High-resolution In Vivo Manual Segmentation Protocol for Human Hippocampal Subfields Using 3T Magnetic Resonance Imaging
Summary
The goal of this manuscript is to study the hippocampus and hippocampal subfields using MRI. The manuscript describes a protocol for segmenting the hippocampus and five hippocampal substructures: cornu ammonis (CA) 1, CA2/CA3, CA4/dentate gyrus, strata radiatum/lacunosum/moleculare, and subiculum.
Abstract
The human hippocampus has been broadly studied in the context of memory and normal brain function and its role in different neuropsychiatric disorders has been heavily studied. While many imaging studies treat the hippocampus as a single unitary neuroanatomical structure, it is, in fact, composed of several subfields that have a complex three-dimensional geometry. As such, it is known that these subfields perform specialized functions and are differentially affected through the course of different disease states. Magnetic resonance (MR) imaging can be used as a powerful tool to interrogate the morphology of the hippocampus and its subfields. Many groups use advanced imaging software and hardware (>3T) to image the subfields; however this type of technology may not be readily available in most research and clinical imaging centers. To address this need, this manuscript provides a detailed step-by-step protocol for segmenting the full anterior-posterior length of the hippocampus and its subfields: cornu ammonis (CA) 1, CA2/CA3, CA4/dentate gyrus (DG), strata radiatum/lacunosum/moleculare (SR/SL/SM), and subiculum. This protocol has been applied to five subjects (3F, 2M; age 29-57, avg. 37). Protocol reliability is assessed by resegmenting either the right or left hippocampus of each subject and computing the overlap using the Dice's kappa metric. Mean Dice's kappa (range) across the five subjects are: whole hippocampus, 0.91 (0.90-0.92); CA1, 0.78 (0.77-0.79); CA2/CA3, 0.64 (0.56-0.73); CA4/dentate gyrus, 0.83 (0.81-0.85); strata radiatum/lacunosum/moleculare, 0.71 (0.68-0.73); and subiculum 0.75 (0.72-0.78). The segmentation protocol presented here provides other laboratories with a reliable method to study the hippocampus and hippocampal subfields in vivo using commonly available MR tools.
Introduction
The hippocampus is a widely studied medial temporal lobe structure that is associated with episodic memory, spatial navigation, and other cognitive functions10,31. Its role in neurodegenerative and neuropsychiatric disorders such as Alzheimer’s disease, schizophrenia, and bipolar disorder is well-documented4,5,18,24,30. The goal of this manuscript is to provide additional detail to the manual segmentation protocol published previously34 for human hippocampal subfields on high-resolution magnetic resonance (MR) images acquired at 3T. Additionally, the video component accompanying this manuscript will provide further assistance to researchers who wish to implement the protocol on their own datasets.
The hippocampus can be divided into subfields based on cytoarchitectonic differences observed in histologically-prepared post-mortem specimens12,22. Such post-mortem specimens define the ground truth for the identification and study of hippocampal subfields; however preparations of this nature require specialized skills and equipment for staining, and are limited by the availability of fixed tissue, especially in diseased populations. In vivo imaging has the advantage of a much larger pool of subjects, and also presents the opportunity for follow-up studies and observing changes in populations. Although it has been shown that signal intensities in T2-weighted ex vivo MR images reflect cellular density13, it is still difficult to identify undisputed borders between subfields using solely MR signal intensities. As such, a number of different approaches for identifying histology-level detail on MR images have been developed.
Some groups have made efforts to reconstruct and digitize histological datasets and then use these reconstructions along with image registration techniques to localize hippocampal subfield neuroanatomy on in vivo MR1,2,8,9,14,15,17,32. Although this is an effective technique for mapping a version of the histological ground truth directly onto MR images, reconstructions of this nature are difficult to complete. Projects such as these are limited by the availability of intact medial temporal lobe specimens, histological techniques, data loss during histological processing, and the fundamental morphological inconsistencies between fixed and in vivo brains. Other groups have used high-field scanners (7T or 9.4T) in an effort to acquire in vivo or ex vivo images with a small enough (0.20-0.35 mm isotropic) voxel size to visualize spatially localized differences in image contrast that are used to infer boundaries between subfields35,37. Even at 7T-9.4T and with such a small voxel size, the cytoarchitectonic characteristics of hippocampal subfields are not visible. As such, manual segmentation protocols have been developed that approximate the known histological boundaries on MR images. These protocols determine subfield boundaries by interpreting local image contrast differences and defining geometric rules (such as straight lines and angles) relative to visible structures. Although images taken at a high field strength are able to offer detailed insight into hippocampal subfields, high-field scanners are not yet common in clinical or research settings, so 7T and 9.4T protocols currently have limited applicability. Similar protocols have been developed for images collected on 3T and 4T scanners11,20,21,23,24,25,28,33. Many of these protocols are based on images with sub-1mm voxels voxel dimensions in the coronal plane, but have large slice thicknesses (0.8-3 mm)11,20,21,23,25,28,33 or large inter-slice distances20,28, both of which result in a significant measurement bias in the estimation of volumes of the individual subfields. Additionally, many of the existing 3T protocols exclude subfields in all or part of the hippocampal head or tail20,23,25,33 or do not provide detailed segmentations of important substructures (i.e., combine the DG with CA2/CA3 or do not include the strata radiatum/lacunosum/moleculare of the CA)11,20,21,23,24,25,28,33. There is therefore a need in the field for a detailed description of a protocol that can reliably identify relevant subfields throughout the head, body, and tail of the hippocampus that is based on a scanner commonly available in clinical and research settings. Efforts are currently underway by the Hippocampal Subfields Group (www.hippocampalsubfields.com) to harmonize the hippocampal subfield segmentation process between laboratories, similar to an existing harmonization effort for whole hippocampal segmentation6, and an initial paper comparing 21 existing protocols was recently published38. The work from this group will further elucidate optimal segmentation procedures.
This manuscript provides detailed written and video instructions for reliably implementing the hippocampal subfield segmentation protocol described previously by Winterburn and colleagues34 on high-resolution 3T MR images. The protocol has been implemented on five images of healthy controls for the whole hippocampus and five hippocampal subfields (CA1, CA2/CA3, CA4/dentate gyrus, strata radiatum/lacunosum/moleculare, and subiculum). These segmented images are available to the public online (cobralab.ca/atlases/Hippocampus). The protocol and the segmented images will be useful for groups who wish to study detailed hippocampal neuroanatomy in MR images.
Protocol
Study Participants
The protocol in this manuscript was developed for five representative high-resolution images collected from healthy volunteers (3F, 2M; age 29-57, avg. 37) who were free of neurological and neuropsychiatric disorders and cases of severe head trauma. All subjects were recruited at the Centre for Addiction and Mental Health (CAMH). The study was approved by the CAMH Research Ethics Board and was conducted in keeping with the Declaration of Helsinki. All subjects provided written, informed consent for data acquisition and sharing. For details about the acquisition sequence used to collect these images, please refer to Winterburn et al., 2013 and Park et al., 2014.26,34 Images for all five subjects were checked for quality and retained. The hippocampus spanned an average of 118 coronal slices in these images.
1. Software Set-up
- Open Display: From the terminal using the following command: Display image_name.mnc -label label_name.mnc. The program will open 3 windows: 3D visualization window, 3-orientation image viewing window, and a navigation window. The terminal will also be used to run the program. Enlarge the coronal view, as the segmentations will be performed coronally. Zoom in on the hippocampus. Select F (Segmenting) in the navigation window. Select F (XY Radius:0.1). The terminal window will prompt for the user to “Enter xy brush size: ”. Set to 0.1. This will set the size of your paintbrush. The user can now begin drawing the hippocampus onto the MR image.
2. Whole Hippocampus Manual Segmentation
- Set-up: Using a T1-weighted image, scroll to the anterior-most coronal slice of the hippocampus. To advance slices in the anterior direction, use the '+' key; use the '-' key to move in the posterior direction.
- Slice A: Anterior-Most Slice: Using the right-click on the mouse, draw the outer-most border of the hippocampal grey matter where it meets the surrounding temporal lobe white matter and use the high-intensity white matter of the alveus to assist with the superior border, where the hippocampus meets the amygdala12,22. Use the E (Label Fill) key in the segmentation menu of the navigation window to fill in the label inside the border. Continue to apply these borders throughout the anterior hippocampal head.
- Slice B: Hippocampal Head 1 (Figure 1B):
- Superior, inferior, lateral, medial borders: Continue to draw the borders as described in step 2.2, using the white matter of the temporal lobe and alveus as a guide.
- Supero-medial border: For this, using the axial view, draw a horizontal line from the anterior edge of the lateral hippocampus29, and include anything below this line as hippocampus. NOTE: The supero-medial border becomes more ambiguous in these slices, where the grey matter of the hippocampus blends with the grey matter of the amygdala.
- Slice C: Hippocampal Head 2 with Dentations: Depending on the subject, the dentations of the hippocampus may be visible for 3-4 slices (typically, they are more visible on T2-weighted versus T1-weighted images). In these slices, continue to use the white matter of the alveus and temporal lobe to guide border segmentation12,22. For further details, follow steps 2.5.1-2.5.2.
- Slice D: Hippocampal Head 3:
- Superior, inferior, lateral, medial borders: Draw the inferior border of the hippocampus at the white matter of the temporal lobe, the lateral border at the inferior horn of the lateral ventricle, the superior border, following the curve of the dentations, at the white matter of the alveus/fimbria, and the medial border at the hypointense region of the ambient cistern12,22.
- Supero-medial and infero-medial borders: Continue to define the supero-medial border as described in step 2.3.2. Draw the inferior portion of the medial border where the hippocampus thins slightly and extends into the mildly hyperintense grey matter of the entorhinal cortex12,22.
- Slice E: Hippocampal Head 4 with Uncus: Continue to draw the inferior, lateral, and superior borders described in steps 2.5.1-2.5.2. Include the uncus (which is located medal to the main body of the hippocampus and is surrounded by low-intensity CSF) in the hippocampal segmentation12,22.
- Slice F: Hippocampal Body: Continue to draw the inferior, lateral, medial, and superior borders described in steps 2.5.1-2.5.2. Draw the infero-medial border at the point where the hippocampus thins as it transitions to entorhinal cortex/para-hippocampal gyrus12,22. Do not include the low-intensity CSF of the vestigial hippocampal sulcus in the segmentation.
- Slice G: Hippocampal Tail 1: Begin segmenting hippocampal tail-type slices when the crus of the fornix is first visible. Exclude the fascicular gyrus (a grey matter structure which blends with the hippocampus in parts of the hippocampal tail) from the segmentation by extrapolating the shape of the fascicular gyrus into the hippocampal tail from more anterior slices12,22. This extrapolation is only possible for 2-3 slices, after which the two structures cannot be accurately distinguished; at this point, treat all visible grey matter in this area as hippocampus .
- Slice H: Hippocampal Tail 2: Segment the low-intensity grey matter of the posterior hippocampal tail from the surrounding high-intensity white matter.
- Slice I: Posterior-Most Slice: Segment the small remaining area of hippocampal grey matter from the surrounding white matter of the temporal lobe.
3. Hippocampal Subfield Manual Segmentation
- Set-up: Using a T2-weighted image, scroll to the anterior-most coronal slice of the hippocampus (as in step 2.1). To change the color of the paintbrush, select D (Set Paint Lbl:) on the segmenting menu in the navigation window. The command terminal will prompt: “Enter current paint label:”. Enter a number between 1 and 255. Each number corresponds to a different label color.
- Slice A: Anterior-Most Slice: Since subfield divisions are not yet visible in the anterior-most slice, draw a line dividing the visible hippocampal grey matter along its longest visible axis (which is not necessarily parallel to any of the cardinal axes) into two equal sections to approximate the true anatomy12,22. Label the superior of these two sections as CA1 and the inferior section as subiculum by choosing a different colored label for each subfield23,35.
- Slice B: Hippocampal Head 1: Label the low-intensity area in the middle of the hippocampal formation as SR/SL/SM13,37. When the bend along the inferior edge of the hippocampus becomes clear, use this landmark as the lateral border separating the subiculum from the CA112,22. Continue to follow the longest axis of the hippocampus to draw the CA1-subiculum border on the supero-medial tip37.
- Slice C: Hippocampal Head 2 with Dentations:
- SR/SL/SM, CA4/DG, and subiculum: Label the SR/SL/SM, CA4/DG, and subiculum as described for slice D (step 3.5.1).
- CA2/CA3 and CA1: Define the border between CA1 and CA2/CA3 as a 45º angle line extending in the supero-lateral direction from the most supero-lateral edge of the SR/SL/SM12,22. Extend the CA2/CA3 medially along the superior edge to the trough between the dentations12,22. Label the rest of the superior edge as CA112,22.
- Slice D: Hippocampal Head 3
- SR/SL/SM, CA4/DG, and subiculum: Label the dark SR/SL/SM band first, which will follow the curve of the CA137. Label any high-intensity grey matter inside of the SR/SL/SM as CA4/DG12,22,23,35,37. This may not be a continuous region, as in Figure 2C. Continue to define the subiculum-CA1 border using the bend in the inferior hippocampus12,22.
- CA2/CA3 and CA1: Continue to define the CA1 and CA2/CA3 border as in step 3.4.2. Extend the CA2/CA3 medially halfway along the superior edge of the hippocampus12,22 and label the other half of the superior edge as CA112,22.
- Supero-medial hippocampal head: In this slice, divide the supero-medial hippocampal head vertically in half. Label the medial half as SR/SL/SM12. Divide the lateral half in half again, this time horizontally. Label the superior portion as CA4/DG and the inferior portion as CA2/CA312.
- Slice E: Hippocampal Head 4 with Uncus
- Lateral hippocampal head (subiculum): In the lateral portion of these slices, define the subiculum-CA1 border as a vertical line extending in the inferior direction from the most medial edge of the CA4/DG12,22.
- Lateral hippocampal head (CA1, CA2/CA3, CA4/DG, SR/SL/SM.): Define the CA1-CA2/CA3 border in the same way as in step 3.4.2. Continue to label the SR/SL/SM as the low intensity region following the curve of the CA regions. Label the CA4/DG as the center cavity inside the SR/SL/SM, as in step 3.5.1.
- Uncal hippocampal head (SR/SL/SM): Label the uncus of the hippocampus for approximately 10 slices as the hippocampal head transitions into the hippocampal body. In the uncus, label the low intensity region in the center as SR/SL/SM (when this is difficult to see, approximate the anatomy by segmenting a line 2-3 voxels wide up the center of the uncus)12.
- Uncal hippocampal head (CA2/CA3, CA4/DG): Draw a line at the superior edge of the SR/SL/SM section along infero-lateral/supero-medial axis of the uncus. Label all grey matter above this line as CA2/CA312. Label any unlabeled grey matter below this line (on either side of the SR/SL/SM) as CA4/DG12.
- Slice F: Hippocampal Body: Continue to apply the borders described in step 3.6.1-3.6.2.
- Slice G: Hippocampal Tail 1: Continue to apply the rules described in step 3.6.1-3.6.2. The subiculum-CA1 border becomes a 45º angle line extending in the infero-medial direction from the medial edge of the CA4/DG12,22.
- Slice H: Hippocampal Tail 2: Once the fascicular gyrus can no longer be distinguished from the hippocampal formation, label the entire outer layer as CA1, the low-intensity area inside of this as SR/SL/SM (as in previous slices), and any remaining grey matter in the middle as CA4/DG12,22.
- Slice I: Posterior-Most Slice: Once the dark SR/SL/SM is no longer visible in the center of the hippocampal formation, label the entire structure as CA112,22.
4. Protocol Reliability
- Resegment either the right or left hippocampus of each subject after waiting approximately one month from performing the original segmentation. Segment all of the subfields along the entire anterior-posterior length of the hippocampus, trying to follow the protocol rules as consistently as is possible.
- Calculate the Dice’s kappa between the original and resegmented volumes:
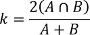
where k=Dice’s kappa and A and B are label volumes.
Representative Results
Results from the protocol reliability test are summarized in Table 2. For the whole bilateral hippocampus, mean spatial overlap as measured by Dice’s kappa is 0.91 and ranges from 0.90 – 0.92. Subfield kappa values range from 0.64 (CA2/CA3) to 0.83 (CA4/dentate gyrus). Mean volumes for all subfields and the whole hippocampus are reported in Table 3. Volumes for the whole hippocampus range from 2456.72-3325.02 mm3. The CA2/CA3 is the smallest subfield at 208.33 mm3, while the CA1 is the largest at 857.46 mm3.
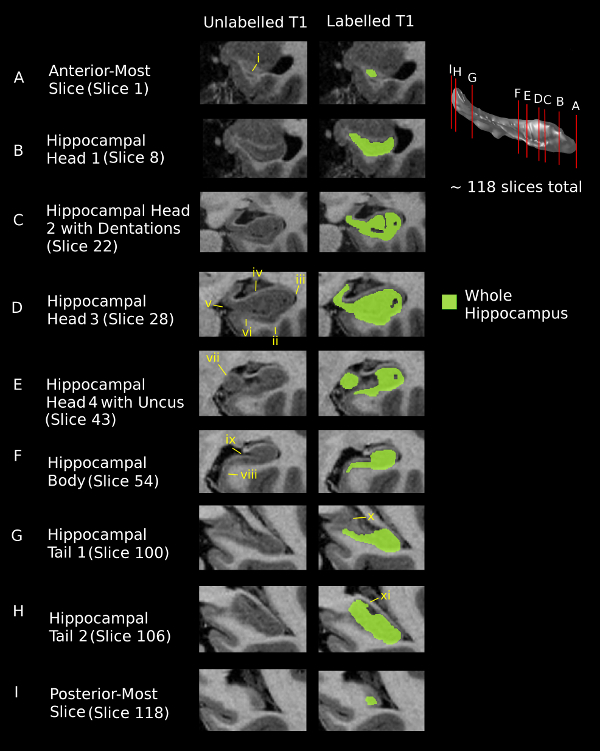
Figure 1. Segmentation of the whole hippocampus for 9 coronal slices (A-I) using T1-weighted images. The vertical red lines on the hippocampal surface illustrate the location of each coronal slice. The hippocampus was present in an average of 118 coronal slices in each of the five subjects included in this study. Images progress from anterior (slice 1) at the top to posterior (slice 118) at the bottom. Images are shown in the left column without segmentation and with segmentation in the right column. The scale bar shows 3 mm for reference. Roman numerals point to specific features identified in the protocol manuscript. i. The alveus distinguishes the hippocampal grey matter from the grey matter of the amygdala in the anterior-most slice. ii. The white matter of the temporal lobe defines the inferior border of the hippocampus in the hippocampal head. iii. The lateral border of the hippocampus in the hippocampal head is the inferior horn of the lateral ventricle. iv. The superior border is defined by the white matter of the alveus/fimbria. v. The medial border of the hippocampal head is the ambient cistern. vi. The infero-medial hippocampus extends into the entorhinal cortex, which shows up as a mildly hyper-intense band in T1-weighted images. vii. The uncus of the hippocampus is present in the hippocampal head and can be easily distinguished from the surrounding CSF. viii. In the infero-medial direction, the border between the subiculum and the para-hippocampal gyrus is defined by a slight thinning of the hippocampal grey matter. ix. The CSF of the vestigial hippocampal sulcus is not included in the segmentation. x. The fascicular gyrus is not included in the segmentation of the hippocampal tail when it is possible to differentiate it. xi. When it is no longer possible to distinguish between the fascicular gyrus and the hippocampal tail, the fascicular gyrus is included in the segmentation. Please click here to view a larger version of this figure.
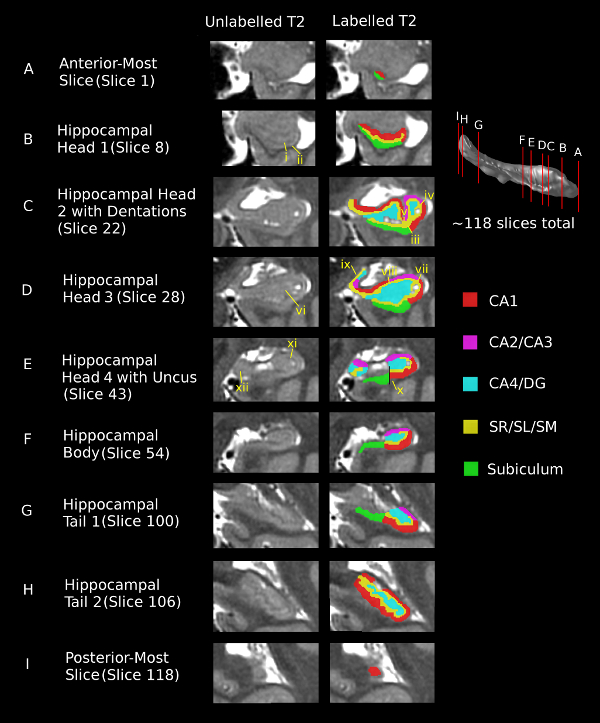
Figure 2. Segmentation of the hippocampal subfields for 9 coronal slices (A-I) using T2-weighted images. The vertical red lines on the hippocampal surface illustrate the location of each coronal slice. The hippocampus was present in an average of 118 coronal slices in each of the five subjects included in this study. Images progress from anterior (slice 1) at the top to posterior (slice 118) at the bottom. Images are shown in the left column without segmentation and with segmentation in the right column. The scale bar shows 3 mm for reference. Roman numerals point to specific features identified in the protocol manuscript. i. The low intensity region in the center of the hippocampal head is the SR/SL/SM. ii. The uncal-shaped bend on the infero-lateral edge of the hippocampus marks the border between the CA1 and the subiculum. iii. The subiculum-CA1 border continues to be defined at the ‘bend’ in the inferior hippocampus in the hippocampal head. iv. The border between CA1 and CA2/CA3 is defined as a 45° angle extending in the supero-lateral direction from the most supero-lateral edge of the SR/SL/SM. v. The CA2/CA3 extends halfway along the superior edge of the hippocampus, to the trough of the dentations, medial to which it is labeled as CA1. vi. The grey matter in the center of the hippocampal head is labeled as CA4/DG. vii. Continue to define the CA1-CA2/CA3 border as a 45° angle extending in the supero-lateral direction from the most supero-lateral edge of the SR/SL/SM. viii. The CA2/CA3 continues to extend halfway along the superior edge of the hippocampus, medial to which it is labeled as CA1. ix. In slice D, the supero-medial hippocampal head is divided into subfields (see step 3.5.3). x. The subiculum-CA1 border is defined as a vertical line extending from the most medial border of the CA4/DG. xi. The SR/SL/SM continues to be the low-intensity region following the curve of the CA regions. xii. In the uncal portion of the hippocampal head, the SR/SL/SM is the low-intensity region in the center of the uncus. If this cannot be seen, draw a line 2-3 pixels wide up the center of the uncus. Please click here to view a larger version of this figure.
Table 1. Superior, inferior, medial, and lateral borders for hippocampal subfields for nine representative slices along anterior-posterior extent of the hippocampus. Borders are described for T2-weighted images. WM = White Matter; GM = Grey Matter; MTL = Medial Temporal Lobe.
| Structure | Slice | Superior Border | Inferior Border | Medial Border | Lateral Border |
| CA1 | Anterior-Most Slice | WM of the alveus | Mid-line of hippocampal grey matter, along longest axis (borders subiculum) | WM of the alveus | WM of the alveus |
| Hippocampal Head 1 | WM of the alveus | SR/SL/SM; inferolateral border with subiculum at the 'bend' of the hippocampus | WM of the alveus | WM of the alveus | |
| Hippocampal Head 2 (with Dentations) | |||||
| Lateral | Follows the curve of the SR/SL/SM; supero-lateral border with CA2/CA3 | WM of the MTL | SR/SL/SM; inferomedial with subiculum at the 'bend' of the hippocampus | WM of the alveus | |
| Medial | WM of the alveus; supero-medial border with CA2/CA3 | Low-intensity SR/SL/SM | Low-intensity SR/SL/SM | CA2/CA3 | |
| Hippocampal Head 3 | |||||
| Lateral | Follows the curve of the SR/SL/SM; supero-lateral border with CA2/CA3 | WM of the MTL | SR/SL/SM; inferomedial with subiculum at the 'bend' of the hippocampus | WM of the alveus | |
| Medial | WM of the alveus; supero-medial border with CA2/CA3 | Low-intensity SR/SL/SM | Low-intensity SR/SL/SM | CA2/CA3 | |
| Hippocampal Head 4 (with Uncus) | Follows the curve of the SR/SL/SM; supero-lateral border with CA2/CA3 | WM of the MTL | SR/SL/SM; inferomedial border with subiculum vertical line along medial edge of CA4/DG | WM of the alveus | |
| Hippocampal Body | Follows the curve of the SR/SL/SM; supero-lateral border with CA2/CA3 | WM of the MTL | SR/SL/SM; inferomedial border with subiculum vertical line along medial edge of CA4/DG | WM of the alveus | |
| Hippocampal Tail 1 | SR/SL/SM; superolateral border with CA2/CA3 | WM of the MTL | Follows the curve of the SR/SL/SM; supero-medial border with the subiculum along line parallel to edge of CA4/DG | WM of the alveus | |
| Hippocampal Tail 2 | Supero-lateral border with the WM of the alveus/fimbria | WM of the MTL | WM of the MTL | WM of the MTL | |
| Posterior-Most Slice | Supero-lateral border with the WM of the alveus/fimbria | Rest of structure is bordered by the WM of the temporal lobe | WM of the MTL | WM of the alveus/fimbria | |
| Subiculum | Anterior-Most Slice | Mid-line of hippocampal grey matter, along longest axis (borders CA1) | WM of the MTL | WM of the alveus | WM of the alveus |
| Hippocampal Head 1 | SR/SL/SM; CA1 on supero-medial edge | WM of the MTL | WM of the alveus | CA1, at 'bend' in hippocampus | |
| Hippocampal Head 2 (with Dentations) | SR/SL/SM | WM of the MTL | Entorhinal cortex (low intensity area medial to inferior hippocampus) | CA1, at 'bend' in hippocampus | |
| Hippocampal Head 3 | SR/SL/SM | WM of the MTL | Entorhinal cortex (low intensity area medial to inferior hippocampus) | CA1, at 'bend' in hippocampus | |
| Hippocampal Head 4 (with Uncus) | CSF of the ambient cistern | WM of the MTL; infero-medial border at entorhinal cortex where cortical band thins slightly and signal intensity drops | CSF of the ambient cistern | CA1 along line parallel to edge of CA4/DG | |
| Hippocampal Body | CSF of the ambient cistern | WM of the MTL; infero-medial border at entorhinal cortex where cortical band thins slightly and signal intensity drops | CSF of the ambient cistern | CA1 along line parallel to edge of CA4/DG | |
| Hippocampal Tail 1 | GM of the fascicular gyrus (where can be separated from hippocampal GM) | WM of the MTL | Difficult to determine; extrapolate from more anterior/posterior slices | CA1 along line parallel to edge of CA4/DG | |
| Hippocampal Tail 2 | NA | ||||
| Posterior-Most Slice | NA | ||||
| CA2/CA3 | Anterior-Most Slice | NA | |||
| Hippocampal Head 1 | NA | ||||
| Hippocampal Head 2 (with Dentations) | |||||
| Lateral | WM of the alveus | Low-intensity SR/SL/SM | CA1 halfway along superior edge of hippocampus; if dentations visible, try to estimate halfway | Infero-lateral border with CA1 along 45° angle from most supero-lateral edge of SR/SL/SM | |
| Medial | CA4/DG halfway along superiorinferior extension of hippocampus | CA1 at base of superior-inferior extension of hippocampus | SR/SL/SM halfway along width of superior-inferior extension of hippocampus | WM of alveus | |
| Hippocampal Head 3 | |||||
| Lateral | WM of the alveus | Low-intensity SR/SL/SM | CA1 halfway along superior edge of hippocampus | Infero-lateral border with CA1 along 45° angle from most supero-lateral edge of SR/SL/SM | |
| Medial | CA4/DG halfway along superiorinferior extension of hippocampus | CA1 at base of superior-inferior extension of hippocampus | SR/SL/SM halfway along width of superior-inferior extension of hippocampus | WM of alveus | |
| Hippocampal Head 4 (with Uncus) | |||||
| Lateral | WM of the alveus | CA4/DG | CSF of the ambient cistern | Infero-lateral border with CA1 along 45° angle from most supero-lateral edge of SR/SL/SM | |
| Medial | CSF of the ambient cistern | Line parallel to superior edge of SR/SL/SM | CSF of the ambient cistern | CSF of the ambient cistern | |
| Hippocampal Body | WM of the alveus | CA4/DG | CSF of the ambient cistern | Infero-lateral border with CA1 along 45° angle from most supero-lateral edge of SR/SL/SM | |
| Hippocampal Tail 1 | WM of the alveus | Inferior border horizontal line extending from most lateral point of SR/SL/SM, following pattern of more anterior slices; inferomedial border with CA4/DG | WM of the fimbria | WM of the fimbria | |
| Hippocampal Tail 2 | NA | ||||
| Posterior-Most Slice | NA | ||||
| CA4/DG | Anterior-Most Slice | NA | |||
| Hippocampal Head 1 | NA | ||||
| Hippocampal Head 2 (with Dentations) | Follows curve of low-intensity SR/SL/SM | Low-intensity SR/SL/SM | CSF of the ambient cistern | Low-intensity SR/SL/SM | |
| Lateral | Low-intensity SR/SL/SM | Low-intensity SR/SL/SM | CSF of the ambient cistern | Low-intensity SR/SL/SM | |
| Medial | Use the axial view to draw a horizontal line medially from the anterior edge of the lateral hippocampus | CA2/CA3 halfway along superiorinferior extension of hippocampus | SR/SL/SM halfway along width of superior-inferior extension of hippocampus | WM of alveus | |
| Hippocampal Head 3 | |||||
| Lateral | Low-intensity SR/SL/SM | Low-intensity SR/SL/SM | CSF of the ambient cistern | Low-intensity SR/SL/SM | |
| Medial | CSF of the ambient cistern | CA2/CA3 halfway along superiorinferior extension of hippocampus | SR/SL/SM halfway along width of superior-inferior extension of hippocampus | WM of alveus | |
| Hippocampal Head 4 (with Uncus) | |||||
| Lateral | Low-intensity SR/SL/SM | Low-intensity SR/SL/SM | CSF of the ambient cistern | Low-intensity SR/SL/SM | |
| Medial | Line parallel to superior edge of SR/SL/SM | CSF of ambient cistern | CSF of ambient cistern; lowintensity SR/SL/SM | CSF of ambient cistern; low intensity SR/SL/SM | |
| Hippocampal Body | CA2/CA3 | Low-intensity SR/SL/SM | CSF of ambient cistern | Low-intensity SR/SL/SM | |
| Hippocampal Tail 1 | CA2/CA3 and fimbria | Low-intensity SR/SL/SM | CSF of lateral ventricle | Low-intensity SR/SL/SM | |
| Hippocampal Tail 2 | NA | ||||
| Posterior-Most Slice | NA | ||||
| SR/SL/SM | Anterior-most slice | NA | |||
| Hippocampal Head 1 | Low-intensity SR/SL/SM in center of CA1 and subiculum | ||||
| Hippocampal Head 2 (with Dentations) | Use the axial view to draw a horizontal line medially from the anterior edge of the lateral hippocampus | Low-intensity SR/SL/SM surrounding CA4/DG | CSF of ambient cistern | CA2/CA3 and CA4/DG halfway along width of superior-inferior extension of hippocampus | |
| Hippocampal Head 3 | Use the axial view to draw a horizontal line medially from the anterior edge of the lateral hippocampus | Low-intensity SR/SL/SM surrounding CA4/DG | CSF of ambient cistern | CA2/CA3 and CA4/DG halfway along width of superior-inferior extension of hippocampus | |
| Hippocampal Head 4 (with Uncus) | |||||
| Lateral | Low-intensity SR/SL/SM surrounding CA4/DG | ||||
| Medial | Low-intensity SR/SL/Smin middle (when difficult to see, approximate with a line 203 voxels wide) | CSF of ambient cistern | CA4/DG | CA4/DG | |
| Hippocampal Body | Low-intensity SR/SL/SM surrounding CA4/DG | ||||
| Hippocampal Tail 1 | Low-intensity SR/SL/SM surrounding CA4/DG | ||||
| Hippocampal Tail 2 | Low-intensity SR/SL/SM surrounding CA4/DG | ||||
| Posterior-Most Slice | NA | ||||
Table 2. Protocol reliability results for all five subfields and the whole hippocampus from the five manually segmented subjects. Resegmentations were performed on either the right or left hippocampus of each subject. Mean Dice’s kappa reflects the mean across the five subjects.
| Structure | Mean Dice’s kappa (range) |
| CA1 | 0.78 (0.77-0.79) |
| CA2/CA3 | 0.64 (0.56-0.73) |
| CA4/dentate gyrus | 0.83 (0.81-0.85) |
| SR/SL/SM | 0.71 (0.68-0.73) |
| Subiculum | 0.75 (0.72-0.78) |
| Whole hippocampus | 0.91 (0.90-0.92) |
Table 3. Mean subfield and whole hippocampal volumes.
| Structure | Mean volume (range) (mm3) |
| CA1 | 857.46 (720.17-981.68) |
| CA2/CA3 | 208.33 (155.10-281.57) |
| CA4/dentate gyrus | 615.50 (500.16-763.01) |
| SR/SL/SM | 687.22 (576.61-895.59) |
| Subiculum | 390.79 (277.21-445.95) |
| Whole hippocampus | 2759.31 (2456.72-3325.02) |
Discussion
Hippocampal subfield segmentation in MR images is well-represented in the literature. However, existing protocols exclude portions of the hippocampus20,23,33,35, apply only to fixed images37, or require ultra-high field scanners for image acquisition35,37. This manuscript offers a segmentation protocol that includes five major subdivisions (CA1, CA2/CA3, CA4/dentate gyrus, SR/SL/SM, and subiculum) of the hippocampus and spans the entire anterior-posterior length of the structure. The complete segmented atlases are available to the public online (cobralab.ca/atlases/Hippocampus). This work is applicable to many groups within the neuroimaging field, and will help to limit some of the existing discrepancies in hippocampal subfield segmentation.
Reliability testing of the protocol shows a high degree of spatial overlap between original and resegmented labels, which reflects a high intra-rater reliability (Table 2). A kappa value of 0.91 for the whole hippocampus compares favorably with other values reported in the literature35,37. The intra-rater reliabilities of many of the subfields also compare well with other similar segmentation protocols; however, some structures have lower reliabilities25,33,35,37.This may be a result of including the SR/SL/SM subfield in the present protocol where other groups do not, which results in adjacent subfields (the subiculum, CA1, and CA2/CA3) being thinner, and therefore more heavily penalized by the Dice’s kappa metric33,35. Additionally, the retest process used in this protocol is perhaps more rigorous and therefore more reflective of true protocol reliability than those used by other groups. The entire anterior-posterior length of one hemisphere of each subject was resegmented, whereas other groups with higher reliabilities segment only a few coronal slices23,33,37.The subfield with the lowest kappa (0.64) is the CA2/CA3, which is a small, thin structure. It has previously been shown that the intra-rater error for all subfields in this protocol is higher than a simulated 0.3 mm translational error in every cardinal direction, or a simulated 1% expansion/shrinkage of labels34. In other words, the manual resegmentation error is smaller than introducing a small systematic error, which supports the high manual reproducibility of the protocol.
The expert manual rater studied each of the five high-resolution images in detail to determine which of the subfields present in Duvernoy’s histology could be seen12. It was determined that it was not possible to reliably differentiate the CA2 from the CA3, so to increase protocol reliability, they were combined into one structure. This rule follows the precedent of previous groups33,37. It was also not possible to distinguish the CA4 from the stratum moleculare, stratum granulosum, and polymorphic layer of the dentate gyrus in the images, or to distinguish among the dentate gyrus layers themselves. The CA4 and all dentate gyrus layers were therefore combined into one label (CA4/DG). There is, in fact, a debate in the hippocampal subfield segmentation community as to whether the CA4 region should be considered a part of the cornu ammonis, as with Duvernoy12, or as a part of the dentate gyrus, as with Amaral3. The method presented in this manuscript accommodates both of these views, and follows the work of previous MR segmentation groups23,28,33,35,37. The strata radiatum, lacunosum, and moleculare of the cornu ammonis also could not be distinguished separately, so were combined into one label, as with previous groups37.
The most accurate analysis of neuroanatomy is through histological sectioning and staining, but this type of analysis suffers from a number of issues: limited access to fixed specimens (which results in very small sample sizes); the expertise required to prepare samples; distortions of the brain after fixation; and difficulties in applying a fixed atlas to digital, in vivo data1,2,8. In ex vivo imaging, long acquisition times of a fixed brain in an MR scanner also provides a detailed picture of neuroanatomy, but as with histology, sample number is limited and there are morphometric differences between the fixed and in vivo brain37. In vivo MR imaging has a limited resolution, but presents the possibility for much larger sample sizes, as well as the potential for imaging a single subject at multiple time points. By lengthening the acquisition time on standard field strength scanners (within the confines of subject comfort), the level of detail available in in vivo images becomes sufficient to resolve sub-structure-level neuroanatomy. The acquisition used for the images segmented in this protocol therefore offers a reasonable trade-off between sample availability and image resolution.
This protocol was developed for high-resolution MR images such as those used to illustrate the protocol steps in this manuscript26,34. High-resolution images were acquired on a 3T scanner by taking advantage of long scan times and image averaging. The total scan time for both of the FSPGR-BRAVO and FSE-CUBE acquisitions together was just under 2 hours. It is recognized that this is a prohibitive scan length for clinical applications: this sequence was performed here for illustrative purposes for the segmentation protocol. The authors believe that the segmentation protocol described in this manuscript could be adapted to images with a shorter scan time, for example a single 3T acquisition (as opposed to 3 acquisitions for each contrast type, as used by Winterburn et al., 201334 and Park et al., 201426) or a 7T image. Even with a slightly lower resolution, many of the rules in the protocol would still apply, such as those using a geometrical approach (ex. 45° angle line between CA1 and CA2/CA3; vertical line from medial edge of CA4/DG to separate subiculum and CA1 in hippocampal body). Other rules would perhaps have to be adapted (ex. it may not be possible to differentiate the SR/SL/SM, in which case it could be included as a part of the CA subfields). Additionally, it has previously been show that high-resolution atlases can be applied to lower-resolution images using an automated segmentation pipeline7,27.
The protocol was designed for and implemented on images of healthy subjects, but could also be applied (either manually or using an automated segmentation pipeline7,16,27) to images of diseased populations such as Alzheimer’s disease patients, for whom severe atrophy makes the hippocampus a structure of particular interest.5,30 In spite of this atrophy, landmarks surrounding the hippocampus and intensity contrast in the images would mean the segmentation protocol would still be largely viable. However, such clinical images would likely be acquired on a scanner with a much lower field strength, such as 1.5T, where the resolution would be too low to be able to see substructures.
The type of software used to perform the segmentations is relevant, as it is important to be able to look at the structure from multiple viewpoints (i.e., coronal, sagittal, axial). In addition, the use of a 3D visualization of the surface of structure can be used to smooth out the overall topology of the hippocampus. Often errant voxels or illogical shapes will not be obvious in the 2-dimensionsal cardinal planes, but will be very clear on a 3D surface. On high-resolution images, the protocol applies to approximately 118 coronal slices and requires upwards of 40 hours of work per subject by a previously-trained expert manual rater. This amount of manual labor limits the applicability of the full protocol to a large subject set. It would be possible to implement a modified version of the protocol as a time-saving measure: for example, every other coronal slice could be segmented to provide an estimation of subfield volumes, or subfields could be combined, for example all cornu ammonis subfields (CA1, CA2/CA3, and SR/SL/SM).
In conclusion, this manuscript presents a detailed manual segmentation protocol for the whole hippocampus and five hippocampal subfields (CA1, CA2/CA3, CA4/dentate gyrus, strata radiatum/lacunosum/moleculare, and subiculum). This protocol has been applied to five subjects, and the atlases have been made available publically (cobralab.ca/atlases/Hippocampus). These atlases allow other laboratories interested in hippocampal segmentation to perform reliable, repeatable segmentations of hippocampal subfields on new image datasets.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors would like to acknowledge support from the CAMH Foundation, thanks to Michael and Sonja Koerner, the Kimel Family, and the Paul E. Garfinkel New Investigator Catalyst Award. This project was funded by the Fonds de Recherches Santé Québec, the Canadian Institutes of Health Research (CIHR), the Natural Sciences and Engineering Research Council of Canada, the Weston Brain Institute, the Alzheimer's Society of Canada, and the Micheal J. Fox Foundation for Parkinson's Research (MMC), as well as CIHR, the Ontario Mental Health Foundation, NARSAD, and the National Institute of Mental Health (R01MH099167) (ANV). The authors would also like to thank Anusha Ravichandran for assistance acquiring the images.
Materials
| Discovery MR750 3T | GE | NA | Or equivalent 3T scanner |
| Minc Tool Kit | McConnell Brain Imaging Center, Montreal Neurological Institute | NA | Open source: http://www.bic.mni.mcgill.ca/ServicesSoftware/ServicesSoftwareMincToolKit |
References
- Adler, D. H., et al. Reconstruction of the human hippocampus in 3D from histology and high-resolution ex-vivo MRI. IEEE Intl. Symp. on Biomed. Img. , 294-297 (2012).
- Adler, D. H., et al. Histology-derived volumetric annotation of the human hippocampal subfields in postmortem MRI. NeuroImage. 84 (1), 505-523 (2014).
- Amaral, D. G. A golgi study of cell types in the hilar region of the hippocampus in the rat. J. Comp. Neurol. 182 (4 Pt 2), 851-914 (1978).
- Blumberg, H. P., et al. Amygdala and Hippocampal Volumes in Adolescents and Adults With Bipolar Disorder. Arch Gen Psychiatry. 60 (12), 1201-1208 (2003).
- Braak, H., Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol . 82 (4), 239-259 (1991).
- Boccardi, M., et al. Survey of protocols for the manual segmentation of the hippocampus: preparatory steps towards a joint EADC-ADNI harmonized protocol. J. Alzheimer’s Dis. 26 (3), 61-75 (2011).
- Chakravarty, M. M., et al. Performing label-fusion-based segmentation using multiple automatically generated templates. Hum. Brain Mapp. 34 (10), 2635-2654 (2013).
- Chakravarty, M. M., Bertrand, G., Hodge, C. P., Sadikot, A. F., Collins, D. L. The creation of a brain atlas for image guided neurosurgery using serial histological data. NeuroImage. 30 (2), 359-376 (2006).
- Collins, D. L., Neelin, P., Peters, T. M., Evans, A. C. Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J. Comput. Assist. Tomogr. 18 (2), 192-205 (1994).
- Heijer, F. V., et al. Structural and diffusion MRI measures of the hippocampus and memory performance. NeuroImage. 63 (4), 1782-1789 (2012).
- Duncan, K., Tompary, A., Davachi, L. Associative encoding and retrieval are predicted by functional connectivity in distinct hippocampal area ca1 pathways. The Journal of Neuroscience. 34 (34), 11188-11198 (2014).
- Duvernoy, H. M. . The Human Hippocampus: Functional Anatomy Vascularization, and Serial Sections with MRI. , (2005).
- Fatterpekar, G. M., et al. Cytoarchitecture of the human cerebral cortex: MR microscopy of excised specimens at 9.4 Tesla. Am. J. Neuroradiol. 23 (8), 1313-1321 (2002).
- Frey, S., Pandya, D. N., Chakravarty, M. M., Bailey, L., Petrides, M., Collins, D. L. An MRI based average macaque monkey stereotaxic atlas and space (MNI monkey space). NeuroImage. 55 (4), 1435-1442 (2011).
- Goubran, M., Crukley, C., de Ribaupierre, S., Peters, T. M., Khan, A. R. Image registration of ex-vivo. MRI to sparsely sectioned histology of hippocampal and neocortical temporal lobe specimens. NeuroImage. 83, 770-781 (2013).
- Heckemann, R. A., Hajnal, J. V., Aljabar, P., Rueckert, D., Hammers, A. Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. NeuroImage. 33 (1), 115-126 (2006).
- Holmes, C. J., Hoge, R., Collins, L., Woods, R., Toga, A. W., Evans, A. C. Enhancement of MR images using registration for signal averaging. J. Comput. Assist. Tomogr. 22 (2), 324-333 (1998).
- Karnik-Henry, M. S., Wang, L., Barch, D. M., Harms, M. P., Campanella, C., Csernansky, J. G. Medial temporal lobe structure and cognition in individuals with schizophrenia and in their non-psychotic siblings. Schizophrenia Research. 138 (2-3), 128-135 (2012).
- Kim, J. S., et al. Automated 3-D extraction and evaluation of the inner and outer cortical surfaces using a Laplacian map and partial volume effect classification. NeuroImage. 27 (1), 210-221 (2005).
- La Joie, R., et al. Differential effect of age on hippocampal subfields assessed using a new high-resolution 3T MR sequence. NeuroImage. 53 (2), 506-514 (2010).
- Libby, L. A., Ekstrom, A. D., Ragland, J. D., Ranganath, C. Differential connectivity of perirhinal and parahippocampal cortices within human hippocampal subregions revealed by high-resolution functional imaging. The Journal of Neuroscience. 32 (19), 6550-6560 (2012).
- Mai, J. K., Paxinos, G., Voss, T. . Atlas of the Human Brain. , (2008).
- Mueller, S. G., et al. Measurement of hippocampal subfields and age-related changes with high resolution MRI at 4T. Neurobiol Aging. 28 (5), 719-726 (2006).
- Narr, K. L., et al. Regional specificity of hippocampal volume reductions in first-episode schizophrenia. NeuroImage. 21 (4), 1563-1575 (2004).
- Olsen, R. K., Palombo, D. J., Rabin, J. S., Levine, B., Ryan, J. D., Rosenbaum, R. S. Volumetric Analysis of Medial Temporal Lobe Subregions in Development Amnesia using High-Resolution Magnetic Resonance Imaging. Hippocampus. 23 (10), 855-860 (2013).
- Park, M. T. M., et al. Derivation of high-resolution MRI atlases of the human cerebellum at 3T and segmentation using multiple automatically generated templates. NeuroImage. 95, 217-231 (2014).
- Pipitone, J., et al. Multi-atlas Segmentation of the Whole Hippocampus and Subfields Using Multiple Automatically Generated Templates. NeuroImage. 101, 494-512 (2014).
- Pluta, J., Yushkevich, P., Das, S., Wolk, D. In vivo analysis of hippocampal subfield atrophy in mild cognitive impairment via semi-automatic segmentation of T2-weighted MRI.Journal of Alzheimer’s Disease. 31 (1), 85-99 (2012).
- Pruessner, J. C., et al. Volumetry of hippocampus and amygdala with high-resolution MRI and three- dimensional analysis software: minimizing the discrepancies between laboratories. Cereb Cortex. 10 (4), 433-442 (2000).
- Sabuncu, M. R., et al. The dynamics of cortical and hippocampal atrophy in Alzheimer disease. Archives of Neurology. 68 (8), 1040-1048 (2011).
- Scoville, W. B., Milner, B. Loss of recent memory after bilateral hippocampal lesions. J. Neuropsych. and Clin. Neurosci. 12 (1), 103-113 (1957).
- Toga, A. W., Thompson, P. M., Mori, S., Amunts, K., Zilles, K. Towards multimodal atlases of the human brain. Nat. Rev. Neurosci. 7 (12), 952-966 (2006).
- van Leemput, K., et al. Automated segmentation of hippocampal subfields from ultra-high resolution in vivo. MRI. Hippocampus. 19 (6), 549-557 (2009).
- Winterburn, J. L., et al. A novel in vivo atlas of human hippocampal subfields using high-resolution 3 T magnetic resonance imaging. NeuroImage. 74, 254-265 (2013).
- Wisse, L. E. M., Gerritsen, L., Zwanenburg, J. J. M., Kuijf, H. J. Subfields of the hippocampal formation at 7 T MRI: in vivo. volumetric assessment. NeuroImage. 61 (4), 1043-1049 (2012).
- Yelnik, J., et al. A three-dimensional, histological and deformable atlas of the human basal ganglia. I. Atlas construction based on immunohistochemical and MRI data. NeuroImage. 34 (2), 618-638 (2007).
- Yushkevich, P. A., et al. A high-resolution computational atlas of the human hippocampus from postmortem magnetic resonance imaging at 9.4 T. NeuroImage. 44 (2), 385-398 (2009).
- Yushkevich, P. A., et al. Quantitative Comparison of 21 Protocols for Labeling Hippocampal Subfields and Parahippocampal Subregions in In Vivo MRI: Towards a Harmonized Segmentation Protocol. NeuroImage. , (2015).

