was generated from the RHΔhxgprt strain from the Roos Lab14. The PruΔku801 parental strain was generated from PruΔhxgprt (Prugniaud strain BSG-422) that contains a stably integrated CAT selectable marker and a green fluorescent protein (GFP) reporter under the control of the bradyzoite stage specific promoter LDH2.
- Inoculate a 25 cm2 flask containing confluent HFF cells with 1 x 106 viable type I RH Δku80Δhxgprt parasites, or inoculate two 25 cm2 flasks with 2 x 106 viable type II Prugniaud (Pru) Δku80Δhxgprt parasites.
Note: Viable parasites are defined as extracellular parasites that have freshly lysed out.
Note: This scale provides a sufficient number of viable parasites for three transfection experiments.
- Inspect the Δku80Δhxgprt infected cultures ~68 – 72 hr post-infection. Verify the presence of viable parasites and ~90 – 95% lysis of HFF cells to plan the precise timing of transfection.
Note: The isolation of freshly egressed parasites is essential for the success of this protocol because low parasite viability will abolish gene-targeting efficiency.
- Agitate viable parasites into solution by closing the plug-seal lid on the 25 cm2 flask and vigorously shake the flask back and forth to agitate the parasites off of the growth surface without splashing liquid onto the inside of the lid or the neck of the flask.
Note: Parasite harvest does not require a syringe-needle release protocol for the type I or type II Δku80 strains.
- Transfer the parasite solution to a 10 ml syringe attached to a filter holder containing a 3 μm membrane and place the filter unit on top of an open 15 ml screw-cap tube. Syringe filter the parasites into the 15 ml screw-cap tube.
Note: Filtering the parasites through the membrane removes infected cells and cellular debris.
- Determine the parasite concentration (tachyzoite forms per ml) using a hemocytometer.
Note: Optional: Using the parasite solution, set up a plaque forming unit (PFU) assay21 that will be read 7 – 8 days later to determine adequate viability of the transfected parasites (PFU to tachyzoite ratio of ≥0.2).
- Pellet parasites for 7 min at 1400 x g and aspirate supernatant to ~0.4 ml without disturbing the parasite pellet. Spin for 2 min at 1,400 x g and aspirate remaining supernatant to ~0.01 ml without disturbing the parasite pellet.
- Resuspend the parasite pellet by flicking the bottom of the tube and immediately add cytomix23 buffer (1.33x concentration) to the parasite pellet to obtain a parasite concentration of 4 – 5 x 107 parasites/ml cytomix.
Note: Omit the addition of ATP and glutathione to cytomix since these additions do not improve the efficiency of gene targeting.
- Thaw the 100 μl aliquot of the linearized pΔGOI from protocol 1.1 (step 26).
- Transfer 0.3 ml of the parasite cytomix solution (~1.3 – 1.6 x 107 parasites) to the 100 μl of linearized pΔGOI from protocol 1.1 (step 26), and immediately transfer the entire 0.4 ml parasite + pΔGOI mix to a chilled 2 mm gap electroporation cuvette.
- Electroporate the parasites at 1.4 kV and 24 Ω.
- Rest the transfection cuvette at room temperature for ~5 min then transfer the entire contents of the transfection cuvette into a 150 cm2 flask containing confluent HFF cells with 30 ml infection medium and incubate the infected culture overnight.
- Begin selection in mycophenolic acid + xanthine (MPA + X) ~20 hr post-transfection (Figure 2A) by replacing the infection medium with MPA + X selection medium (MPA (25 μg/ml) and xanthine (50 μg/ml) in infection medium).
Note: Do not disturb the incubating MPA + X selection flask.
- Estimate the total number of PFUs in the MPA + X selection flask ~8 days post-transfection by visually inspecting the monolayer. Verify the presence of developing PFUs and healthy zones of infection by light microscopy.
Note: If plaques are not visible by day 8, maintain the selection and monitor for signs of infection since mutant strains may have a reduced replication rate.
Note: The Δku80Δhxgprt parasite strains have an extremely low background of undesired nontargeted events, which allows for the successful isolation of mutant strains with quite severe, but not lethal growth defects. If a gene is essential and it cannot be deleted, the primary transfection, or subsequently passed parasite population, may reveal a phenotype where the parasites cease to replicate during the selection as the population resolves to targeted knockouts.
- Shake the flask as in step 3 to dislodge extracellular parasites from the monolayer [after visible plaques have developed] to generate a parasite solution. Transfer ~0.5 ml of the parasite solution from the MPA + X selection flask to a 25 cm2 MPA + X selection flask containing confluent HFF cells (designate this culture pass 1A).
- Dislodge parasites in the original 150 cm2 MPA + X selection flask ~3 days later and transfer ~0.5 ml parasite solution to a second 25 cm2 MPA + X selection flask containing confluent HFF cells (designate this culture pass 1B).
- Allow zones of infection to develop in pass 1A and/or pass 1B flasks for ~5 days. Visually verify by light microscopy that pass 1A and 1B flasks contain a minimum of 25 viable PFUs with healthy zones of infection.
- Choose either the pass 1A or pass 1B flask for continued passage in MPA + X selection medium in 25 cm2 flasks. Maintain passage under MPA + X selection.
Note: Parasites must be cultured for a sufficient number of generations to delete nonintegrated persisting episomes from the population prior to subcloning. Do not perform subcloning prior to 20 days post-transfection for type I RH Δku80Δhxgprt, or prior to 25 days post-transfection for type II Pru Δku80Δhxgprt parasites to allow sufficient time to dilute nonintegrated episomes1,2.
- Subclone parasites in a 96-well tray containing confluent HFF cells with MPA + X selection medium. Prepare one tray at a concentration of 1 parasite/well and a second tray at a concentration of 2 parasites/well.
Note: Subclone parasites that have recently lysed (~90 – 95% of the HFF cells in the 25 cm2 flask) to ensure that the parasites have high viability.
Note: The optimal time frame post-transfection for subcloning was established by measuring how rapidly successfully targeted parasites arise at a high frequency in the selected population1.
- Continue to passage the primary population of transfected parasites in MPA + X selection medium using a weekly passage schedule of infecting a 25 cm2 HFF flask with 10 μl of the parasite solution.
Note: If clones carrying the targeted gene deletion (knockouts) are not obtained from the first subcloning in step 18, resubclone the parasites from the continuously maintained population ~10 – 12 weeks post-transfection.
Note: One of the reasons a gene deletion is not obtained arises from the episomal persistence of some pΔGOI plasmids. Episomal persistence is determined by the sequences contained in the 5' and the 3' genomic targeting flanks and does not appear to arise from the HXGPRT genetic element. Approximately 5 – 10% of targeting plasmids exhibit significant episomal persistence that necessitates a later time of subcloning to allow the parasite population a sufficient number of generation times to dilute the persisting episomes and resolve the population to primarily stable integrants.
- Score the 96-well tray 6 – 7 days post-subcloning (type I parasites) or 7 – 8 days post-subcloning (type II parasites) for wells that contain a single PFU, identified as a single zone of infection in light microscopy at either 40X or 60X power. Mark a small dot on the 96-well tray lid to designate the location of the PFU in the well.
- Mix the contents of each well containing a single PFU using a pipette set at 50 μl (200 μl tip) by directing fluid flow over the PFU to disperse parasites in the well.
Note: Mixing the well accelerates parasite lysis of the HFF monolayer. Type I RH strains will lyse the well ~4 days after mixing, type II Pru parasites will lyse the well ~5 days after mixing.
- Select a dozen lysed wells (clones) and scratch across the bottom of each of these lysed wells using a 0.5 – 10 μl pipette tip while simultaneously drawing-up 6 μl of parasite solution. Transfer the 6 μl of parasite solution to a well in a 24-well tray containing confluent HFF cells in 1 ml MPA + X selection medium.
Note: It is essential to scratch the bottom of the well to keep the opening bore of the pipette tip in constant contact with parasites residing at the bottom of the well.
- Monitor the 24-well tray for lysis of the HFF cells.
Note: Type I RH parasites will typically lyse the well in the 24-well format in ~4 days. Type II Pru parasites will typically lyse the well in ~5 days.
- Transfer 2 μl of viable parasite solution scratched from the bottom of each well in the 24-well format to the corresponding well of a new 24-well tray containing confluent HFF cells and 1 ml MPA+X selection medium per well.
Note: All parasites clones and lines can be continuously maintained by passing 2 μl of viable parasite solution every 7 days in this 24-well tray format.
- Verify that parasites were successfully transferred to a new 24-well tray by visually inspecting with light microscopy ~18 hr post-passage.
- Harvest parasites from the lysed wells of the 24-well tray from step 23 – 24 using either a 1 ml or 10 ml pipette and transfer the parasite solution to an Eppendorf tube.
- Pellet the parasites at 1,400 x g for 7 min, rinse once in 1 ml phosphate buffered saline (PBS), and pellet again at 1,400 x g for 3 min.
- Aspirate PBS from the pellet, then add 200 μl of PBS to the pellet to resuspend the parasites. Freeze the parasite solution at -80 °C until DNA isolation.
- Isolate parasite DNA from each clone using a tissue DNA isolation minikit.
- Validate the targeted gene deletion (knockout) by PCR using validation primers (Figures 2A-C). Test for the absence of the functional coding region of the gene of interest and using the upstream CxF and downstream CxR genomic DNA primers (Figures 2A-B) test for the presence of PCR products that span the genomic targeting flanks and HXGPRT to demonstrate correct 5' and 3' targeted integration of the HXGPRT gene into the deleted gene locus.
Note: Primers for the validation must be unique to the gene of interest or a false positive result can be obtained. Primer design can be validated on http://www.toxodb.org by blasting primer sequences to the genome(s) to verify primers are unique.
- Passage parasite clones on a weekly schedule as described in step 24. Once the targeted deletion has been validated, continuing selection in MPA+X is optional.
- Archive parasite clones by preparing freezer stocks. Pellet extracellular parasites from lysed HFF cells in a 25 cm2 culture flask, remove media and gently resuspend parasite pellet in cell culture freezing medium at a parasite concentration >4 x 107 parasites per ml. Transfer aliquots to cryovials and store parasites indefinitely in liquid nitrogen, or at -80 °C.
2. Deletion of HXGPRT
This protocol is designed for removing the HXGPRT marker from its integration site in the genome of a genetically manipulated Δku80 strain. Removal of HXGPRT allows for marker recovery and the generation of strains with multiple genetic manipulations using only selections based on HXGPRT1-3. While the protocol detailed below describes the method to re-target a locus to delete HXGPRT, it should be noted that removal of HXGPRT at the gene locus of interest also permits simultaneous re-integration of the wild-type gene (complementation), re-integration of a mutant gene, as well as re-integration of a tagged gene (N- or C-terminal GFP, HA tag, etc,), allowing for a variety of genetic manipulations. The mechanisms of action24 and targeting protocols using 6-thioxanthine selections have been previously described1-3,15.
2.1 Delete HXGPRT
- Excise the HXGPRT selectable marker from pΔGOI by digesting with RE.Z to cut at the unique restriction enzyme sites flanking the HXGPRT (Figure 1B).
- Verify complete digestion of DNA by agarose gel electrophoresis. Isolate the larger of the two bands from the agarose gel.
Note: The larger of the two bands contains the plasmid along with the 5' and 3' DNA target flanks, but not the HXGPRT gene.
- Place the agarose section containing the larger DNA band in a spin column laying the gel flat on the membrane. Spin the column at 13,000 x g for 4 min at RT. Add sterile H2O to the flow-through to bring the volume to 100 μl.
Note: Other commercial methods are available to isolate DNA from agarose.
- Concentrate DNA by EtOH precipitation, resuspending the DNA in a final volume of 18 μl sterile H2O.
- Mix 1 μl of concentrated DNA with 7 ml H2O, 1 μl 10x ligation buffer and 1 μl T4 DNA ligase (5 units) and place the reaction at 4 °C overnight to generate pΔGOIc (pΔGOIclean) (Figure 3A).
Note: DNA fragments with RE.Z containing DNA ends can be designed and included at this step to create plasmids suitable for targeted re-integration of the wild-type gene (complementation), targeted re-integration of a mutant gene as well as targeted re-integration of a tagged gene (N- or C-terminal GFP, HA tag, etc.).
- Dilute the reaction 2x with sterile H2O prior to electroporation.
- Transform pΔGOIc into E. coli as in protocol 1.1 to subclone, isolate, and validate the targeting plasmid. Then linearize pΔGOIc in preparation for transfection (see steps 1.1.11 to 1.1.26).
- Repeat the parasite transfection protocol as outlined in protocol 1.2 (steps 1 to 11).
Note: Prior to conducting steps 1 to 8, verify that targeted removal of HXGPRT is feasible in the mutant strain. Infect a 150 cm2 flask of confluent HFF cells with 1 x 106 tachyzoites of the mutant strain using 6-thioxanthine (6TX) selection medium (200 μg/ml 6TX in infection medium) and place the flask in a PFU assay. Inspect the flask 8 days later to verify that no (or very few; <10) PFU are visible, which is essential to establish that the potential gene targeting efficiency will exceed any potential spontaneous reversion of the mutant strain to a phenotype with reduced HXGPRT expression (a 6TX resistance phenotype).
Note: Approximately 5 – 10 % of HXGPRT integration sites exhibit a significant and spontaneous frequency of 6TX resistance even though the HXGPRT marker is still integrated at the targeted site (see Representative Results section for explanation).
- Begin 6TX selection ~20 hr post-transfection by changing the medium in the 150 cm2 flask to 6TX selection medium.
Note: Do not disturb the flask for ~10 days post-transfection.
- Inspect the flask for PFU formation at day 10 – 12 post-transfection (type I parasites) or day 10 – 16 post-transfection (type II parasites).
Note: Parasites being targeted for the deletion of HXGPRT will not begin to grow under 6TX selection until the HXGPRT gene and mRNA is deleted, and the residual HXGPRT protein is inactivated.
- Shake the selection flask to create a parasite solution, and transfer 0.5 – 1.0 ml of the parasite solution to a new 25 cm2 flask containing confluent HFF cells and 5 ml 6TX selection medium. Repeat the transfer from the primary 150 cm2 to new 25 cm2 flasks once a day for two more days.
Note: Multiple sampling increases the chances of capturing viable targeted parasites that have lost the HXGPRT gene.
- Inspect the 25 cm2 flasks ~5 days after infection to verify the presence of developing PFUs with zones of healthy replicating parasites. Pick one or two flasks containing zones of infection.
Note: Parasites deleted of the HXGPRT gene replicate at a normal growth rate in 6TX selection.
- Continue to passage the parasites in 6TX selection medium.
- Subclone the parasite population after 25 – 30 days of selection. Set up one 96-well tray with ~ 1 parasite/well and another with ~ 2 parasites/well.
- Prepare parasite DNA from isolated clones and maintain clones in a 24-well format culture according to protocol 1.2 (steps 19 to 29).
- Validate deletion of HXGPRT by PCR using the strategy outlined in Figures 3A-B.
3. C-terminal Tagging of Proteins
This protocol is designed for C-terminal tagging of proteins by targeting the tag for integration via double cross over homologous recombination at the genomic locus of the gene using MPA + X selection2,4. This protocol works efficiently because the 5' dhfr sequence of the HXGPRT marker is a fully validated functional 3' untranslated region for other genes2,4.
3.1 Direct C-terminal tagging of proteins at endogenous genetic loci
- Create targeting pΔGOItag plasmid construct using yeast recombinational cloning (Figure 4) and methods described in protocol 1.1. The 5' genomic targeting flank contains the last 800 to 1,200 bp of coding region (or genomic DNA) of the GOI, except the termination codon is moved to a position 3' to the tag of choice (HA tag, Myc tag, His tag, etc.)
- Create the targeted insertion of the C-terminal tag at the endogenous locus of the protein-coding gene by following the steps in protocol 1.1 and 1.2 using the strategy outlined (Figure 4).
- Verify the insertion of the C-terminal tag at the endogenous gene locus using a PCR strategy.
- Retarget the gene locus using methods described in protocol 1 and protocol 2 to delete the HXGPRT selectable marker to create a precisely regulated endogenous gene locus that expresses a tagged protein. This protocol also recovers the HXGPRT selectable marker that can be used again to target another locus in the tagged strain (see protocol 1).
Representative Results
A detailed template is provided for constructing a targeting plasmid to delete a gene, including the placement of restriction enzyme sites and generation of the primers that facilitate genetic targeting and validation of gene targeting, as well as plasmid construction for the subsequent deletion of HXGPRT in a single-step process (Figures 1A-C, Figure 2A, Figure 3A). A general schematic is presented for making a targeted gene deletion (Figure 2A), the primer pairs used to validate the deletion of a knockout, for example, type I rop18 (Figure 2B), and representative results of the PCR validation are shown (Figure 2C). This representative result is shown to illustrate the range of results that can be obtained at genetic loci that are relatively difficult to target. A successfully targeted gene deletion will result in the absence of the gene of interest PCR product (PCR 1), the presence of the 3′ genomic targeting flank (PCR2), and the presence of the HXGPRT selectable marker properly integrated between the 5′ and 3′ genomic targeting flanks that define the deletion (PCR3 and PCR4). Clones 3, 4, 5, 6, 8, 9, and 11 are validated targeted gene deletions (knockouts) where HXGPRT has replaced the GOI (Figure 2C).
A clone that does not contain a gene deletion can be represented by a variety of banding patterns. Typically, a clone without a gene deletion will mirror the parental strain (parental pattern) with bands observed for PCR1 and PCR2, but not for PCR3 or PCR4 as seen for clones 1 and 2 (Figure 2C). This “parental pattern” arises from a few potential mechanisms. Occasionally, MPA resistant selected parasites carry nonintegrated persisting episomes of the pΔGOI targeting plasmid, and we note that continued selection often forces these episomes to integrate. We also observe some rare background in the type I Δku80 strain due to unintended integration of the pΔGOI targeting plasmid, which contains a HXGPRT cDNA expressed via DHFR 5′ and 3′ promoter elements, into either the DHFR locus or into the partially deleted HXGPRT locus14. While this rare event is a targeted integration, it was not the intended integration and serves as a key point to remember in designing any gene replacement strategy. DNA homology greater than 120 bp carried on your pΔGOI targeting plasmid may provide an alternative site for recombination in Δku80 strains2 that could give back an undesired background. In the type II Pru Δku80 strain this background is reduced or nonexistent compared to type I because the HXGPRT selectable marker is based on type I sequences that have nucleotide polymorphisms when compared with type II DNA which greatly reduces this rare background in experiments using the type II Pru Δku80 strain1. If a gene locus is essential (cannot be deleted), or has an extremely low gene targeting frequency at the locus, or if persisting [nonintegrated] episomes are not easily eliminated via growth and selection, this parental pattern will dominate the pattern observed in MPA resistant clones. Alternatively, the targeting DNA molecule is sometimes observed to integrate at only the 5′ or the 3′ genomic targeting flank and a band is observed for either PCR3 or PCR4 (but not both) along with PCR1 and PCR2 as seen for clone 10 (5′ integration) and clone 7 (3′ integration) (Figure 2C). These patterns suggest the infrequent occurrence of a single cross over integration of the targeting plasmid at the locus that integrates HXGPRT but this targeted integration does not delete the gene of interest. This pattern (lanes 7 & 10 in Figure 2C) emphasizes the need to report PCR data to verify that targeted integration occurred instead of a single homologous cross over and a second nonhomologous integration of the targeting molecule. Rarely, we observe a genetic mixture represented by clone 12 (Figure 2C) that represents both a parental pattern and a targeted deletion pattern. This pattern most likely arises on occasion from two parasite genotypes that are present at the same location in a cloning well.
A general schematic for the removal of HXGPRT from Δku80Δgoi::HXGPRT to delete HXGPRT from the strain is shown (Figure 3A). The primer pair used to validate the removal of HXGPRT from Δku80Δgra2::HXGPRT (Figure 3B) amplifies a unique ~1.2 Kbp band in PCR5 as seen in clones 4, 7, 9, 11 and 12 (Figure 3C). If HXGPRT is not removed from the locus, a ~3.4 Kbp band is observed (clones 1, 2, 3, 6, 8, 10, and the parental control). Several mechanisms act to make the removal of HXGPRT (6TX selection) more challenging and less efficient than the integration of HXGPRT (MPA + X selection). MPA selection is simply more efficient than 6TX selection14,16. In addition, higher levels of HXGPRT expression are needed for 6TX selection than for MPA selections16,24. Consequently, mutations that may reduce HXGPRT enzyme activity or mutations or epigenetic changes that reduce the expression level of HXGPRT at a gene’s locus can potentially abolish the effectiveness of 6TX selection. Even with these challenges of 6TX selection, the success rate of 6TX selection in Δku80 strains is greater than 90% on the first attempt1-3.
A general scheme for direct C-terminal tagging of protein-coding genes is shown (Figure 4). This scheme uses direct integration via double cross over homologous recombination of HXGPRT 3′ of the tagged gene and also employs validation strategies similar to those described above. When a gene is suspected to be an essential gene this can be verified using a second transfection with an independently isolated targeting DNA plasmid. Alternative methods, such as schemes for regulated gene expression, are available for further verifying if a gene is essential25.
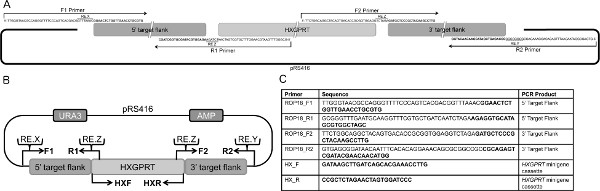
Figure 1. Overview of the design of a targeting DNA plasmid. A. Schematic for designing overlap primers used to PCR amplify the 5′ and 3′ target flanks with 33 bp overlaps for the pRS416 shuttle vector and HXGPRT minigene cassette. Primers depicted in the schematic were used to generate the 5′ and 3′ target flanks of pΔROP1810. B. Overall strategy for designing a targeting DNA molecule. The backbone of pΔGOI is the pRS416 shuttle vector containing uracil (URA) and ampicillin (AMP) selectable markers. Inserted into pRS416 is a ~1 Kbp 5′ DNA target flank amplified from genomic DNA with overlaps for the HXGPRT minigene cassette and pRS416 using the F1 and R1 primers, a ~1 Kbp 3′ DNA target flank is amplified from genomic DNA with overlaps for the HXGPRT minigene cassette and pRS416 using the F2 and R2 primers and the HXGPRT minigene cassette. The following unique restriction enzyme digest sites are added to the primers: RE.X (restriction enzyme X cut site) in the F1 primer to cut the plasmid at the 5′ end of the 5′ DNA target flank, RE.Y in the R2 primer to cut the plasmid at the 3′ end of the 3′ DNA target flank and RE.Z in the R1 and F2 primers to excise the HXGPRT minigene cassette. C. Primer sequences used to generate the targeting construct for the deletion of rop18. The bold regions correspond to T. gondii genomic sequence, and the nonbold regions correspond to the restriction enzyme sites and sequences that overlap with pRS416 and the HXGPRT minigene cassette. The HX_F and HX_R primer pair amplifies the HXGPRT minigene cassette14. Primers read from 5′ to 3′. Table adapted from Fentress et al10. Click here to view larger figure.
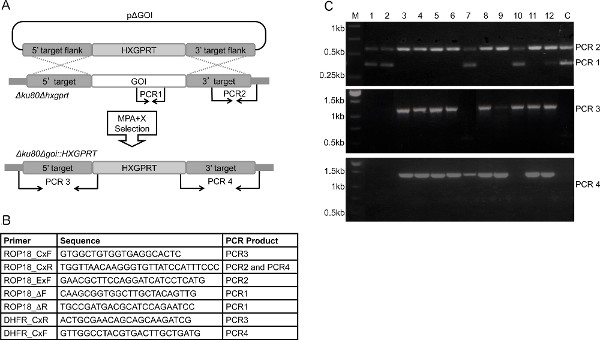
Figure 2. Overview of the protocol for the deletion of a gene using HXGPRT. A. Disruption of a GOI in the Δku80Δhxgprt strain by a double crossover homologous recombination event using a ~1 Kbp 5′ DNA target flank and a ~1 Kbp 3′ DNA target flank on plasmid pΔGOI. A PCR strategy using PCR1, PCR2, PCR3, and PCR4 (not to scale) is shown that enables genotype verification to validate clones with targeted integration of HXGPRT and deletion of the GOI locus. B. Representative primer pairs designed to validate the deletion of rop18. ROP18_DF and ROP18_DR amplify PCR1, ROP18_ExF and ROP18_CxR amplify PCR2, ROP18_CxF and DHFR_CxR amplify PCR3 and DHFR_CxF and ROP18_CxR amplify PCR4 (see Figure 2A). Primers read from 5′ to 3′. Table adapted from Fentress et al10. C. Representative results of validation of a targeted gene deletion (rop18) using HXGPRT. Following transfection of plasmid pΔROP18 into Δku80Δhxgprt and selection in MPA+X, MPA+X resistant clones were assayed for the deletion of rop18. The parental strain control is positive for PCR 1(~400 bp) and PCR 2 (~650 bp) product and negative for PCR 3 (~1,200 bp) and PCR 4 (~1,300 bp) product. A targeted GOI knockout is positive for the PCR2, PCR3 and PCR4 products and is negative for the PCR 1 product (see Figure 2A). Shown are representative panels of the results of PCR1 and PCR2 (top panel), PCR3 (middle panel), and PCR4 (bottom panel). Clones 3, 4, 5, 6, 8, 9 and 11 exhibit the correct banding pattern of PCR 1, PCR2, PCR3 and PCR4 that defines a targeted gene deletion event in the clone. Clones 1, 2, 7, 10 and 12 are not gene deletions, and correspond to other potential representative results. (C = parental control strain Δku80Δhxgprt, M = size markers). Click here to view larger figure.
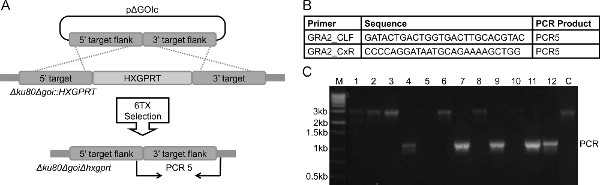
Figure 3. Overview of the protocol for re-targeting the gene of interest to delete HXGPRT. A. Strategy for removal of the HXGPRT selectable marker by a double crossover homologous recombination event in the strain Δku80Δgoi::HXGPRT using a ~1 Kbp 5′ DNA target flank and a ~1 Kbp 3′ DNA target flank on plasmid pΔGOIc. The PCR strategy for genotype verification is depicted using a primer pair to assay for a PCR product (PCR5) that spans the deletion (not to scale). B. A representative primer pair designed to validate the removal of the HXGPRT selectable marker from the gra2 locus in strain Δku80Δgra2::HXGPRT. Primers GRA2 _CLF and GRA2_CxR amplify PCR5. Primers read from 5′ to 3′. C. Representative panel of 6TX-resistant clones that may be obtained following transfection of plasmid pΔGOIc into Δku80Δgoi::HXGPRT and selection in 6TX. Clones 4, 7, 9, 11 and 12 exhibit the correct banding pattern of PCR5 that corresponds to targeted deletion of the HXGPRT marker (~1.2 Kbp product that spans the 3′ flank with slight overlap with the 5′ flank and outside the 3′ flank). Clones 1, 2, 3, 6, 8, and 10 (this band is light) represent the expected banding pattern of around ~3.4 Kbp that corresponds to the parental clone genotype, even though these clones exhibited a degree of resistance to 6TX that permitted their selection (this phenotype can occasionally arise spontaneously by shut-down of HXGPRT expression (see note in protocol 2.1 (step 8) and Representative Results section). (C = parental control strain Δku80Δgoi::HXGPRT, M = size markers). Click here to view larger figure.
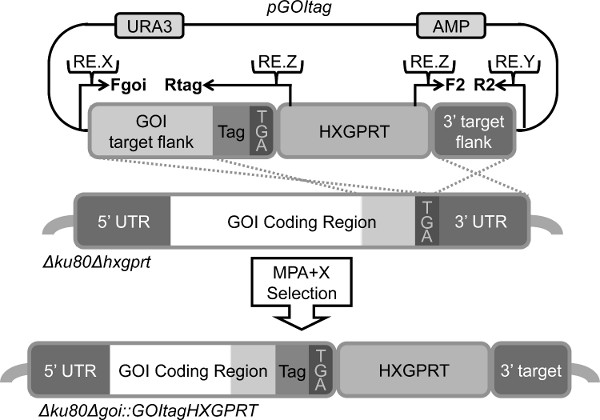
Figure 4. Design of a targeting DNA molecule to tag the C-terminal end of a protein. The strategy is based on the ability of the HXGPRT marker to also function as a 3′ downstream regulatory region. The backbone of pΔGOItag is the pRS416 shuttle vector containing uracil (URA) and ampicillin (AMP) selectable markers. Inserted into pRS416 is a ~1 Kbp 5′ DNA target flank amplified from genomic DNA with overlaps for the HXGPRT minigene cassette and pRS416 using the Fgoi and Rtag primers, a ~1 Kbp 3′ DNA target flank is amplified from genomic DNA with overlaps for the HXGPRT minigene cassette and pRS416 using the F2 and R2 primers and the HXGPRT minigene cassette. The 5′ DNA target flank replaces the termination codon with the tag (HA, Myc, His, etc.) followed by a replacement termination codon. MPA + X selection integrates the C-terminal tag and a downstream HXGPRT, and moves the 3′ UTR to a position following the HXGPRT marker. The following unique restriction enzyme digest sites are added to the primers: RE.X (restriction enzyme X cut site) in the Fgoi primer to cut the plasmid at the 5′ end of the 5′ DNA target flank, and RE.Y in the R2 primer to cut the plasmid at the 3′ end of the 3′ DNA target flank, and RE.Z in the Rtag and F2 primers to excise the HXGPRT minigene cassette for use for plasmid construction to re-target the tagged gene locus to delete the HXGPRT marker which will restore the placement of the 3′ UTR.
Discussion
Here we provide a protocol for efficient gene targeting in Δku80 parasite strains to enable the efficient recovery of genetically manipulated progeny that possess targeted gene deletions, gene replacements, and/or tagged genes. The sequential execution of these methods provides a reliable approach for the isolation of parasite mutants that contain single or multiple targeted genetic manipulations1-3. While the generation of a targeted deletion depends on multiple factors, the use of the Δku80 strain with the presented strategy and protocol greatly increases the efficiency and ease of making precisely defined mutant strains of T. gondii for functional genomic studies.
The genome of T. gondii during asexual stages is haploid. Thus a consideration to make when planning to delete a gene is that the gene must not be essential in vitro. Nonetheless, the efficiency of targeting gene knockouts in Δku80 strains is extremely high, and this enables the isolation of mutant strains with significantly impaired replication rates. If a targeted gene is essential, parasites often cease to replicate during selection. Another critical factor in targeted genetic manipulation is that perfect homology with at least 620 bp of the genomic targeting flank is required to obtain detectable targeted integrations2. Longer regions of homology produce higher efficiencies of targeting and using targeting DNA flanks of approximately 1,000 bp is a reliable and efficient approach1-4. For this reason it is important that genomic targeting flanks are generated from the same strain of T. gondii (RH, Pru) as the strain that is being genetically manipulated. Lack of sufficient homology can frequently result in the targeted integration of one genomic targeting flank, but not the other. Incomplete integration or failure to obtain a knockout may also be due to episomal persistence. Immediately after transfection, all of the targeting molecules are nonintegrated episomes, and sometimes targeting molecules can persist as episomes for many generations. Using target DNA flanks of ~1 Kbp and continuing to pass parasites under selection prior to re-subcloning frequently resolves problems associated with episomal persistence.
Once a knockout is validated and the HXGPRT marker is removed, the gene targeting strategy can be repeated on a second GOI to generate multiple gene deletions in a single parasite strain1-3. The generation of deletions, replacements, or tagged genes can also be accomplished by using different selectable markers (bleomycin, CAT, pyrimethamine resistant DHFR, etc.). Although the CAT selectable marker currently is present in the type II Δku80 Pru genome1, this marker can be removed using the HXGPRT selection protocol described here. In our hands, HXGPRT selection is the safest and the most efficient marker with the highest targeting efficiency where a single-copy targeted insertion is sufficient for robust selection in MPA + X medium. HXGPRT also provides the only currently useful approach for targeted deletion of a selectable marker (Figure 3) using 6-thioxanthine (6TX) selection2,3. Thus this protocol provides the only current approach for developing defined mutant strains with several targeted genetic manipulations.
This protocol provides a valuable and efficient method for targeted genetic manipulation in Δku80 strains of Toxoplasma gondii and is applicable to analysis of a single gene, a family of genes, or genome-wide functional genomic studies. Prior to the availability of Δku80 strains1,2, these approaches were not widely feasible due to the inefficiency of these methods. Consequently, this protocol is widely applicable for targeted genetic manipulation of Toxoplasma gondii and is accessible to all investigators interested in addressing a range of questions on parasite biology, host response, and functional genomics.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by grants from the NIH to DJB (AI041930, AI073142, AI075931, and AI091461).
Materials
| Name of Reagent/Material | Company | Catalog Number | Comments |
| Overlap Primers | Integrated DNA Technologies | 4 nmole Ultramer DNA oligos | |
| Validation Primers | Integrated DNA Technologies | 100 nmole DNA Oligo | |
| Yeast Strain #90845 | ATCC | Designation FY834 | |
| Shuttle Vector pRS416 | ATCC | 87521 | |
| DH10B E. coli | Invitrogen | 12033-015 | SOC broth in kit with E. coli |
| Resuspension Buffer | Qiagen | Buffer P1 in QIAprep Spin Miniprep Kit | |
| Miniprep Kit | Qiagen | 27104 | QIAprep Spin Miniprep Kit |
| Glass Beads | Scientific Industries | SI-BG05 | 0.5 mm acid-washed |
| Qiacube Automated Robotic Work Station | Qiagen | ||
| Electroporation Cuvette | USA Scientific | 9104-5050 | 2 mm-gap |
| BTX600 electroporator | BTX | ||
| Maxiprep Kit | Qiagen | 12662 | QIAprep Spin Maxiprep Kit |
| 25 cm2 Canted neck plug seal flask | Corning | 430168 | |
| 150 cm2 Canted Neck plug seal flask | Corning | 430823 | |
| Human Foreskin Fibroblasts (HFF) cells | ATCC | SCRC1041 | |
| Swin-lok Filter Holder | Whatman | 420200 | 25-mm-diameter |
| Membrane | Whatman | 110612 | Nucleopore Track-etched Membrane 3 mm pore size, 25-mm-diameter |
| Mycophenolic acid (MPA) | Sigma | ||
| Xanthine (X) | Sigma | ||
| 96-well Tissue Culture Tray | Corning Costar | ||
| 24-well Tissue Culture Tray | Corning Costar | ||
| MEM Eagle Media | Lonza Biowhittaker | 12-611F | |
| Fetal Bovine Serum | Gibco | 26140-111 | |
| Antibiotic-Antimycotic (Anti-Anti) | Gibco | 15240-062 | |
| Spin Column | Primm Labs | PAE-100 | Easy Clean DNA Extraction Spin Kit |
| T4 DNA Ligase | New England Biolabs | ||
| 6-thioxanthine | Acros Organics | ||
| Tissue DNA Minikit | Qiagen | 51104 | QIAamp DNA Blood Mini Kit |
| Cell Culture Freeze/Recovery Media | Gibco | 126-48-010 | |
| Phosphate Buffered Saline | Hyclone | SH30028.02 | minus calcium, minus magensium |
References
- Fox, B. A., et al. Type II Toxoplasma gondii KU80 knockout strains enable functional analysis of genes required for cyst development and latent infection. Eukaryot. Cell. 10, 1193-1206 (2011).
- Fox, B. A., Ristuccia, J. G., Gigley, J. P., Bzik, D. J. Efficient gene replacements in Toxoplasma gondii strains deficient for nonhomologous end joining. Eukaryotic Cell. 8, 520-529 (2009).
- Fox, B. A., Bzik, D. J. Avirulent uracil auxotrophs based on disruption of orotidine-5′-monophosphate decarboxylase elicit protective immunity to Toxoplasma gondii. Infection and Immunity. 78, 3744-3752 (2010).
- Hortua Triana, M. A., et al. Biochemical and molecular characterization of the pyrimidine biosynthetic enzyme dihydroorotate dehydrogenase from Toxoplasma gondii. Molecular and Biochemical Parasitology. 184, 71-81 (2012).
- Kim, K., Weiss, L. M. Toxoplasma gondii: the model apicomplexan. Int. J. Parasitol. 34, 423-432 (2004).
- Gajria, B., et al. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids research. 36, 553-556 (2008).
- Kim, K., Weiss, L. M. Toxoplasma: the next 100 years. Microbes and infection / Institut Pasteur. 10, 978-984 (2008).
- Butcher, B. A., et al. Toxoplasma gondii rhoptry kinase ROP16 activates STAT3 and STAT6 resulting in cytokine inhibition and arginase-1-dependent growth control. PLoS pathogens. 7, e1002236 (2011).
- Daher, W., Klages, N., Carlier, M. F., Soldati-Favre, D. Molecular characterization of Toxoplasma gondii formin 3, an actin nucleator dispensable for tachyzoite growth and motility. Eukaryot. Cell. 11, 343-352 (2012).
- Fentress, S. J., et al. Phosphorylation of immunity-related GTPases by a Toxoplasma gondii-secreted kinase promotes macrophage survival and virulence. Cell Host Microbe. 8, 484-495 (2010).
- Musiyenko, A., Majumdar, T., Andrews, J., Adams, B., Barik, S. PRMT1 methylates the single Argonaute of Toxoplasma gondii and is important for the recruitment of Tudor nuclease for target RNA cleavage by antisense guide RNA. Cellular microbiology. 14, 882-901 (2012).
- Straub, K. W., Peng, E. D., Hajagos, B. E., Tyler, J. S., Bradley, P. J. The moving junction protein RON8 facilitates firm attachment and host cell invasion in Toxoplasma gondii. PLos Pathog. 7, e1002007 (2011).
- Szatanek, T., et al. Cactin is essential for G1 progression in Toxoplasma gondii. Molecular Microbiology. 84, 566-577 (2012).
- Donald, R. G., Carter, D., Ullman, B., Roos, D. S. Insertional tagging, cloning, and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene. Use as a selectable marker for stable transformation. J. Biol. Chem. 271, 14010-14019 (1996).
- Donald, R. G., Roos, D. S. Gene knock-outs and allelic replacements in Toxoplasma gondii: HXGPRT as a selectable marker for hit-and-run mutagenesis. Molecular and Biochemical Parasitology. 91, 295-305 (1998).
- Pfefferkorn, E. R., Borotz, S. E. Toxoplasma gondii: characterization of a mutant resistant to 6-thioxanthine. Exp. Parasitol. 79, 374-382 (1994).
- Gietz, R. D., Schiestl, R. H. Frozen competent yeast cells that can be transformed with high efficiency using the LiAc/SS carrier DNA/PEG method. Nature Protocols. 2, 1-4 (2007).
- Oldenburg, K. R., Vo, K. T., Michaelis, S., Paddon, C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25, 451-452 (1997).
- Fox, B. A., Gigley, J. P., Bzik, D. J. Toxoplasma gondii lacks the enzymes required for de novo arginine biosynthesis and arginine starvation triggers cyst formation. Int. J. Parasitol. 34, 323-331 (2004).
- Roos, D. S. Molecular genetic tools for the identification and analysis of drug targets in Toxoplasma gondii. Curr. Top. Microbiol. Immunol. 219, 247-259 (1996).
- Roos, D. S., Donald, R. G., Morrissette, N. S., Moulton, A. L. Molecular tools for genetic dissection of the protozoan parasite Toxoplasma gondii. Methods in Cell Biology. 45, 27-63 (1994).
- Singh, U., Brewer, J. L., Boothroyd, J. C. Genetic analysis of tachyzoite to bradyzoite differentiation mutants in Toxoplasma gondii reveals a hierarchy of gene induction. Molecular Microbiology. 44, 721-733 (2002).
- vanden Hoff, M. J., Moorman, A. F., Lamers, W. H. Electroporation in ‘intracellular’ buffer increases cell survival. Nucleic Acids Research. 20, 2902 (1992).
- Pfefferkorn, E. R., Bzik, D. J., Honsinger, C. P. Toxoplasma gondii: mechanism of the parasitostatic action of 6-thioxanthine. Exp. Parasitol. 99, 235-243 (2001).
- Mital, J., Meissner, M., Soldati, D., Ward, G. E. Conditional expression of Toxoplasma gondii apical membrane antigen-1 (TgAMA1) demonstrates that TgAMA1 plays a critical role in host cell invasion. Mol. Biol. Cell. 16, 4341-4349 (2005).

