2D and 3D Chromosome Painting in Malaria Mosquitoes
Summary
Chromosome painting is a useful method for studying organization of the cell nucleus and evolution of the karyotype. Here, we demonstrate an approach to isolate and amplify specific regions of interest from single polytene chromosomes that are subsequently used for two- and three-dimensional fluorescent in situ hybridization (FISH).
Abstract
Fluorescent in situ hybridization (FISH) of whole arm chromosome probes is a robust technique for mapping genomic regions of interest, detecting chromosomal rearrangements, and studying three-dimensional (3D) organization of chromosomes in the cell nucleus. The advent of laser capture microdissection (LCM) and whole genome amplification (WGA) allows obtaining large quantities of DNA from single cells. The increased sensitivity of WGA kits prompted us to develop chromosome paints and to use them for exploring chromosome organization and evolution in non-model organisms. Here, we present a simple method for isolating and amplifying the euchromatic segments of single polytene chromosome arms from ovarian nurse cells of the African malaria mosquito Anopheles gambiae. This procedure provides an efficient platform for obtaining chromosome paints, while reducing the overall risk of introducing foreign DNA to the sample. The use of WGA allows for several rounds of re-amplification, resulting in high quantities of DNA that can be utilized for multiple experiments, including 2D and 3D FISH. We demonstrated that the developed chromosome paints can be successfully used to establish the correspondence between euchromatic portions of polytene and mitotic chromosome arms in An. gambiae. Overall, the union of LCM and single-chromosome WGA provides an efficient tool for creating significant amounts of target DNA for future cytogenetic and genomic studies.
Introduction
Chromosome painting is a useful technique for studying evolution of karyotypes 1-5 and for visualizing cytogenetic abnormalities via hybridization of fluorescently labeled chromosomal DNA 1,6-8. This method is also applied to studying the 3D dynamics of chromosome territories in development 9 and pathology 10. Differentially labeled chromosome paints are used for the visualization of individual mitotic 11,12, meiotic 13,14, or interphase non-polytene 15,16 and polytene 9,11,17 chromosomes. A simple and robust protocol to obtain chromosome paints would be very useful in expanding this technique to non-model organisms, such as mosquitoes. The Anopheles gambiae complex is a group of seven morphologically indistinguishable mosquitoes that vary in behavior and adaptation, including the ability to transmit malaria. These mosquitoes serve as an excellent model to better understand the evolution of closely related species that differ greatly in their vectorial capacity. Most cytogenetic studies in malaria mosquitoes have been done using well-developed, highly polytenized chromosomes (reviewed in 18,19). The readable banding patterns of polytene chromosomes allowed researchers to demonstrate the association between polymorphic inversions and ecological adaptations in Anopheles gambiae 20. Also, tissue-specific 21 and species-specific 22 features of 3D polytene chromosome organization have been characterized in the An. macullipennis complex. However, studying mitotic chromosomes could provide additional important information. For example, a higher level of heterochromatin polymorphism observed in mitotic chromosomes has been correlated with reduced mating activity and fertility in An. gambiae 23. The correspondence between euchromatic segments of polytene and mitotic chromosome arms could be difficult to establish only by comparing their relative lengths. This is because heterochromatin makes up a significant portion of mitotic chromosomes, but is underrepresented in polytene chromosomes 24. The availability of chromosome paints for malaria mosquitoes would allow researchers to significantly expand cytogenetic studies by including additional species, while substantially reducing the cost and time in the analyses of karyotypic evolution and 3D dynamics of chromosomes in this group of epidemiologically important insects.
In order to obtain large stretches of chromosomes, microdissection has become an integral technique to manipulate and isolate specific regions of interest of the chromosomal complement. When coupled with whole genome amplification (WGA), microdissection results in powerful downstream applications including FISH 11,12 and next-generation genome sequencing 25-27. Previously, the technique required the use of specialized microdissection needles that had to be manually controlled by an experienced user 28. The advancement of laser capture microdissection (LCM) has resulted in a simplified tool better suited for isolating single cells 29,30 or individual chromosomes 31-33 with a lessened risk of contamination. This approach allows the user to study the genetic heterogeneities and chromosomal abnormalities that occur in single cells, instead of a consensus landscape that results from pooling multiple cells together 34-36. Multiple methods have been used to amplify the DNA produced from microdissection. DOP-PCR, a technique useful for the amplification of highly repetitive sequences, has been used to amplify microdissected chromosomes from species including the grasshopper 37, the spiny eel 38, and the Nile tilapia 39. More recently, the PCR-based GenomePlex WGA4 Single Cell kit and multiple displacement amplification based (MDA) Repli-G Single Cell kit have become valuable tools for experiments involving genetic analysis of single human cells as well as chromosomes40-42, including the B chromosome systems in grasshoppers 37 and locusts 43. These two kits each have advantages and disadvantages, but their apparent superiority above other available amplification systems has been demonstrated 44.
The quantity of DNA that can be obtained from a single chromosome or a chromosome segment is significantly less than that of a whole nucleus. Therefore microdissection, amplification, and subsequent analysis of a single chromosome are far more challenging, especially in organisms with small genomes like in Drosophila or Anopheles. Although paints have been developed from single microdissected chromosomes of a human 33 and a wasp 45, a successful FISH experiment required multiple (at least 10-15) microdissected mitotic chromosomes of a fruit fly 11. However, the ability to microdissect and amplify a single chromosome would be important for (i) reducing the chance of contamination with material from a different chromosome, (ii) minimizing the number of chromosomal preparations needed for microdissection, (iii) lowering nucleotide and structural polymorphism of the microdissected sample in both FISH and sequencing downstream applications. Polytene chromosomes found in many Dipteran species offer a unique opportunity to acquire a much higher starting quantity of DNA. They also provide a higher resolution and chromosome structure that cannot be achieved through the use of mitotic chromosomes. This added resolution can be critical in visualizing chromosomal rearrangements, chromatin structure, and chromosomal segments to be microdissected 28,46.
Here we present a procedure to efficiently isolate a euchromatic segment from a single polytene chromosome arm, amplify the DNA, and use it in downstream FISH applications in malaria mosquitoes. First, we apply LCM to isolate and extract a single chromosome arm from specially prepared membrane slides. Second, WGA is used to amplify the DNA from the microdissected material. Third, we hybridize the amplified DNA in FISH experiments to polytene squash preparations 47, metaphase and interphase chromosome slides 48, as well as 3D ovarian whole mount samples. This procedure has been done to successfully paint a majority of the euchromatin in chromosomal arms of An. gambiae.
Protocol
1) Polytene chromosome slide preparation for laser capture microdissection
- Dissect half-gravid Anopheles females at 25 hr post-blood feeding. Fix ovaries from approximately five females into 500 µl of fresh modified Carnoy’s solution (100% methanol: glacial acetic acid, 3:1) at room temperature for 24 hr. Transfer ovaries to -20 °C for a long-term storage.
- Prepare Carnoy’s solution (100% ethanol: glacial acetic acid, 3:1) and 50% propionic acid just prior to making chromosome slides.
- Place one pair of ovaries in one drop of Carnoy’s solution on a Zeiss 1.0 PET membrane slide. Depending on the size, split ovaries into approximately 2-4 sections with dissecting needles and place them into a drop of 50% propionic acid on clean slides under a dissection microscope.
- Separate follicles and remove remaining tissue using paper towel under a dissection stereomicroscope. Add a new drop of 50% propionic acid to the follicles and allow them to sit for 3-5 min at room temperature.
- Place a siliconized coverslip on top of the droplet. Let the slide stand for approximately 1 min.
- Cover the slide with an absorbent material (filter paper is used for this method), and while using the eraser side of a pencil, apply a generous amount of pressure to the coverslip by tapping on it repeatedly with the eraser.
- Heat the slide to 60 °C on a slide denaturation/hybridization system for 15-20 min to aid in flattening the polytene chromosomes. Place slides into a humid chamber at 4 °C overnight to allow the acid to further flatten chromosomes.
- Place slides in cold 50% ethanol for 10 min. Gently remove the coverslip, and replace in cold 50% ethanol for 10 more min.
- Dehydrate slides in 70%, 90%, 100% ethanol for 5 min each. Air dry slides.
- Prepare a solution of GURR buffer solution by adding a single buffer tablet to 1 L of distilled water. Autoclave.
- Prepare the Giemsa solution by adding 1 ml of Giemsa staining solution to 50 ml of GURR buffer.
- Place air dried slides in Giemsa solution for 10 min and wash three times in 1X PBS. Air dry slides again in a controlled sterile climate to avoid contamination.
2) Laser capture microdissection of a single polytene chromosome arm
This section details the use of the PALMRobo software, which comes with the PALM MicroBeam Laser Microdissection system.
- Clean the microscope with 100% ethanol. Sterilize gloves and tubes with a UV light in a UV-crosslinker.
- Power up the PALM MicroBeam Laser Microdissection microscope and turn on laser. Open the laser dissection suite, PALMRobo, and configure the “Power” and “Focus” settings as necessary.
- Search for polytene chromosome arm of interest.
- Using the “Pencil” tool, outline the selected region.
- Open the “Elements window” from the menu bar.
- Select the “Drawn element,” ensure that you have selected “Cut.”
- Install the adhesive cap tube into the holder and place above the slide, leaving a small gap <1 mm in size, and start the laser cut.
- Place the “Catapult selection” within the cut site, leaving space between the edge and chromosome.
- Select “LPC” from the drop down option and begin catapulting.
- Check to ensure sample was catapulted into cap by pressing the “Eye” icon.
3) Purification of DNA from a single microdissected polytene chromosome arm
Follow the instructions of the QIAamp DNA Micro Kit to release and purify the collected DNA. Step 3.1 was modified to accommodate the inverted tube.
- Add 15 µl Buffer ATL and 10 µl proteinase K to the inverted tube (inside the cap) and incubate at 56 °C for 3 hr.
- Add 25 µl buffer ATL, 50 µl buffer AL, and 1 µl carrier RNA; mix. Add 50 µl 100% EtOH; mix.
- Transfer lysate to QIAamp column; centrifuge. Wash by adding 500 µl buffer AW1; centrifuge. Place column into new collection tube, add 500 µl buffer AW2; centrifuge. Place the column into a new tube; centrifuge to remove excess liquid.
- Place column into a 1.5 µl microcentrifuge tube and add 20 µl of water to elute; centrifuge.
- Evaporate freshly eluted DNA down to a final volume of 9 µl using a vacuumfuge.
4) Amplification of DNA from a single microdissected polytene chromosome arm
Two different protocols were used for the WGA of a single chromosome arm.
- DNA amplification and probe preparation via GenomePlex WGA
- Follow the GenomePlex Single Cell WGA4 Kit protocol to produce the first batch of amplified DNA:
- Add freshly prepared Proteinase K solution to the 9 µl sample; mix. Incubate DNA at 50 °C for 1 hr, then heat to 99 °C for 4 min. Keep on ice.
- Add 2 µl 1X Single Cell Library Preparation Buffer and 1 µl of Library Stabilization Solution; mix. Heat sample to 95 °C for 2 min. Cool on ice and centrifuge.
- Add 1 µl of Library Preparation Enzyme; mix and centrifuge. Incubate as follows:
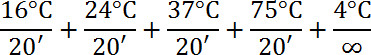
- Add 7.5 µl 10X Amplification Master Mix, 48.5 µl water. 5.0 µl WGA DNA Polymerase; mix and centrifuge.
- Thermocycle as follows:
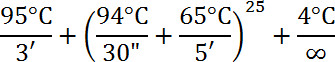
- Purify DNA using the Genomic DNA Clean & Concentrator Kit. The protocol is as follows:
- Add 5:1 DNA binding buffer:DNA sample (specifically for genomic DNA of less than 2 kb. If sample DNA is greater than 2 kb, use a 2:1 ratio) and transfer to provided spin column. Centrifuge.
- Add 200 µl DNA Wash Buffer and centrifuge. Repeat wash step. Then, add 50 µl of water and elute the DNA into a new 1.5 ml tube.
- Re-amplify sample DNA using the GenomePlex WGA3 Reamplification Kit as follows:
- Add 10 µl DNA to PCR tube (the kit recommends 10 ng total of DNA) with 49.5 µl water, 7.5 µl 10x Amplification Master Mix, 3.0 µl 10 mM DNTP mix, and 5.0 µl WGA DNA Polymerase. Mix and centrifuge.
- Use the following profile for the reaction:
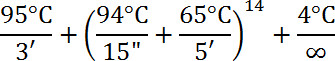
- Store DNA at -20 °C.
- Label DNA for FISH using the GenomePlex WGA3 Reamplification Kit as following:
- Create a master mix from the GenomePlex WGA3 Reamplification Kit by adding 10 µl DNA to PCR tube with 49.5 µl water, 7.5 µl 10X Amplification Master Mix, 3.0 µl 1 mM dNTP mix (1 mM of dATP, dCTP, dGTP, 0.3 µl 1 mM dTTP – if using labeled dUTP), 1 µl of 25 nM labeled dUTP, and 5.0 µl WGA DNA Polymerase.
- Use the following profile for the reaction:
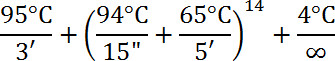
- Ethanol precipitate the labeled probe by adding 1/10 the final reaction volume (7.5 µl for 75 µl reaction) of 3 M Sodium Acetate pH 5.2 and 2-3 volumes of 100% ethanol. Chill DNA sample at -80 °C for at least 30 min.
- Centrifuge sample at 4 °C for 10 min to create labeled pellet and remove supernatant and air-dry pellet.
- Create hybridization buffer as follows:
0.2 g Dextran Sulfate
1200 µl Deionized formamide
580 µl H2O
120 µl 20X SSC - Add 40 µl of hybridization buffer to air-dried pellet.
- Follow the GenomePlex Single Cell WGA4 Kit protocol to produce the first batch of amplified DNA:
- DNA amplification and probe preparation via REPLI-G Single Cell WGA followed by nick-translation
- Follow the REPLI-g Single Cell WGA Kit protocol to produce the amplified DNA:
- Prepare Buffer D2 (3 µl of 1 M DTT + 33 µl Buffer DLB).
- Mix 4 µl of purified microdissected material with 3 µl Buffer D2. Flick tube to mix. Incubate for 10 min at 65 °C. Add 3 µl of Stop solution; mix.
- Add 9 µl H2O, 29 µl REPLI-g Reaction Buffer, and 2 µl of REPLI-g DNA Polymerase to the sample. Incubate at 30 °C for 8 hr. Inactivate DNA polymerase by heating to 65 °C for 3 min. Store DNA at -20 °C.
- Follow the nick translation protocol to label REPLI-g amplified DNA:
- Prepare the following labeling mix:
1 µg of amplified DNA
5 µl 10X DNA Polymerase Buffer
5 µl 10X dNTP
5 µl 1X BSA
1 µl 1 mM Labeled dNTP
4 µl 1 U/µl DNase I
1 µl 10 U/µl DNA Polymerase I
H2O to 50 µl - Incubate at 15 °C for 2 hr. Add 2 µl of 0.5 M EDTA to stop reaction. Check DNA fragment size by running on gel.
- Follow steps from 4.1.4.3-4.1.4.6 to precipitate and solubilize pellet.
- Prepare the following labeling mix:
- Follow the REPLI-g Single Cell WGA Kit protocol to produce the amplified DNA:
5) 2D FISH on polytene and mitotic chromosome squash preparations
Please refer to detailed protocols for FISH on preparations of polytene squashes 47 and mitotic chromosome slides 48. Here we provide brief protocols.
- FISH on polytene chromosome squash preparations
- Immerse slides in 1X PBS for 20 min at RT. Fix slides in 4% Paraformaldehyde in 1X PBS for 1 min.
- Dehydrate slides through ethanol washes: 50%, 70%, 90%, 100% for 5 min each at RT. Air-dry slides.
- Prewarm the probe in hybridization buffer at 37 °C. Add 10-20 µl of probe to slide. Cover with 22 X 22 mm coverslip. Press out any air bubbles using a pipette tip.
- Denature chromosomes and probe at 90 °C for 10 min. Seal edges of coverslip with rubber cement. Transfer slide to humid chamber and incubate at 39 °C overnight.
- Wash the slides with 1X SSC at 39 °C for 20 min. Wash the slides with 1X SSC at RT for 20 min. Rinse slides in 1X PBS, then add Prolong anti-fade with DAPI. Cover with coverslip, and store in slide box at 4 °C for at least an hour before visualization.
- FISH on mitotic chromosome squash preparations
- Extract mitotic chromosomes from imaginal discs of 4th instar larva of An. gambiae.
- Prepare chromosome slides suitable for FISH.
- Add 2-3 µl of labeled DNA probe to hybridization buffer in a tube and mix gently by pipetting.
- Apply 10 µl of the probe mixture to the slide and cover with a 22 X 22 mm coverslip. Press out any air bubbles using a pipette tip.
- For counterstaining and detection, apply Prolong anti-fade with DAPI to the preparation and keep in dark for at least an hour before visualization.
6) 3D FISH on whole-mount ovarian tissue
- Prepare the following Buffer A mix:
60 mM KCl
15 mM NaCl
0.5 mM Spermidine
0.15 mM Spermine
2 mM EDTA
0.5 mM EGTA
15 mM PIPES - Prepare slides for nuclear visualization by adding a layer of nailpolish in a square pattern to match the size of the coverslips. This creates a raised surface to prevent squashing of nuclei when placing on coverslip in the future.
- Dissect fresh ovaries from Christopher’s stage 3 females and keep in a solution of 150-250 µl Buffer A with 0.5% digitonin. Run larger dissection needle over follicles (in tube with Buffer A with 0.5% digitonin) to destroy follicular membrane.
- Vortex for 5-10 min to further disturb follicles. Scrape down any large follicular pieces and centrifuge tube for 30 sec at lowest setting of ~500 revolutions per minute (RPM). Transfer supernatant to a new 2 ml Eppendorf tube, and add 100 µl of Buffer A. Repeat step 6.3 between 5-7 times, until the visible tissue is broken into small particles.
- Spin both tubes for 10 min at 2,000 RPM. Discard supernatant in both tubes. Note: Both tubes will be used for making final nuclear visualization slides. The tube with collected supernatant should contain primarily extracted nuclei, while the original tube with tissue will contain a mixture of tissue and nuclei embedded in nurse cells. Add 200 µl of Buffer A – 0.1% Triton and incubate overnight at 4 °C. Centrifuge 5 min at 10,000 RPM (10,621 x G) and remove supernatant.
- Add 200 µl 4% Paraformaldehyde in PBS. Incubate in thermomixer for 30 min, mixing at 450 RPM. Centrifuge 5 min at 5,000 RPM (2,655 x G) and remove supernatant. Wash with Buffer A with 0.1% Triton for 5 min, mixing at 450 RPM in thermomixer. Centrifuge 5 min at 5,000 RPM (2,655 G) and remove supernatant.
- Add pre-warmed at 37 °C labeled probe to tube. Denature at 95 °C in thermomixer, mixing at 450 RPM, for 10 min. Continue denaturation at 80 °C for 15 min with continued mixing. Incubate at 37 °C in thermomixer, with 450 RPM mixing overnight. Centrifuge 5 min at 5,000 RPM (2,655 x G). Remove supernatant.
- Wash with Buffer A with 0.1% Triton for 5 min. Centrifuge 5 min at 5,000 RPM (2,655 x G). Repeat 2 times. Apply drop of Prolong Anti-fade with DAPI.
- Pipet out nuclei/DAPI solution carefully (avoiding bubbles), apply to slide, and cover with coverslip.
Representative Results
Figure 1 describes the overall flow-through of the protocol described in this article. The user initially starts by microdissecting chromosome DNA samples from membrane slides. Microdissected material is extracted and purified. The purified DNA is then amplified, re-amplified, labeled, and then used for FISH to label chromosome spreads.
The LCM protocol can be broken down into three overall steps: 1) Finding chromosomes of interest and preparing the region for cutting (Figure 2A), 2) Cutting and catapulting the chromosome region of interest via laser (Figure 2B), and 3) Checking to determine if the sample is actually catapulted into adhesive cap (Figure 2C). The process of LCM used on the An. gambiae Sua strain polytene chromosomes is shown in Video 1. The video details the entire process from program start up, including important software functions and tips.
GenomePlex and REPLI-g single cell WGA kits used in this protocol differ greatly in resulting product size as well as overall yield. Figure 3 shows the results of gel electrophoresis ran for the GenomePlex and REPLI-g kits, as well as quantification of the samples by Nanodrop.
Microdissected material has been used to create FISH probes that target euchromatic regions of a chromosome. Figure 4 demonstrates FISH of five probes generated from microdissected material on polytene chromosomes of An. gambiae. Four autosomal arms were labeled with three fluorophores using the WGA3 kit: the 3R chromosome is labeled in green (fluorescein), the 3L chromosome is in a mixture of red (Cy-3) and yellow (Cy-5), the 2R chromosome is in yellow (Cy-5), and the 2L chromosome is in red (Cy-3). The X chromosome was labeled in orange (Cy-3) using nick-translation of the REPLI-g material in a separate experiment. To establish the correspondence between euchromatic portions of polytene and mitotic chromosome arms, chromosome paints have been hybridized to interphase, prophase, prometaphase and metaphase chromosomes of An. gambiae (Figure 5). To visualize the 3D organization of a single polytene chromosome arm in the cell nucleus, whole mount FISH was performed on An. gambiae Sua strain ovarian nurse cells (Video 2). Distinct chromosome arm territories are clearly seen in nuclei with interphase (Figure 5D) and polytene (Video 2) chromosomes.
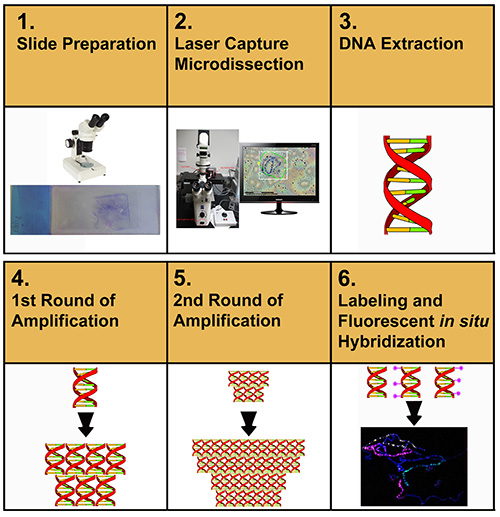
Figure 1. Schematic representation of the experimental procedures toward the preparation of chromosome paints. Click here to view larger image.
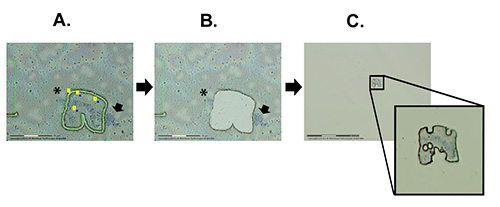
Figure 2. The major steps in chromosome microdissection. A) Laser-assisted cutting of the chromosomal region of interest through the membrane. B) The membrane with a hole after the catapulting is performed. C) The view of the catapulted piece of the membrane with a chromosomal segment in it attached to the adhesive cap. The arrow indicates the heterochromatin of the X chromosome that remained on the slide. The asterisk shows a piece of another chromosome that remained on the slide. Click here to view larger image.
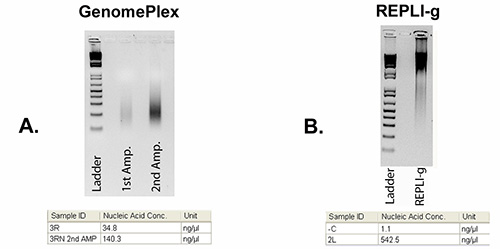
Figure 3. Agarose gel images showing DNA after WGA. A) Low molecular weight (200-500 bp) DNA from arm 3R after using the WGA4 and WGA3 GenomePlex kits. B) High molecular weight (10-20 kb) DNA from arm 2L after REPLI-g amplification. The 100 bp ladder is shown in the left lanes. The tables below gel images show DNA concentrations measured by Nanodrop. Click here to view larger image.
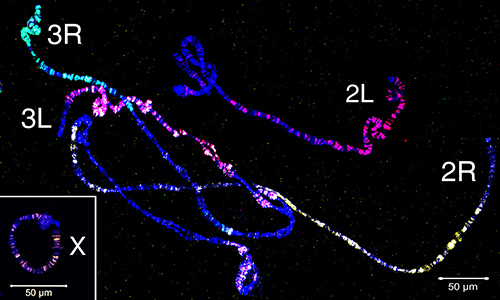
Figure 4. Painting of polytene chromosomes from ovarian nurse cells of An. gambiae using four probes generated from microdissected material. The X chromosome is labeled in orange (Cy-3) by nick-translation of the REPLI-g material. The 2R arm is labeled in yellow (Cy-5); the 2L arm is in red (Cy-3); the 3R arm is labeled in green (fluorescein); the 3L arm is labeled in a mixture of red (Cy-3) and yellow (Cy-5). Autosomes are labeled with the WGA3 amplification kit. Chromatin is stained in blue (DAPI). Chromosome names are placed near telomeric regions. Click here to view larger image.
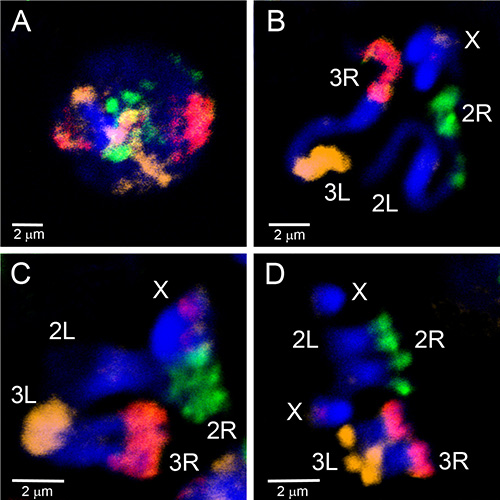
Figure 5. Painting of interphase (A), prophase (B), prometaphase (C) and metaphase (D) chromosomes from larval imaginal discs of the An. gambiae Mopti strain using three probes generated from microdissected material labeled by WGA3. The 2R arm is labeled in green (fluorescein); the 2L arm is unlabeled; the 3R arm is in pink, a mixture of red (Cy-3) and orange (Cy-5); the 3L arm is labeled in orange (Cy-5). The X chromosome has a red label corresponding to the 18S rDNA probe. Chromatin is stained in blue (DAPI). Brightly stained regions of chromosomes correspond to the heterochromatin. Click here to view larger image.
Video 1. The process of LCM of the An. gambiae polytene chromosomes. Click here to view Video 1.
Video 2. Whole mount 3D FISH performed on An. gambiae ovarian nurse cells. The probe is labeled in Cy-3 (depicted in blue) and was made from a microdissected 2R chromosome arm. Chromatin was stained with DAPI and is depicted by cyan pseudo-coloring (light blue). Click here to view Video 2.
Discussion
There are multiple steps critical to successfully amplifying DNA from microdissected polytene chromosome samples. The protocol employs the use of LCM, a method that both increases overall efficiency and reduces exposure to foreign DNA by removing the interaction of physical tools with the sample. However, the amplification of foreign DNA is still the greatest potential pitfall of this experiment. Thus, during the entire process, it is essential to keep the samples protected from contamination. Throughout the slide preparation and microdissection phases, it is essential to ethanol wash dissection needles, slides, cover slips, as well as the workspace. It is also advised to UV treat all applicable equipment (needles, slides, coverslips) prior to use.
The membrane on the dissection slides makes spreading of the chromosomes very difficult. It is important to use more of the ovary (half to a full pair of ovaries) when making these slides, to provide a greater chance of finding a well spread nucleus. It is also recommended to use fresh tissue when making slides, as amplification rates appear to drop as tissues age 49 and chromosome spreading becomes more challenging. The preparation of slides for microdissection is the most time consuming step in this protocol. Chromosomes must be well spread to avoid accidental acquisition of unwanted material. Giemsa staining allows the user to check spread quality with a phase contrast microscope prior to using the microdissection system.
The combination of microdissection and amplification provides the opportunity to extract and analyze chromosomal fragments ranging in size from small regions of interest to a majority of the arms. This protocol allows the user to obtain DNA from morphologically distinct regions such as inversions, specific euchromatic bands and interbands, telomeric, centromeric and intercalary heterochromatin. The user can apply the generated painting probes to examining aberrant chromosomes, studying inter-species homology at particular loci, or characterizing spatial organization of chromosomes in an intact 3D nucleus. For developing chromosome paints, we selected euchromatic segments to avoid hybridization of repetitive DNA with multiple chromosomal regions. As a result, we obtained arm-specific painting without using a competitor, like total genomic DNA or C0t-1 DNA fraction.
We chose to use the GenomePlex WGA and REPLI-G kits based on reviews that compared the efficiency and dropout rate of multiple amplification kits 40,44. Both kits performed the best among the available methods in both drop-out rate (GenomePlex had a 12.5% rate compared to REPLI-g's 37.5%) and percentage of amplified markers (GenomePlex had a 45.24% amplification rate vs. REPLI-g's 30.0%) 40. The GenomePlex kit also provided a higher quantity of DNA, and thus made it a better candidate for multiple downstream techniques. A re-amplification kit for the GenomePlex system is also available, allowing for further amplification of DNA. However, it is important to note that amplification is not perfect. The possibility remains that successful amplification can introduce errors or have a bias toward specific loci in the target DNA. It is important to consider the final fragment size of the available genome amplification methods. GenomePlex fragmentation results in a library with fragments ranging from 200-500 bp, while the REPLI-g kit produces fragments approximately 10-20 kb in size. The intended downstream application of this protocol is FISH, thus making the GenomePlex kit a more feasible option, as it provided the desired fragment size and the ability to label DNA fragments directly through WGA. Long DNA molecules produced by the REPLI-g amplification must be fragmented in the downstream nick-translation labeling reaction.
This protocol has been adapted to successfully amplify DNA from a single polytene chromosome arm. Other protocols require the pooling of many (typically 10-30) chromosome fragments in order to successfully amplify the sample 7,11,28,40. Although pooling of multiple chromosomes is possible using our method, the increased likelihood of sample contamination emphasizes the importance of beginning the experiment with as few chromosomes as possible. If polytene chromosomes are not available, our protocol could be adapted for use in mitotic chromosomes. It may be necessary, however, to pool 10-15 mitotic chromosomes prior to amplification for successful FISH 11. Amplification bias is lower with high quantities of starting template DNA 50. Polytene chromosomes provide approximately 512 copies of a single DNA sequence and 1024 copies of two homologous DNA sequences. Thus, pooling mitotic chromosomes will help to increase overall quality of DNA product after amplification.
This procedure has many potential uses in cytogenetic and genomic studies. Here, we used chromosome paints to establish the correspondence between euchromatic segments of polytene and mitotic chromosome arms in An. gambiae. The same painting probes can be applied to cytogenetic characterization of An. gambiae cell lines. Extensive genomic rearrangements and changes in chromosome numbers are common features of cell lines 51,52. Aneuploidy, polyploidy, and chromosomal rearrangements such as translocations, inversions, deletions, and ring chromosomes have been detected in different mosquito cell lines 53,54. Classical cytogenetic observations 55 and physical mapping 56 have demonstrated the occurrence of whole-arm chromosomal translocations in the evolution of malaria mosquitoes. Therefore, microdissected material can be used in conjunction with FISH experiments to compare homology of chromosomal regions between species. We successfully performed 3D FISH with the 2R-painting probe on polytene chromosomes in whole mount ovarian nurse cells. This method will facilitate the tracing of chromosome trajectories and studying the nuclear architecture in Anopheles. Techniques like DNA genotyping 57,58, next-generation genome sequencing 26,27, physical and linkage map development, and generation of probes for chromosome painting 33,59 all can benefit from arm- and region-specific microdissection. By combining LCM technology and advancing single-cell WGA, we demonstrate an efficient, practical method for producing reasonable quantities of DNA from a single polytene chromosome arm.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by the grant from National Institutes of Health 1R21AI094289 to Igor V. Sharakhov
Materials
| Acetic acid | Fisher Scientific | A491-212 | |
| Methanol | Fisher Scientific | A412-4 | |
| Propionic acid | Sigma-Aldrich | 402907 | |
| Membrane slides 1.0 PET | Zeiss | 415190-9051-000 | |
| Buffer tablets “GURR” | Life Technologies | 10582-013 | |
| KaryoMAX Giemsa Stain | Life Technologies | 10092-013 | |
| ThermoBrite Slide Denaturation/Hybridization System | Abbott Molecular | 30-144110 | |
| Vacufuge vacuum concentrator | Eppendorf | 22820001 | |
| Spectroline Microprocessor-Controlled UV Crosslinker XL-1000 | Fisher Scientific | 11-992-89 | |
| Thermo Scientific NanoDrop | Fisher Scientific | ND-2000 | |
| REPLI-g Single Cell Kit | Qiagen | 150343 | |
| GenomePlex Single Cell Kit (WGA4) | Sigma-Aldrich | WGA4-10RXN | |
| GenomePlex WGA Reamplification Kit (WGA3) | Sigma-Aldrich | WGA3-50RXN | |
| MZ6 Leica stereomicroscope | Leica | VA-OM-E194-354 | A different stereomicroscope can be used |
| Olympus CX41 Phase Microscope | Olympus | CX41RF-5 | A different phase microscope can be used |
| PALM MicroBeam Laser Microdissection Microscope | Zeiss | ||
| Thermomixer | Eppendorf | 22670000 | |
| 10x PBS | Invitrogen | P5493 | |
| 50x Denhardt’s solution | Sigma-Aldrich | D2532 | |
| 99% Formamide | Fisher Scientific | BP227500 | |
| Dextran sulfate sodium salt | Sigma | D8906 | |
| 20x SSC buffer | Invitrogen | AM9765 | |
| ProLong Gold antifade reagent with DAPI | Invitrogen | P-36931 | |
| dATP, dCTP, dGTP, dTTP | Fermentas | R0141, R0151, R0161, R0171 | |
| Cy3-dUTP, Cy5-dUTP | GE Healthcare | PA53022, PA55022 | |
| BSA | Sigma-Aldrich | A3294 | |
| DNA polymerase I | Fermentas | EP0041 | |
| DNase I | Fermentas | EN0521 | |
| Spermine | Sigma-Aldrich | S3256-1G | |
| Spermidine | Sigma-Aldrich | S0266-1G | |
| Potassium Chloride (KCl) | Fisher Scientific | BP366-500 | |
| Sodium Chloride (NaCl) | Fisher Scientific | BP3581 | |
| EGTA | Sigma-Aldrich | E0396-25G | |
| PIPES | Sigma-Aldrich | P6757-25G | |
| EDTA | Fisher Scientific | S311-500 | |
| Digitonin | Sigma-Aldrich | D141-100MG | |
| Triton-X100 | Fisher Scientific | BP151-100 | |
| AdhesiveCap 500 clear | Zeiss | 415190-9211-000 | |
| QIAamp DNA Micro Kit (50) | Qiagen | 56304 | |
| Genomic DNA Clean and Concentrator Kit | Zymo Research | D4010 | |
| 37% Paraformaldehyde | Fisher Scientific | F79-500 |
References
- Ried, T., Schrock, E., Ning, Y., Wienberg, J. Chromosome painting: a useful art. Human molecular genetics 7. , 1619-1626 (1998).
- Yang, F., et al. Reciprocal chromosome painting illuminates the history of genome evolution of the domestic cat, dog and human. Chromosome Res. 8, 393-404 (2000).
- Badenhorst, D., Dobigny, G., Robinson, T. J. Karyotypic evolution of hapalomys inferred from chromosome painting: a detailed characterization contributing new insights into the ancestral murinae karyotype. Cytogenet Genome Res 136. , 83-88 (2012).
- Trifonov, V. A., et al. Chromosomal evolution in Gekkonidae. I. Chromosome painting between Gekko and Hemidactylus species reveals phylogenetic relationships within the group. Chromosome Res. 19, 843-855 (2011).
- Nie, W., et al. Chromosomal rearrangements and karyotype evolution in carnivores revealed by chromosome painting. Heredity (Edinb. , 108-1017 (2012).
- Guan, X. Y., et al. Detection of chromosome 6 abnormalities in melanoma cell lines by chromosome arm painting probes). Cancer Genet Cytogenet. 107, 89-92 (1998).
- Guan, X. Y., et al. Characterization of a complex chromosome rearrangement involving 6q in a melanoma cell line by chromosome microdissection. Cancer genetics and cytogenetics 134. , 65-70 (2002).
- Breen, M., et al. Detection of equine X chromosome abnormalities in equids using a horse X whole chromosome paint probe (WCPP). Vet J. 153, 235-238 (1997).
- Kokhanenko, A. A., Anan’ina, T. V., Stegniy, V. N. The changes in chromosome 6 spatial organization during chromatin polytenization in the Calliphora erythrocephala Mg. (Diptera: Calliphoridae) nurse cells. Protoplasma 250. , 141-149 (2013).
- Timme, S., et al. Nuclear position and shape deformation of chromosome 8 territories in pancreatic ductal adenocarcinoma. Anal Cell Pathol (Amst. 34, 21-33 (2011).
- Drosopoulou, E., et al. Sex chromosomes and associated rDNA form a heterochromatic network in the polytene nuclei of Bactrocera oleae (Diptera: Tephritidae). Genetica. 140, 169-180 (2012).
- Pazian, M. F., Shimabukuro-Dias, C. K., Pansonato-Alves, J. C., Oliveira, C., Foresti, F. Chromosome painting of Z and W sex chromosomes in Characidium (Characiformes). Crenuchidae). Genetica. 141, 1-9 (2013).
- Howe, E. S., Murphy, S. P., Bass, H. W. Three-dimensional acrylamide fluorescence in situ hybridization for plant cells. Methods Mol Biol 990. , 53-66 (2013).
- Lysak, M. A., Mandakova, T. Analysis of plant meiotic chromosomes by chromosome painting. Methods Mol Biol 990. , 13-24 (2013).
- Ji, Z., Zhang, L. Chromosomics: detection of numerical and structural alterations in all 24 human chromosomes simultaneously using a novel OctoChrome FISH assay. J Vis Exp. 10, (2012).
- Idziak, D., et al. Painting the chromosomes of Brachypodium: current status and future prospects. Chromosoma. 120, 469-479 (2011).
- Fuchs, J., Kuhfittig, S., Reuter, G., Schubert, I. Chromosome painting in Drosophila. Chromosome Res. 6, 335-336 (1998).
- Zhimulev, I. F. . Morphology and structure of polytene chromosomes. 34, 1-490 (1996).
- Sharakhov, I. V., Sharakhova, M. V. in Chromosome Mapping Research Developments eds. J.F. Verrity & L.E. Abbington) (Nova Science. , (2008).
- Coluzzi, M., Sabatini, A., Torre, d. e. l. l. a., Di Deco, A., A, M., Petrarca, V. A polytene chromosome analysis of the Anopheles gambiae species complex). Science. 298, 1415-1418 (2002).
- Stegnii, V. N. Systemic reorganization of the architectonics of polytene chromosomes in the onto- and phylogenesis of malaria mosquitoes. Genetika. 23, 821-827 (1987).
- Stegnii, V. N. Systemic reorganization of the architectonics of polytene chromosomes in the onto- and phylogenesis of malarial mosquitoes. II. Species specificity in the pattern of chromosome relations with the nuclear envelope of nutrient ovarian cells. Genetika. 23, 1194-1199 (1987).
- Bonaccorsi, S., Santini, G., Gatti, M., Pimpinelli, S., Colluzzi, M. Intraspecific polymorphism of sex chromosome heterochromatin in two species of the Anopheles gambiae complex. Chromosoma. 76, 57-64 (1980).
- Zhimulev, I. F. Polytene chromosomes, heterochromatin, and position effect variegation. Adv Genet. 37, 1-566 (1998).
- Seifertova, E., et al. Efficient high-throughput sequencing of a laser microdissected chromosome arm. BMC Genomics. 14, 1471-2164 (2013).
- Weise, A., et al. High-throughput sequencing of microdissected chromosomal regions. European journal of human genetics. EJHG. 18, 457-462 (2010).
- Murphy, S. J., et al. Mate pair sequencing of whole-genome-amplified DNA following laser capture microdissection of prostate cancer. DNA research : an international journal for rapid publication of reports on genes and genomes 19. , 395-406 (2012).
- Moshkin, Y. M., et al. Microdissection and sequence analysis of pericentric heterochromatin from the Drosophila melanogastermutant Suppressor of Underreplication. Chromosoma. 111, 114-125 (2002).
- Decarlo, K., Emley, A., Dadzie, O. E., Laser Mahalingam, M. capture microdissection: methods and applications. Methods Mol Biol 755. , 1-15 (2011).
- Iyer, E. P., Cox, D. N. Laser capture microdissection of Drosophila peripheral neurons. J Vis Exp. 10, (2010).
- Kubickova, S., Cernohorska, H., Musilova, P., Rubes, J. The use of laser microdissection for the preparation of chromosome-specific painting probes in farm animals. Chromosome Res. 10, 571-577 (2002).
- Fukova, I., et al. Probing the W chromosome of the codling moth, Cydia pomonella, with sequences from microdissected sex chromatin. Chromosoma. 116, 135-145 (2007).
- Thalhammer, S., Langer, S., Speicher, M. R., Heckl, W. M., Geigl, J. B. Generation of chromosome painting probes from single chromosomes by laser microdissection and linker-adaptor PCR. Chromosome Res. 12, 337-343 (2004).
- Sims, C. E., Allbritton, N. L. Analysis of single mammalian cells on-chip. Lab on a chip 7. , 423-440 (2007).
- Hutchison, C. A., 3rd, J. C., Venter, Single-cell genomics. Nature biotechnology. 24, 657-658 (2006).
- Lasken, R. S., Egholm, M. Whole genome amplification: abundant supplies of DNA from precious samples or clinical specimens. Trends in biotechnology 21. , 531-535 (1016).
- Teruel, M., et al. Microdissection and chromosome painting of X and B chromosomes in the grasshopper Eyprepocnemis plorans. Cytogenet Genome Res. 125, 286-291 (2009).
- Liu, J. D., et al. Sex chromosomes in the spiny eel (Mastacembelus aculeatus) revealed by mitotic and meiotic analysis. Cytogenet Genome Res. 98, 291-297 (2002).
- Harvey, S. C., et al. Molecular-cytogenetic analysis reveals sequence differences between the sex chromosomes of Oreochromis niloticus: evidence for an early stage of sex-chromosome differentiation. Cytogenet Genome Res. 97, 76-80 (2002).
- Hockner, M., Erdel, M., Spreiz, A., Utermann, G., Kotzot, D. Whole genome amplification from microdissected chromosomes. Cytogenet Genome Res. 125, 98-102 (2009).
- Kitada, K., Taima, A., Ogasawara, K., Metsugi, S., Aikawa, S. Chromosome-specific segmentation revealed by structural analysis of individually isolated chromosomes. Genes Chromosomes Cancer. 50, 217-227 (2011).
- Ma, L., et al. Direct determination of molecular haplotypes by chromosome microdissection. Nature methods. 7, 299-301 (2010).
- Teruel, M., et al. Microdissection and chromosome painting of X and B chromosomes in Locusta migratoria. Chromosome Res. 17, 11-18 (2009).
- Treff, N. R., Su, J., Tao, X., Northrop, L. E., Scott, R. T. Single-cell whole-genome amplification technique impacts the accuracy of SNP microarray-based genotyping and copy number analyses. Molecular human reproduction 17. , 335-343 (2011).
- Rutten, K. B., et al. Chromosomal anchoring of linkage groups and identification of wing size QTL using markers and FISH probes derived from microdissected chromosomes. in Nasonia(Pteromalidae : Hymenoptera). Cytogenetic and Genome Research 105. , 126-133 (2004).
- Post, R. J., Kruger, A., Somiari, S. B. Laser-assisted microdissection of polytene chromosomes from Diptera for the development of molecular markers. Molecular Ecology Notes. 6, 634-637 (2006).
- George, P., Sharakhova, M. V., Sharakhov, I. V. High-throughput physical mapping of chromosomes using automated in situ hybridization. Journal of visualized experiments : JoVE. 10, (2012).
- Timoshevskiy, V. A., Sharma, A., Sharakhov, I. V., Sharakhova, M. V. Fluorescent in situ Hybridization on Mitotic Chromosomes of Mosquitoes. J Vis Exp. 10, (2012).
- Frumkin, D., et al. Amplification of multiple genomic loci from single cells isolated by laser micro-dissection of tissues. BMC biotechnology. 8, 10-1186 (2008).
- Raghunathan, A., et al. Genomic DNA amplification from a single bacterium. Applied and environmental microbiology 71. , 3342-3347 (2005).
- Cassio, D. Long term culture of MDCK strains alters chromosome content. BMC Res Notes. 6, 162-1610 (2013).
- Landry, J. J., et al. The Genomic and Transcriptomic Landscape of a HeLa Cell Line. G3. , (1534).
- Steiniger, G. E., Mukherjee, A. B. Insect chromosome banding: technique for G- and Q-banding pattern in the mosquito Aedes albopictus. Can J Genet Cytol. 17, 241-244 (1975).
- Brown, S. E., et al. Toward a physical map of Aedes aegypti. Insect Mol Biol. 4, 161-167 (1995).
- Green, C., Hunt, R. Interpretation of variation in ovarian polytene chromosomes of Anopheles funestus Giles. A. parensis Gillies, and A. aruni? Genetica. 51, 187-195 (1980).
- Sharakhova, M. V., Xia, A., Leman, S. C., Sharakhov, I. V. Arm-specific dynamics of chromosome evolution in malaria mosquitoes. BMC evolutionary biology. 11, 10-1186 (2011).
- Gu, L. H., et al. DNA genotyping of oral epithelial cells by laser capture microdissection]. Fa yi xue za zhi 22. , 196-197 (2006).
- Rook, M. S., Delach, S. M., Deyneko, G., Worlock, A., Wolfe, J. L. Whole genome amplification of DNA from laser capture-microdissected tissue for high-throughput single nucleotide polymorphism and short tandem repeat genotyping. The American journal of pathology 164. , 23-33 (2004).
- Houen, A., Field, B. L., Saunders, V. A. Microdissection and chromosome painting of plant B chromosomes. Methods in cell science : an official journal of the Society for In. Vitro Biology. 23, 115-124 (2001).

