A 3D Human Lung Tissue Model for Functional Studies on Mycobacterium tuberculosis Infection
Summary
Human tuberculosis infection is a complex process, which is difficult to model in vitro. Here we describe a novel 3D human lung tissue model that recapitulates the dynamics that occur during infection, including the migration of immune cells and early granuloma formation in a physiological environment.
Abstract
Tuberculosis (TB) still holds a major threat to the health of people worldwide, and there is a need for cost-efficient but reliable models to help us understand the disease mechanisms and advance the discoveries of new treatment options. In vitro cell cultures of monolayers or co-cultures lack the three-dimensional (3D) environment and tissue responses. Herein, we describe an innovative in vitro model of a human lung tissue, which holds promise to be an effective tool for studying the complex events that occur during infection with Mycobacterium tuberculosis (M. tuberculosis). The 3D tissue model consists of tissue-specific epithelial cells and fibroblasts, which are cultured in a matrix of collagen on top of a porous membrane. Upon air exposure, the epithelial cells stratify and secrete mucus at the apical side. By introducing human primary macrophages infected with M. tuberculosis to the tissue model, we have shown that immune cells migrate into the infected-tissue and form early stages of TB granuloma. These structures recapitulate the distinct feature of human TB, the granuloma, which is fundamentally different or not commonly observed in widely used experimental animal models. This organotypic culture method enables the 3D visualization and robust quantitative analysis that provides pivotal information on spatial and temporal features of host cell-pathogen interactions. Taken together, the lung tissue model provides a physiologically relevant tissue micro-environment for studies on TB. Thus, the lung tissue model has potential implications for both basic mechanistic and applied studies. Importantly, the model allows addition or manipulation of individual cell types, which thereby widens its use for modelling a variety of infectious diseases that affect the lungs.
Introduction
In humans, responses to infection, tissue inflammation, cellular recruitment, tissue remodeling and the regulation of tissue homeostasis are complex events involving different cell types. Hence, these processes are best studied in the local tissue environment. Previously, this has mainly been possible using experimental animal models. However, the widely used experimental animals hold many limits as they often respond to pathogens in a different way than humans and also display a different course of disease 1. A human in vitro lung tissue model holds the possibilities to study specific immune responses in the human lung.
Human tuberculosis infection (TB) is mainly a disease affecting the lungs. Mycobacterium tuberculosis (M. tuberculosis), the causative agent of TB, reaches the lung via aerosol droplets that are transported to the alveolar space, where the bacteria are engulfed by pulmonary dendritic cells and alveolar macrophages as part of the innate immune response to the infection 2,3. Phagocytosis of the pathogen leads to the compartmentalization of the bug within a phagosome and ideally results in the neutralization and killing of the pathogen by the phagocyte. Up to 50% of individuals exposed to M. tuberculosis are believed to be able to clear the infection through the innate immune response 4. Other outcomes of infection are clearance by the adaptive immune system at a later stage, latent infection or in worst cases chronic active disease 5.
Previously there have been no in vitro tissue models for studies of human TB. Single cell cultures of human macrophages or other peripheral blood cells have often been used 6,7. The disadvantage of this approach is that they cannot reflect the dynamics of different cell types operating together in a lung tissue exposed to M. tuberculosis. Thus, there is a need for an in vitro model to be able to perform functional and mechanistic studies on TB. The cell-based in vitro human lung tissue model described here was originally established by our group for studies on dendritic cell functions 8. We have adapted this method for the study of TB.
The human lung tissue model presented here consists of tissue-specific epithelial cells and fibroblasts 8. These cells are cultured in a matrix of collagen on top of a porous membrane in a transwell insert and form structures resembling normal human lung tissue (Figure 1). When exposed to air the cells start to secrete mucus at the apical side 8. By implanting human primary macrophages infected with M. tuberculosis to the model, we have observed how the immune cells migrate in the tissue and form early stages of TB granulomas 9. This is the first human tissue model described for TB and it poses a promising tool for studying innate immune responses to TB and other diseases of the lung. To date, we have used only monocytes and macrophages as immune cells in the model but the level of complexity can be increased by inclusion of additional relevant cell types.

Figure 1. Schematic outline of the lung tissue model. (A) The model is composed of human lung-specific epithelial cells, M. tuberculosis-infected primary macrophages and red dye labeled monocytes seeded onto collagen embedded fibroblasts prepared on a transwell filter. Exposure of the tissue model to air initiates production of extra-cellular matrix proteins, mucus secretion and stratification by the epithelium. The 3D tissue model thus developed is a useful tool to study M. tuberculosis infection in an environment that closely resembles a human lung. (B) Representative microscopic images of the different steps in the preparation of the tissue model. (C) Complete structure of the lung model tissue section. Scale – 100 µm. Please click here to view a larger version of this figure.
Protocol
Note: Human peripheral blood from healthy anonymous blood donors purchased at the blood bank of Linköping University Hospital, Sweden was used as the source of immune cells for this study. This protocol is designed for 24 mm 6-well plate inserts. Direct adaptation to other well formats is not recommended since the tissue model contracts both vertically and horizontally during development.
1. Preparation of Materials, Media and Culture of Bacteria/ Cell Lines
- Culture of bacteria:
- Grow the mycobacterial strain M. tuberculosis H37Rv carrying the pFPV2 plasmid to constitutively express green fluorescent protein (GFP), in Middlebrook 7H9 medium containing 0.05% Tween-80, 0.5% glycerol, kanamycin (20 µg/ml), and supplemented with Middlebrook albumin, dextrose and catalase enrichment (Middlebrook ADC Enrichment), at 37 °C with 5% CO2 for 7-10 days.
Note: All experimental steps involving live virulent M. tuberculosis strains should be performed in a BSL-3 facility.
- Grow the mycobacterial strain M. tuberculosis H37Rv carrying the pFPV2 plasmid to constitutively express green fluorescent protein (GFP), in Middlebrook 7H9 medium containing 0.05% Tween-80, 0.5% glycerol, kanamycin (20 µg/ml), and supplemented with Middlebrook albumin, dextrose and catalase enrichment (Middlebrook ADC Enrichment), at 37 °C with 5% CO2 for 7-10 days.
- Prepare 1x Dulbecco’s Modified Eagle’s Medium (DMEM) complete medium (supplemented with 1 mM sodium pyruvate, 2 mM L-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin, 10 mM HEPES, 0.1 mM non-essential amino acids and 10% heat-inactivated fetal bovine serum (FBS)). Prepare also antibiotic-free DMEM complete medium.
- Prepare 1x Minimum Essential Medium (MEM) complete medium (1 mM sodium pyruvate, 2 mM L-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin, 10 mM HEPES, 0.1 mM non-essential amino acids and 10% heat-inactivated fetal bovine serum (FBS)).
- Preparation of fibronectin/collagen-coated flasks (total 10 ml):
- Pipette 8.8 ml sterile 1x phosphate buffered saline (PBS) into a clean tube. Add 1 ml bovine serum albumin (1 mg/ml), 100 µl type I bovine collagen (3 mg/ml) and 100 µl recombinant human fibronectin (1 mg/ml).
- Mix the solution by turning the tube upside-down 5 times. Flasks are coated with a fibronectin/collagen solution (1 ml for a T-25 and 2 ml for a T-75 flask). Leave it O/N at 37 °C. After incubation, remove the solution and store the coated flasks at RT.
Note: The collected solution can be stored at 4 °C and reused for three times. Storage for more than 2 weeks may cause the liquid to turn brown (formation of crystals), where upon it must be discarded.
- Culture of fibroblasts:
- Grow and maintain MRC-5, (a human lung fibroblast cell line derived from normal lung tissue of a 14-week old male foetus), in complete DMEM in 5% CO2 at 37 °C. Use the fibroblasts at passages 24-26 and grow until 70-80% confluent.
Note: MRC-5 cell line at passage >30 tend to lose the morphology and is not recommended for use in the tissue model.
- Grow and maintain MRC-5, (a human lung fibroblast cell line derived from normal lung tissue of a 14-week old male foetus), in complete DMEM in 5% CO2 at 37 °C. Use the fibroblasts at passages 24-26 and grow until 70-80% confluent.
- Culture of epithelial cells:
- Obtain 16HBE14o- (16HBE), an immortalized human bronchial epithelial cell line that retains the differentiated morphology and function of normal human airway epithelia, (this was a gift from Dr. Dieter Gruenert, Mt. Zion Cancer Center, University of California, San Fransisco, USA 10. ). Culture 16HBE cells in fibronectin/collagen-coated flasks and maintain the cells in complete MEM in 5% CO2 at 37 °C.
- Preparation of 5x DMEM
- Prepare 5x DMEM by dissolving 13.4 g of DMEM powder and 3.7 g of sodium bicarbonate in 150 ml of sterile distilled water. Adjust the pH of the medium to 7.3, make up the volume to 200 ml and filter it using 0.22 µm membrane filter. Collect the filtered medium in a sterile container and stored until use at RT.
2. Preparation of Collagen-embedded Fibroblasts
- Thaw frozen aliquots of FBS and L-glutamine in a 37 °C water bath. After thawing keep the samples on ice. Place Sodium bicarbonate (71.2 mg/ml) and Gentamicin (50 mg/ml) at 4 °C. Pre-cool 50 ml centrifuge tubes and 10 ml sterile pipettes to 4 °C.
Note: All materials used (except the 5x DMEM) are chilled on ice prior to use and all steps are performed on ice. Type I Bovine Collagen (1.1 mg/ml) must be kept cold, as this prevents solidification of collagen. - Prepare the fibroblast cells:
- Warm Trypsin in a 37 °C water bath and incubate sufficient quantity with Lung fibroblasts (MRC-5) cells for 10 min at 5% CO2 at 37 °C. Neutralize the trypsin by adding 1x DMEM complete. Aspirate the cell suspension and centrifuge at 300 x g for 5 min. Aspirate the supernatant and resuspend the cells at 2.3 x 105 cells/ml in DMEM complete. Place the cells on ice until ready for use.
- Prepare the pre-mix:
- Add the following to a tube labeled “pre-mix”; 395 µl of 5x DMEM, 40 µl L-Glutamine, 120 µl NaHCO3 (71.2 mg/ml), 440 µl FBS , 5 µl Gentamicin (50 mg/ml), Total volume 1,000 µl, then swirl the pre-mix well and put on ice.
Note: The volumes given are for one 24 mm 6-well culture insert. Calculate the specific amounts required for the total number of inserts but add one extra to be sure, enough pre-mix is prepared.
- Add the following to a tube labeled “pre-mix”; 395 µl of 5x DMEM, 40 µl L-Glutamine, 120 µl NaHCO3 (71.2 mg/ml), 440 µl FBS , 5 µl Gentamicin (50 mg/ml), Total volume 1,000 µl, then swirl the pre-mix well and put on ice.
- Prepare the acellular collagen mixture:
- To each culture add 1 ml of acellular collagen mixture. Add the following to a 50-ml conical tube on ice in the given order; 686 µl of 1.1 mg/ml collagen, 250 µl Pre-mix and 64 µl 1x complete DMEM to a total volume of 1,000 µl. Mix the solution well ensuring no air bubbles. Work fast and add the collagen on the wall of the tube to avoid air bubbles.
- Add 1 ml of the acellular layer mixture to the insert placed in the 6 well plate. Do not add any medium to the well outside the insert. Incubate for 30 min in a 37 °C incubator. Make sure the acellular mixture covers the entire insert without any air bubbles.
- Prepare the cellular collagen mixture:
- Mix components of the cellular layer in a 50 ml conical tube kept on ice in the following order; 2 ml Collagen, 615 µl Pre-mix, 58 µl of 1x complete DMEM and 327 µl cell suspension Lung fibroblasts (MRC-5) to make up the total volume to 3,000 µl. Each culture requires 3 ml of cellular collagen mixture.
CRITICAL STEP: Make sure to mix the collagen and premix carefully before addition of the cell suspension. This will neutralize the pH of the collagen to avoid toxic effects on the fibroblasts. - Add the cellular layer (3 ml) on top of the acellular collagen layer and incubate for 2 hr in a 37 °C incubator. Work fast and add the collagen on the wall of the tube to avoid air bubbles.
- Following polymerization, add 2 ml of DMEM complete to the bottom of the 6 well plate (under the insert) and incubate for 24 hr.
NOTE: If polymerization did not occur, discard the plate inserts containing the fibroblast-collagen matrix and start-over again. The most likely cause is an error in addition or incorrect volume of one of the above listed reagents.
- Mix components of the cellular layer in a 50 ml conical tube kept on ice in the following order; 2 ml Collagen, 615 µl Pre-mix, 58 µl of 1x complete DMEM and 327 µl cell suspension Lung fibroblasts (MRC-5) to make up the total volume to 3,000 µl. Each culture requires 3 ml of cellular collagen mixture.
3. Continuous Culture of the Fibroblast-collagen Matrix
- Carefully lift the insert using a clean forceps and aspirate the culture media from the bottom of the well. Add 2 ml complete DMEM to the bottom of the well followed by 2 ml complete DMEM within the insert. Avoid introducing air bubbles under the insert, as this will prevent the diffusion of nutrients between the outer and inner chambers. Remove air bubbles with a micropipette tip.
- Change the culture media (inside and below the insert) every second day and culture for approximately 5-7 days. Take care in removing the media from the insert. To avoid contact with the fibroblast-collagen matrix, slightly tilt the insert using a clean forceps and aspirate the media from the insert walls.
CRITICAL STEP: The fibroblasts in the collagen matrix should get an elongated phenotype, and remodel the collagen, which then contracts. In about 5-7 days, the matrix has contracted to form a platform (10-14 mm in diameter) in the center of the insert. The contracted matrix is ready for use in the next step. To obtain a uniform contraction of the matrix it is critical that the fibroblasts are well mixed with the collagen prior to seeding (Step 2.6.1).
4. Seeding of Immune Cells (Infected/Uninfected Monocyte-macrophage Mixture)
Note: The following experimental steps involve virulent mycobacteria and therefore must be performed in a BSL-3 facility.
- Preparation of primary monocytes and macrophages:
- Isolate peripheral blood monocytes from donor blood using an established protocol. Isolate monocytes on the same day as setting up the tissue model and culture and differentiate them into macrophages for approximately 7 days before infection with M. tuberculosis.
NOTE: This will ensure both the macrophages and the contracted fibroblast-collagen matrix are available after 7 days. Also isolate fresh monocytes that will be added together with the infected macrophages.
- Isolate peripheral blood monocytes from donor blood using an established protocol. Isolate monocytes on the same day as setting up the tissue model and culture and differentiate them into macrophages for approximately 7 days before infection with M. tuberculosis.
- Preparation of M. tuberculosis -infected macrophages
- Harvest the cultured bacteria, wash with 1x PBS containing 0.05% Tween-80, resuspend in antibiotic-free complete DMEM, pass through a truncated sterile 27 G needle to disperse bacterial clumps and measure the optical density.
Note: Pre-determine the M. tuberculosis colony forming unit equivalents of optical density in the laboratory. This will give an estimate of the number of bacteria to be used for infection. - Incubate the macrophages for 4 hr with M. tuberculosis at the multiplicity of infection (MOI) 10). After infection, wash 3x with 1x PBS to remove the extracellular bacteria. Use uninfected macrophages cultured in the same way but without M. tuberculosis, as controls.
- Detach the macrophages from the culture plate by treatment with 2 mM EDTA for 10 min at 37 °C and resuspended the cells in antibiotic-free complete DMEM.
- Harvest the cultured bacteria, wash with 1x PBS containing 0.05% Tween-80, resuspend in antibiotic-free complete DMEM, pass through a truncated sterile 27 G needle to disperse bacterial clumps and measure the optical density.
- Labelling of monocytes
Note: Isolation and labeling of monocytes can be performed in the BSL-2 bench and then taken into BSL-3 facility for further processing.- Stain freshly prepared monocytes (2 x 107 cells) with a final concentration of 2 µM PKH26 red dye for 5 min, according to the manufacturer’s instructions. Wash 3x and resuspend the cells with antibiotic-free complete DMEM at a density of 1 x 107 cells/ml.
- Addition of immune cells to fibroblast-collagen matrix
- After 5-7 days of culture of the fibroblast-collagen matrix, aspirate the culture media from the outer and inner chambers and add 1.5 ml of fresh antibiotic-free DMEM complete to the outer chamber.
- Prepare a labeled monocyte-macrophage (uninfected/infected) mixture with a ratio of MO:MQ (5:1) in 50 µl DMEM complete. For 50,000 macrophages, take 250,000 labeled monocytes.
- Add 50 µl MO:MQ mixture to the fibroblast-collagen matrix and incubate for 1 hr in 5 % CO2 at 37 °C. After incubation, gently add 2 ml of culture media into the insert and incubate for an additional 24 hr in 5% CO2 at 37 °C.
Note: As cells added are loosely attached, addition of media should be slow by gently adding on the walls of the insert.
5. Seeding of Lung Epithelial Cells (16HBE)
Note: The following steps must be performed in a BSL-3 facility.
- Seed lung epithelial cells (16HBE) on top of the MO:MQ-fibroblast-collagen layer. To perform this, first dissociate 16HBE cells from the flask by treating with trypsin (as in 2.2.1.) and resuspend at 4 x 106 cells/ml in antibiotic-free DMEM.
- Aspirate the culture media inside and outside of the insert. Then add 1.5 ml antibiotic-free DMEM complete in the bottom of the well outside the insert.
- Add 50 µl of 16HBE on top of the immune cell-fibroblast collagen matrix. Leave for 2 min in the hood and incubate for 1 hr in 37 °C incubator with 5% CO2. After incubation, add gently 2 ml of antibiotic-free DMEM complete within the insert and culture at 37 °C for 3 days. The culture step facilitates proliferation of epithelial cells in the tissue model.
Note: As cells added are loosely attached, addition of media should be slow and gentle by sliding through the walls of the insert.
6. Air-exposure of the 3D Lung Model
Note: After day 5 post addition of infected macrophages, the tissue models are air-exposed and the following steps must be performed in a BSL-3 facility.
- Aspirate the culture media inside and outside of the insert.
Note: In this step supernatants can be collected for detection of secreted factors. Centrifuge, sterile-filter and store supernatants at -70 °C. - Add 1.8 ml antibiotic-free DMEM complete in the outer chamber and incubate in 5% CO2 at 37 °C incubator for 2 days. Do not add culture media within the insert.
NOTE: Air-lifting of the tissue model facilitates the formation of stratified epithelia and mucus secretion, which provides the strength to the tissue and physiological resemblance to human lung tissue.
7. Harvesting and Mounting the 3D Lung Tissue Model
Note: The following steps must be performed in a BSL-3 facility.
- On day 7 post implantation of infected macrophages, the tissue models are ready for harvesting. Remove the culture media entirely from the tissue model. Fix the tissue model with 4% paraformaldehyde for 30 min in the dark at RT. This step kills the bacteria and fixes the tissue/ cell morphology for further processing.
- Using a scalpel, separate the membrane from the well insert. Transfer the membrane containing the tissue to a well containing 1x PBS.
- Cut and remove the sides of the tissue model using a clean scalpel. Then slice the tissue model into 4 approximately equal square pieces. Transfer a piece of tissue onto a superfrost glass slide. Store the tissue pieces in 1x PBS at 4 °C.
- Dry the tissue for 5 min and mount using ProLong Gold antifade with DAPI and coverslip. Leave the slides without disturbing in the dark at RT until dry.
Note: The thickness of the tissue may vary between the center and periphery, causing slight tilting of the coverslip. To avoid this, a spacer (for instance parafilm) may be placed on the corner of the cover slip. - Apply nail polish to the edges of the coverslip and allow it to dry. Immerse the slides in 70% ethanol to make them safe to bring out of the BSL-3 facility.
8. Visualization, Acquisition and Quantitative 3D Analysis
- Visualize the tissue slides using a confocal microscope system with lasers emitting at 488 nm for excitation of GFP (green channel), 420 nm for DAPI (blue) and 555 nm for the PKH26-labeled monocytes (red) respectively.
- Acquire 3D images at a 512×512 resolution with Z-stacks covering at a minimum of 20 µm thickness and having 1 – 1.5 µm separation between stacks. Acquire 5-10 different fields covering the entire piece of tissue.
Note: Use the configuration such as Nyqvist for optimal settings of optical resolution (wavelength, laser power/exposure, pixel size and zoom). Avoid under or over saturation of pixels. - Analyze the confocal images with 3D image processing software. For 3D quantification of cell clusters, the following steps are recommended for optimal analysis.
- Open the 3D image processing software and load the image. Measure the dimensions of objects to be analysed in the image, for example, the size of a nucleus, individual monocyte and single bacteria. These observations are useful for defining or filtering the objects.
- Using a display adjustment tool, optimise volume rendering by adjusting each channel for image contrast, brightness and blend opacity. This step is to minimise the interference of noise in volume rendering.
Note: Gamma correction may lead to manipulation of images and thus should be avoided. - Create surfaces by choosing the red channel (monocytes) and set the threshold (automatic or manual selection). If required, use filters to restrict the selection of red monocytes or to exclude the background. Similarly, create surfaces for green (M. tuberculosis) and blue (nuclei) channels as described above.
- Export the data into Ms-Excel files. It is possible to export a particular parameter or all the data. The parameters that are relevant for analysis of cell clustering are volume, intensity, number of objects, number of voxels and sphericity.
- Save and export the images in a suitable picture format preferably TIFF.
NOTE: Animations can also be made using the animation menu and saved as a media file. - Save the analysis settings of each channel using the Add parameters option and can be retrieved later using the Rebuild function. Simultaneously analyse more files using the Batch processing tool.
Note: All the images to be compared must be acquired, processed and analyzed in the same manner. For instance a 8 bit image (256 integer) should not be compared with a 12 bit image (4096 integer).
Representative Results
A 3D lung tissue model for human TB can be effectively used to study the host-pathogen interactions in M. tuberculosis infection. The basic steps of this method, representative microscopic images of different steps and an overall microscopic structure of a tissue section are outlined in Figure 1. The model has several components of human lung tissue, including lung fibroblasts, bronchial epithelial cells and primary monocytes/macrophages embedded in the 3D tissue environment. Besides incorporating components of human lung tissue, the model resembles physiological conditions namely stratification of epithelia and mucus secretion.
An example for the use of the lung tissue model in monitoring a TB infection is presented in Figure 2. For visualizing the M. tuberculosis-immune cell migration and interaction, we introduced macrophages infected with M. tuberculosis that express GFP (green) together with the freshly isolated PKH26-labelled monocytes (red) into the tissue model (blue, DAPI stained for nuclei). On day 7 post addition of M. tuberculosis-infected cells to the tissue model, confocal microscopy reveals clustering of red monocytes at the site of infection (green) (Figure 2), which mimics the hallmark lesions of human TB 9.
A series of representative images for 3D visualization of M. tuberculosis-infected tissue model and quantification of cell clusters is shown in Figure 3. The 3D visualization gives the user flexibility to interact, examine and quantify several features in a 3D image. The spatial arrangement of green bacteria and red monocyte clusters can be seen from the apical, rotational and lateral view as illustrated in Figure 3B, which reveals clustering of monocytes at the site of M. tuberculosis. The clusters were not observed in uninfected tissues (Figure 3A). We quantified the size and number of monocyte cell clusters and found that the size (volume) of cell clusters is enhanced (p<0.001), while the number of individual monocytes decreased (p<0.01) in M. tuberculosis infected tissues as compared to uninfected tissue models (Figure 3C and 3D). This data validates our previous finding of early granuloma formation in M. tuberculosis infection observed in lung tissue models analyzed in 2D tissue sections 9.
Our data suggests that the tissue model provides a natural 3D habitat to investigate the complex host cell-M. tuberculosis communication network. We also found that 3D visualization and quantitative analysis are better tools for studying the features in the tissue model (Figure 3). Quantification of a cell cluster (granuloma for instance) often stretches to several cell layers and can be completely captured by a 3D quantitative analysis. Moreover, visualization of exact spatial and temporal features of individual cells or bacteria in the model allow live-imaging, migration and tracking studies in a designated laboratory.
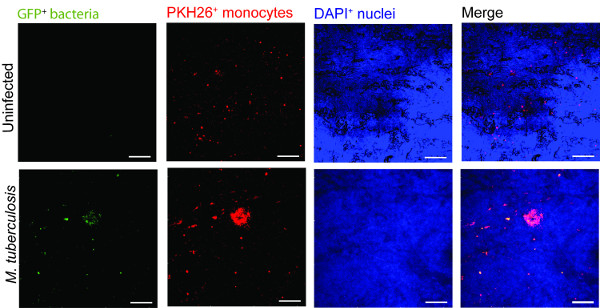
Figure 2. Monocyte in the tissue model cluster around virulent M. tuberculosis. Representative confocal images of uninfected and M. tuberculosis infected tissue model is presented. Panels from green (M. tuberculosis-GFP), red (PKH26-labelled monocytes), blue (DAPI-stained nuclei) and merged channels show the recruitment of monocytes in the infected tissue as compared to uninfected tissues. Scale – 100 µm. Please click here to view a larger version of this figure.
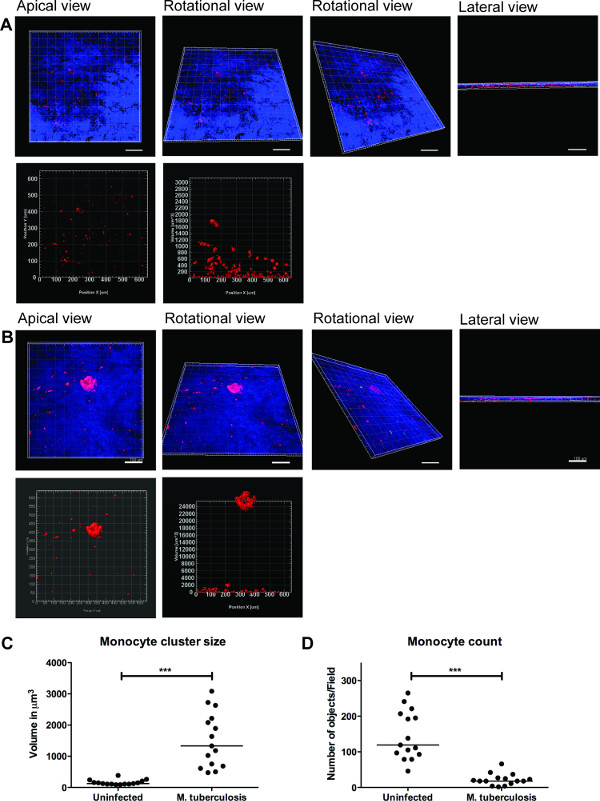
Figure 3. 3D visualization and quantitative analysis of tissue model provide useful information. Representative images of 3D visualization of the entire tissue model (A) uninfected tissue, (B) infected with M. tuberculosis, through optical sectioning using Zeiss LSM700 confocal microscope and quantitative analysis by Imaris image processing software (version 7.6.8). These images were acquired at 20X magnification, 14 z-stacks covering a tissue thickness of 19.5 µm with 1.5 µm interval, allowing the visualization from apical, rotational horizontal, rotational vertical and lateral view (A and B). (C) Quantitative analysis of monocyte cell clusters reveal enhanced (p<0.0001) size of early granuloma clusters after M. tuberculosis infection when compared to absence of infection. (D) Quantification of number of monocytes showed a decline (p<0.01) in infected tissue as compared to uninfected tissue, reiterating more clusters in the infected tissue. Green –M. tuberculosis-GFP, Red – PKH26-labelled monocytes, Blue – cell nuclei, Scale – 100 µm. Please click here to view a larger version of this figure.
Discussion
The ability to recruit and form organized cell clusters at the site of infection is the hallmark of human TB 11. These dynamic structures known as tubercle granulomas primarily consist of immune cells (macrophages, monocytes, T-cells and B-cells) and multi-nucleated giant cells surrounding M. tuberculosis. The role of the granuloma has long been considered to wall off the infection, preventing local spread of bacteria. However, more recent studies show that granuloma formation is critical for early bacterial survival, growth and dissemination 12. A strategy of new studies is to identify molecules or pathways that could efficiently be targeted to inhibit the cellular migration in granuloma formation and/or TB dissemination.
A caveat for novel studies on TB is the lack of models that recapitulate human TB. The most widely used experimental animals do not form true granuloma upon M. tuberculosis infection, and are therefore not appropriate choices for studies of TB 13-16. Non-human primates have the closest resemblance to human TB 17, but are not the preferred choice owing to high operational costs and ethical issues. Human TB is a complex immunological process and is difficult to model in vitro. Cell cultures of monolayers or co-cultures lack the 3D environment and tissue responses. Therefore, we have developed an innovative lung tissue model based on human primary immune cells and human lung-specific cell lines 8,9. The model displays characteristic features of human lung tissue, including epithelia with evenly integrated macrophages, formation of extracellular matrix, stratified epithelia and mucus secretion 9.
The 3D human lung tissue model has several benefits over the in vitro single or co-cultures seeded on tissue culture plates or transwell inserts. First, the human lung-specific cells (fibroblasts and epithelial cells) are not commonly included in the in vitro single or co-cultures. Second, the immune cells and lung-specific cells are embedded in a 3D physiological context (collagen rich extra-cellular matrix products). The response of cells to a stimulus/infection and the migratory behaviour of cells, for instance formation of a granuloma, differ significantly between a 2D and 3D environment. Furthermore, the described method enables the 3D visualization and robust 3D quantitative analysis that provides pivotal information on spatial distribution and intricate cellular interactions.
Experimental infection in the model tissue with M. tuberculosis resulted in clustering of macrophages at the site of infection, reminiscent of early TB granuloma (Figure 2 and 3). We have recently demonstrated that mutant strains defective in the ability to secrete the virulence factor ESAT-6 or Mycobacterium bovis BCG that lacks ESAT-6 did not induce the clustering of monocytes (no early granuloma), in contrast to the virulent M. tuberculosis 9. These data are consistent with the observations made from Mycobacterium marinum-infected zebrafish embryos, whose transparency allows for elegant live imaging of granuloma formation 12. As there is no gold-standard model for TB, we took advantage of the surgically resected tissue biopsies from TB patients for validation of the method 9. Our in vitro tissue model shares several characteristics with the lung and lymph node biopsies from TB patients, including the aggregation of macrophages in granuloma, the presence of both intra- and extracellular bacteria 18 and induction of necrosis 11.
Although the described model has physiological relevance to human TB and has several advantages over other in vitro models, it has some limitations. For instance, out of more than 20 collagen proteins identified in humans, only type I is included to the model to mimic the extra-cellular matrix. However, type I collagen is a complex mixture of extra-cellular matrix products and is the most abundant collagen in the human body. Further, we have demonstrated the presence of collagen IV and several extra-cellular matrix proteins such as tropoelastin, vimentin and laminin, which are produced by the epithelial cells and fibroblasts in the tissue model, indicating the synthesis of new collagen 8. Presently, the lung tissue model only has monocytes and macrophages, besides lung-specific cells. It lacks neutrophils and lymphocytes that are also known to be present in the granuloma. Remarkably the model is not limited to the introduction of additional immune cells and is of interest to explore how they contribute to the complex cellular interactions in human TB. Implantation of primary alveolar macrophages, skin-specific cells and lung carcinoma cells has already been tested in the model. Since our objective was to use a model that closely resembles human TB, introduction of mouse cells have not been attempted.
In summary, the lung tissue model has implications for both basic mechanistic and applied studies. Potential applications of the lung model include the study of innate immunity, investigating mechanistic aspects of host defences such as phagosomal maturation, autophagy, production of cytokines, chemokines and anti-microbial peptides, and functional characterization of individual cell types. Strikingly, the in vitro tissue model allows manipulation of one or more cells types and provides a relevant tissue micro-environment, not only for studies on TB, but for a variety of infectious and non-infectious diseases that affect the lungs.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors acknowledge the Microscopy core facility at the Faculty of Health Sciences, Linköping University for providing access to advanced imaging systems; Karl-Eric Magnusson (Emeritus Scientist) at the Dept. of Clinical and Experimental Medicine, Linköping University for providing access to Imaris 3D/4D image processing software (Bitplane, Switzerland); and S. Braian for his help with the lung model cartoon. This work was supported by funds from the Swedish Research Council (Alternatives to animal research, 2012-1951) and Swedish Research Council (2012-3349) to M.L. and Swedish Foundation for Strategic Research to S.B. S.B. receive grants from the Karolinska Institutet, Swedish Research Council, the Swedish International Development Cooperation Agency (Sida) and the Swedish Civil Contingencies Agency (MSB), and the Swedish Heart and Lung Foundation (HLF). M.S. received grants from the Karolinska Institutet and Stockholm County Council.
Materials
| Cell culture inserts | BD Falcon | 353092 | |
| 6-well culture plates | BD Falcon | 353046 | |
| MRC-5 cells, lung fibroblasts | ATCC#CCL-171 | ||
| 16HBE cells, lung epithelial cells | Gift from Dr. Dieter Gruenert, Mt. Zion Cancer Center, University of California, San Fransisco, USA | ||
| 5 x Dulbecco’s modified Eagle’s medium (5 x DMEM) | Gibco | 12800-082 | Made from powder but add 5 times less water. Adjust pH to 7.3 and filter it using a 0.2 µm filter. |
| Dulbecco’s modified Eagle’s medium with glucose (DMEM) 1x | Gibco | 41965-039 | |
| Minimum Essential Medium (MEM) 1x with Earle’s salts | Sigma | M4655 | |
| Non-Essential Amino Acids Solution, 100x | Life Technologies | 11140-035 | |
| L-glutamine 200 mM (100x) | Gibco | 25030-024 | |
| Sodium Pyruvate | Life Technologies | 11360-039 | |
| NaHCO3 (71.2 mg/ml) | Prepared in house | ||
| Heat inactivated Fetal Bovine Serum (FBS) | Gibco | 10270-106 | Heat inactivated for 30 min, 56 °C |
| Gentamicin (50 mg/ml) | Gibco | 15750-060 | |
| Hepes buffer solution 1M | Gibco | 15630-056 | |
| Penicillin Streptomycin (Pen Strep) | Gibco | 15140-122 | |
| Lymphoprep | Axis-Shield | 7801 | |
| Ultrapure 0.5 M EDTA | Gibco | 15575 | |
| Bovine Collagen PA treated (500 ml) | Organogenesis | 200-055 | |
| Pure col purified Bovine Collagen solution (100 ml) | Advanced biomatrix | 5005-B | |
| Extracellular matrix protein, Fibronectin (1 mg) | BD | 354008 | |
| Primary human monocytes/macrophages | Isolated from human whole blood or buffy coats. | ||
| PKH26 Red fluorescent cell linker | Sigma | MINI26 | |
| Mycobacterium tuberculosis H37Rv expressing green fluorescent protein | M. tuberculosis H37Rv wild type was transformed with the pFPV2 plasmid constitutively expressing GFP. | ||
| Middlebrook 7H9 medium | Difco | 271310 | |
| BBL Middlebrook ADC Enrichment | BBL | 211887 | |
| Tween-80 | |||
| Glycerol | |||
| Kanamycin B sulfate (20 µg/ml) | Sigma | B5264 | |
| Prolong Gold anti=-fade reagent with DAPI | Invitrogen | P36935 | |
| Trypsin -EDTA | |||
| Bovine serum albumin | |||
| Paraformaldehyde | |||
| DAPI | |||
| LSM700 Confocal microscope | Zeiss | ||
| iMaris Scientific 3D/4D image processing software, version 7.6.8 | Bitplane AG |
References
- Sakamoto, K. The pathology of Mycobacterium tuberculosis infection. Veterinary pathology. 49, 423-439 (2012).
- Saunders, B. M., Britton, W. J. Life and death in the granuloma: immunopathology of tuberculosis. Immunol Cell Biol. 85, 103-111 (2007).
- Kaufmann, S. H. New issues in tuberculosis. Ann Rheum Dis. Ann Rheum Dis. 63, ii50-ii56 (2004).
- Morrison, J., Pai, M., Hopewell, P. C. Tuberculosis and latent tuberculosis infection in close contacts of people with pulmonary tuberculosis in low-income and middle-income countries: a systematic review and meta-analysis. Lancet Infect Dis. 8, 359-368 (2008).
- Barry 3rd, C. E., et al. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat Rev Microbiol. 7, 845-855 (2009).
- Puissegur, M. P., et al. An in vitro dual model of mycobacterial granulomas to investigate the molecular interactions between mycobacteria and human host cells. Cell Microbiol. 6, 423-433 (2004).
- Kapoor, N., et al. Human Granuloma In Vitro Model, for TB Dormancy and Resuscitation. PLoS One. 8, e53657 (2013).
- Nguyen Hoang, ., T, A., et al. Dendritic cell functional properties in a three-dimensional tissue model of human lung mucosa. Am J Physiol Lung Cell Mol Physiol. 302, L226-L237 (2012).
- Parasa, V. R., et al. Modeling Mycobacterium tuberculosis early granuloma formation in experimental human lung tissue. Dis Model Mech. 7, 281-288 (2014).
- Cozens, A. L., et al. CFTR expression and chloride secretion in polarized immortal human bronchial epithelial cells. Am J Respir Cell Mol Biol. 10, 38-47 (1994).
- Brighenti, S., Andersson, J. Local immune responses in human tuberculosis: learning from the site of infection. J Infect Dis. 205, S316-S324 (2012).
- Davis, J. M., Ramakrishnan, L. The role of the granuloma in expansion and dissemination of early tuberculous infection. Cell. 136, 37-49 (2009).
- Gupta, U. D., Katoch, V. M. Animal models of tuberculosis. Tuberculosis (Edinb). 85, 277-293 (2005).
- Kashino, S. S., Napolitano, D. R., Skobe, Z., Campos-Neto, A. Guinea pig model of Mycobacterium tuberculosis latent/dormant infection. Microbes Infect. 10, 1469-1476 (2008).
- Singhal, A., et al. Experimental tuberculosis in the Wistar rat: a model for protective immunity and control of infection. PLoS One. 6, e18632 (2011).
- Subbian, S., et al. Phosphodiesterase-4 inhibition alters gene expression and improves isoniazid-mediated clearance of Mycobacterium tuberculosis in rabbit lungs. PLoS Pathog. 7, e1002262 (2011).
- Lin, P. L., et al. Tumor necrosis factor neutralization results in disseminated disease in acute and latent Mycobacterium tuberculosis infection with normal granuloma structure in a cynomolgus macaque model. Arthritis Rheum. 62, 340-350 (2010).
- Rahman, S., et al. Compartmentalization of immune responses in human tuberculosis: few CD8+ effector T cells but elevated levels of FoxP3+ regulatory t cells in the granulomatous lesions. Am J Pathol. 174, 2211-2224 (2009).

