Novel RNA-Binding Proteins Isolation by the RaPID Methodology
Summary
RNA-protein interactions lie at the heart of many cellular processes. Here, we describe an in vivo method to isolate specific RNA and identify novel proteins that are associated with it. This could shed new light on how RNAs are regulated in the cell.
Abstract
RNA-binding proteins (RBPs) play important roles in every aspect of RNA metabolism and regulation. Their identification is a major challenge in modern biology. Only a few in vitro and in vivo methods enable the identification of RBPs associated with a particular target mRNA. However, their main limitations are the identification of RBPs in a non-cellular environment (in vitro) or the low efficiency isolation of RNA of interest (in vivo). An RNA-binding protein purification and identification (RaPID) methodology was designed to overcome these limitations in yeast and enable efficient isolation of proteins that are associated in vivo. To achieve this, the RNA of interest is tagged with MS2 loops, and co-expressed with a fusion protein of an MS2-binding protein and a streptavidin-binding protein (SBP). Cells are then subjected to crosslinking and lysed, and complexes are isolated through streptavidin beads. The proteins that co-purify with the tagged RNA can then be determined by mass spectrometry. We recently used this protocol to identify novel proteins associated with the ER-associated PMP1 mRNA. Here, we provide a detailed protocol of RaPID, and discuss some of its limitations and advantages.
Introduction
RNA-binding proteins (RBPs) represent about 10% of S. cerevisiae proteins1,2 and about 15% of mammalian proteins3-5. They are implicated in many cellular processes such as mRNA post-transcriptional processing and regulation, translation, ribosome biogenesis, tRNA aminoacylation and modification, chromatin remodeling, and more. An important subgroup of RBPs is the mRNA-binding proteins (mRNPs)6,7. In the course of mRNA maturation, different RBPs bind the transcript and mediate its nuclear processing, export out of the nucleus, cellular localization, translation and degradation6-8. Thus, the distinct set of RBPs bound to a particular transcript at any time point determines its processing and ultimately its fate.
The identification of RBPs associated with an mRNA could significantly improve our understanding of processes underlying their post-transcriptional regulation. Diverse genetic, microscopic, biochemical and bioinformatics methods have been used to identify proteins involved in mRNA regulation (reviewed in9-11). However, only a few of these methods enable the identification of proteins associated with a particular target mRNA. Of note is the Yeast Three Hybrid system (Y3H), which utilizes the mRNA of interest as bait to screen an expression library in yeast cells. Positive clones are usually observed through a growth selection or reporter expression12-14. The key advantage of this method is the large number of proteins that can be scanned in a cellular environment and the ability to measure the strength of the RNA-protein interaction. Drawbacks include the relatively large number of false positive results due to non-specific binding, and the high potential for false negative results due, in part, to misfolding of the fusion protein prey or the bait RNA.
An alternative to the genetic approach is affinity purification of RNA with its associated proteins. Poly A-containing mRNAs can be isolated through the use of oligo dT columns, and their associated proteins are detected by mass spectrometry. The RNA-protein interaction is conserved in its cellular context by crosslinking, which makes short-range covalent bonds. The use of the oligo dT column yields a global view of the entire proteome that is associated with any poly A-containing mRNA3,5,15. However, this does not provide a list of proteins that are associated with a particular mRNA. Very few methods are available to accomplish such an identification. The PAIR method entails the transfection of nucleic acid with complementarity to the target mRNA16,17. The nucleic acid is also attached to a peptide, which allows crosslinking to RBPs in close vicinity to the interaction site. After crosslinking, the RBP-peptide-nucleic acid can be isolated and subjected to proteomics analysis. Recently, an aptamer-based methodology was successfully applied to extracts from mammalian cell lines18. An RNA aptamer with improved affinity to streptavidin was developed and fused to a sequence of interest (AU-rich element (ARE) in this case). The aptamer-ARE RNA was attached to streptavidin beads and mixed with cell lysate. Proteins that associated with the ARE sequence were purified and identified by mass spectrometry (MS). Although this method detected associations that occur outside the cellular settings (i.e., in vitro), it is likely to be modified in the future so as to introduce the aptamers into the genome and thus enable the isolation of proteins associated with the mRNA while in the cellular milieu (i.e., in vivo). In yeast, where genetic manipulations are well established, the RaPID method (developed in Prof. Jeff Gerst's lab) provides a view of in vivo associations19. RaPID combines the specific and strong binding of the MS2 coat protein (MS2-CP) to the MS2 RNA sequence, and of the streptavidin-binding domain (SBP) to streptavidin conjugated beads. This enables efficient purification of MS2-tagged mRNAs through streptavidin beads. Moreover, expression of 12 copies of MS2 loops allows up to six MS2-CPs to bind simultaneously to the RNA and increase the efficiency of its isolation. This protocol was therefore suggested to enable the identification of novel mRNA-associated proteins once the eluted samples are subjected to proteomics analysis by mass spectrometry.
We recently utilized RaPID to identify novel proteins associated with the yeast PMP1 mRNA20. PMP1 mRNA was previously shown to be associated with the ER membrane and its 3' untranslated region (UTR) was found to be a major determinant in this association21. Thus, RBPs that bind PMP1 3' UTR are likely to play an important role in its localization. RaPID followed by liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS) resulted in the identification of many new proteins that interact with PMP120. Herein, we provide a detailed protocol of the RaPID methodology, important controls that need to be done, and technical tips that may improve yield and specificity.
Protocol
Note: Insert a sequence consisting of 12 MS2-binding sites (MS2 loops; MS2L) into the desired genomic locus, usually between the open reading frame (ORF) and the 3' UTR. A detailed protocol for this integration is provided elsewhere22. Verify proper insertion and expression by PCR, northern analysis or RT-PCR20,23. It is important to verify that the integration did not intervene with the synthesis of the 3'UTR. In addition, a plasmid-expressing MS2-CP fused to SBP under the expression of an inducible promoter (methionine depletion) should also be introduced into the cells24. An identical strain, excluding the introduced MS12 loops, should be used as a control for the detection of non-specific signals.
1. RaPID Purification
- Grow 500 ml of yeast cells (use 250-1,000 ml of yeast cells depending on the expression level of the gene of interest) expressing MS2-tagged mRNA20 and MS2-CP-GFP-SBP fusion protein24 to OD600 0.8 – 1.0 at 30 oC in the appropriate growth medium (e.g., Synthetic Dextrose (SD) selective medium).
- Centrifuge cells at 3,000 x g for 4 min at RT and discard the supernatant.
- Wash cells in 1x phosphate buffered saline (PBS), centrifuge as before (step 1.2), resuspend in an equal volume (step 1.1) of SD without methionine, and incubate for 45 – 60 min at 30 oC to induce expression of the MS2-CP-GFP-SBP fusion protein.
Note: Longer induction time may cause aggregation of GFP. - Set aside 10 ml of cells and extract RNA from this sample using the simple "hot phenol" method25. Caution: Phenol is highly toxic and should be handled according to its safety instructions.
Note: This RNA is a good reference for the quality of isolated RNA (Figure 1A). - For 500 ml cells, add 1.35 ml 37% formaldehyde to a final concentration of 0.1% to crosslink the RNA-protein complexes. Continue incubation at 30 oC for 10 min. Caution: Formaldehyde is highly toxic and should be handled according to its safety instructions.
- Add 26.5 ml of 2.5 M glycine to a final concentration of 0.125 M and incubate for 3 min to stop crosslinking. From this step on, put everything on ice to minimize RNA degradation.
- Centrifuge cells at 3,000 x g for 4 min at 4oC and wash with 45 ml cold 1x PBS.
- Resuspend cells in 1 ml RaPID Lysis buffer per 100 ml of initial cell culture.
- Split into aliquots of 500 µl in screw-capped microfuge tubes and add ~ 400 mg of chilled glass beads to each aliquot (roughly a full 0.2 ml PCR tube).
- Lyse cells in a bead beater for 3 min. Immediately put the lysate on ice.
Note: Cell lysis releases cellular RNases, which are the major cause of RNA degradation. RaPID Lysis buffer contains a high concentration of RNase inhibitors, such as the nonspecific RNase inhibitor heparin and specific inhibitors. Working quickly and keeping everything cold is also recommended. - Transfer the lysate to new tubes. Pierce a small hole in the bottom of each microfuge tube with a hot needle (0.8 mm x 40 mm) and place the microfuge tubes on top of the 15 ml tubes. Use the head of a 5 ml syringe as an adaptor between the microfuge tubes and the 15 ml tubes. Centrifuge the tubes assembly at 3,000 x g for 1 min at 4 oC.
- Transfer the flow-through from the 15 ml tube to a new 1.5 ml tube and centrifuge at 10,000 x g for 10 min at 4 oC to clear cell debris.
- Pool supernatants from all 1.5 ml tubes into a single 15 ml tube, and set aside 1/50 and 1/100 volume of the lysate for RNA and protein isolation, respectively.
Note: These will serve as the "Input" samples for subsequent analysis (Figure 1). - Add 300 µg of avidin per 500 ml of initial yeast culture and incubate for 30 min (this can be reduced to 10 min) at 4 oC under constant rolling (10 rpm).
Note: Avidin blocks biotin and biotinylated proteins from binding to the streptavidin beads. - Pre-wash the streptavidin beads before incubation with the cell lysate.
- Transfer about 300 µl of the streptavidin beads slurry to a 1.5 ml microcentrifuge tube and remove the upper supernatant. This will result in 250 µl of beads. Wash the beads twice with 1 ml of RaPID lysis buffer. Centrifuge the streptavidin beads at 660 x g for 2 min at 4oC between each step.
- Block the beads by incubating for 1 hr with 0.5 ml of RaPID lysis buffer, 0.5 ml of 10 mg/ml BSA and 10 µl of 10 mg/ml yeast tRNA. Beads can be left O/N in blocking solution at 4oC with rotation if necessary.
- Wash twice with 1 ml of RaPID Lysis buffer.
Note: Magnetic beads can also be used to reduce time and background. These, however, have lower capacity than sepharose beads.
- Add 250 µl of the pre-washed streptavidin beads to the avidin-containing lysate (from step 1.14) and incubate 1 hr at 4 °C with constant rotation (10 rpm).
Note: This incubation time is a compromise between providing sufficient binding time and minimizing chances for RNA degradation. - Centrifuge at 660 x g for 2 min at 4 oC to clear the streptavidin beads and transfer the supernatant to a new 15 ml tube. Set aside 1/50 and 1/100 volume of the lysate for RNA and protein isolation, respectively.
Note: These will be the "Unbound" samples. - Wash the beads with 1 ml of RaPID Lysis buffer, shake gently, transfer to a new 1.5-ml microcentrifuge tube, and centrifuge as in step 1.17. Repeat this step three times.
- Wash the beads with 1 ml of RaPID Wash buffer, and this time rotate (10 rpm) for 5 min at 4 °C. Repeat this step twice.
- Wash the beads with 1 ml of 1x PBS and centrifuge as above.
- Wash the beads again with 120 µl of 1x PBS. Centrifuge as above, and take the entire supernatant for RNA and protein extraction as the "Wash" sample.
Note: This last wash sample will indicate the stringency of the washes and should be of the same volume as the Elution sample. This will simplify processing and loading in subsequent assays. - To elute RNA and RNA-associated proteins from the column, add 120 µl of 6 mM biotin in 1x PBS and incubate for 45 min at 4 °C with constant rotation (10 rpm).
- Centrifuge the beads as above and transfer the eluted material to a new 1.5 ml tube. Spin the eluted sample again and transfer the upper phase to a new 1.5 ml tube to ensure no carryover of beads.
- Take samples for RNA or protein extraction.
Note: The volume taken should depend on the following assay and expression level of the RNA or protein of interest. As a guideline, for northern analysis26, take 80% of the sample, for Western blot23,27 or RT-PCR28, take 20%, and for mass spectrometry29 to discover new proteins, take the entire sample.
2. RNA Extraction
- Add an equal volume of 2x Crosslinking Reversal Buffer and incubate for 45 min at 65 °C. Continue directly for RNA extraction.
Note: The Crosslinking Reversal Buffer contains ethylenediaminetetraacetic acid (EDTA), which significantly improves RNA extraction20. - Use the standard phenol:chloroform method26 to extract RNA from the "input" and "unbound" samples (steps 1.13 and 1.17).
Note: These samples contain a large amount of the RNase inhibitor heparin, which can inhibit subsequent Reverse Transcriptase-(RT)utilizing assays (e.g., RT-PCR), therefore, LiCl precipitation30 is advised to remove it. The "wash" and "elution" samples (steps 1.21 and 1.24) contain low amounts of RNA and are precipitated through the following steps. - Add an equal volume of 8M Guanidinium-HCl (GuHCl) and two volumes of 100% ethanol to the "wash" and "elution" samples. Incubate at -80 °C for at least 2 hr.
Note: Guanidinium will efficiently denature proteins and thereby inhibit RNases and proteases in the sample. - Centrifuge at 20,000 x g, 4 °C for 30 min and discard the supernatant. Wash the pellet with cold 80% ethanol and centrifuge again at 20,000 x g, 4 °C for 15 min.
- Dissolve the RNA pellet in 400 µl of RNase free water and precipitate again to remove any leftover GuHCl or other contaminants. Add 40 µl of 3 M sodium acetate, pH 5.2 and 800 µl of 100% ethanol, incubate at -20 °C for at least 1 hr.
- Centrifuge and wash as in step 2.4 and remove all ethanol with a 10 µl tip. Resuspend the RNA pellet in 10 µl RNase free water. Take extra care as the RNA pellet is usually not visible.
Note: Samples can be used for northern analysis as described in23 (Figure 1).
3. Protein Preparation for Western Blot or Mass Spectrometry Analysis
- Add 1/3 volume of 4x Laemmli Sample Buffer (LSB) to protein samples and incubate 45 min at 65 °C to reverse the crosslinking of RNA and proteins.
Note: Samples can be stored at -20 °C. - Run proteins on a SDS polyacrylamide gel (SDS-PAGE)31. Adjust the percentage of the gel according to the size of proteins to be analyzed.
Note: 10% acrylamide gel gives a wide range of separation and is good as a first approximation. - Transfer proteins to the membrane as in the standard immunoblot analysis27 for control of isolation efficiency (Figure 1C). Stain with either Coomassie Blue R250 or silver stain before mass spectrometry analysis.
Note: Silver stain is the preferred choice because it is more sensitive and allows better detection (Figure 1B). Staining can be done with either manually prepared solutions or commercial kits. The incubation time of the gel in the final dye solution should be determined experimentally. Long incubation may reveal low abundant proteins, but can cause highly expressed proteins such as MS2-CP-SBP-GFP to be over stained and may mask surrounding proteins in their vicinity. Additionally, a high background might occur. Follow the reaction carefully, and add Stop Solution at the appropriate time. - Cut out bands from the gel and transfer them to 1.5 ml tubes. Store samples at -20 °C, or send for protein extraction and trypsin digestion followed by LC-MS/MS analysis32.
- As an alternative proteomic approach to exclude the gel separation step, digest eluted material from step 1.24 in solution using trypsin and subject to LC-MS/MS analysis33.
Note: Omitting the SDS-PAGE separation simplifies the protocol and increases yield. However, samples might contain traces of the detergent NP-40, which inhibits mass spectrometry analysis. To overcome this problem, perform the following steps:- Run protein on SDS-PAGE31 for 10 – 15 min.
- Cut out the entire portion of the lane (i.e., where the proteins are located).
Note: The protein sample might include a significant amount of the MS2-CP-SBP protein that may obscure signals of low abundance proteins.
Representative Results
RaPID enables the isolation of a specific target RNA with its associated proteins. Critical for its success is keeping the RNA intact as much as possible, thereby obtaining a sufficient amount of proteins. To determine the isolation efficiency and quality of RNA, northern analysis is performed (Figure 1A). Northern analysis has the advantage of directly reporting the efficiency and quality of RaPID. Thus, the relative amounts of full length and degradation products can be determined in a single run. Ribosomal RNAs (rRNAs) are detected easily in the ethidium bromide staining, and the lack of rRNAs in the elution sample indicates the stringency of purification. The stringency and specificity of the purification are further demonstrated by the lack of a signal of an untagged mRNA in the elution samples (ACT1 bottom panel). The apparent signal in the FPR1 lane, which is not of the size of ACT1, is a leftover from previous hybridization with the MS2L probe. The northern analysis also reveals that the input RNA is somewhat more degraded compared to RNA that was purified by the hot phenol protocol (c/o ACT1 probe panel). This is attributed to the lengthier and more complicated RaPID protocol. Nevertheless, a significant amount of full-length, tagged RNA is isolated specifically, as revealed by the strong signal in the elution fraction (MS2L probe).
Protein samples can be run on SDS-PAGE and stained by silver stain prior to proteomics analysis (Figure 1B). A sample isolated from control, untagged cells (-MS2L) is helpful in distinguishing non-specific associations from RNA-dependent ones. Therefore, only bands that are stronger in the tagged sample (+MS2L) are cut out of the gel and taken for LC-MS/MS. Western analysis with general RBPs is also recommended to indicate the efficiency of protein co-purification (Figure 1C). Herein, we used Yef3, which is known to interact with PMP120, or GFP, which indicates the overall efficiency and specificity of the isolation. Interestingly, the efficiency of Yef3 isolation appears to differ between PMP1 and FPR1, and is much lower compared to GFP.
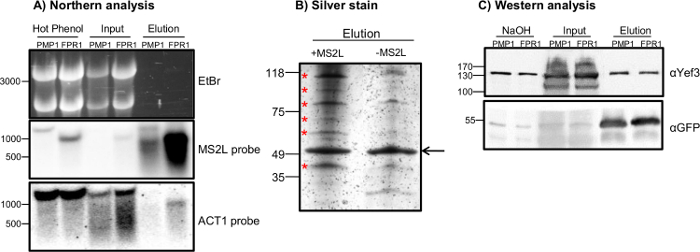
Figure 1. Specific Isolation of MS2L-tagged RNAs and Identification of Associated Proteins. Results of two different RNAs that were subjected to RaPID (PMP1 and FPR1) are presented. (A) Northern blot analysis of RNA samples purified from a RaPID experiment. RNA was run on an agarose gel and stained with ethidium bromide (upper panel). The gel was blotted to nylon membrane and subjected to hybridization with the indicated probes (lower panels). The samples analyzed are: fast-cell lysis (hot phenol [step 1.4]), RaPID cell lysis (input [step 1.13]) and after elution with biotin (elution [step 1.24]). (B) Silver stain of proteins from RaPID with MS2-tagged and untagged cells. Protein samples from elution fractions of MS2-tagged PMP1 (+MS2L) or untagged (-MS2L) control were run in SDS-PAGE and silver stained. Bands with differential intensity (indicated by asterisks) were cut out of the gel and determined by mass spectrometry analysis. The arrow indicates the MS2-CP-GFP-SBP fusion protein, which demonstrates equal protein loading. Irrelevant lanes were cropped out for clarity. (C) Western analysis of positive controls. Western blot with antibodies that recognize the Yef3 or GFP moiety of the MS2-CP fusion protein was conducted. Irrelevant lanes were cropped out for clarity. Please click here to view a larger version of this figure.
Discussion
Various methods use the isolation of specific mRNAs to identify their associated proteins11,34 35. These methods apply in vitro and in vivo strategies to probe RNA-protein interactions. In vitro methods incubate exogenously transcribed RNA with cell lysate to capture RBPs and isolate RNP complexes36,37. An effective approach of this type was presented recently, which enabled the identification of novel proteins that bind a regulatory RNA motif18. A drawback of these methods is the binding of non-specific RBPs because the association occurs outside of the cellular environment. In vivo methods, in which proteins are crosslinked while in their natural settings, may provide a better view of RNA-protein association. Furthermore, crosslinking allows higher stringency during isolation and therefore higher specificity. The PAIR approach17 utilizes the transfection of the antisense sequence to the target RNA in order to direct a peptide moiety to the vicinity of RBPs. The peptide-RBPs can then be crosslinked and isolated through beads that are linked to a sequence that is complementary to the antisense sequence. The PAIR methodology is very advantageous due to its simple application to many cell types without the need for complex genomic manipulations. It is, however, dependent on the efficiency of transfection, and cases where a low percentage of cells accept the antisense will have low efficacy. Furthermore, the isolation of the RBP-peptide-antisense sequence is obtained through magnetic streptavidin beads that are coupled with biotinylated oligonucleotide complementary to the antisense nucleic acid. The multiplicity of interactions (magnetic beads, streptavidin-biotin and base pairing) may limit the stringency and efficiency of the isolation. The RaPID method24 described here provides an alternative to these limitations in several aspects. First, the integration of the MS2 loops into the genomic loci ensures that all cells express it, thus increasing its yield. Second, it uses the high affinity interaction of the MS2-CP-GFP-SBP to twelve MS2 loop sequences and allows them to bind in vivo. Third, the fixation of the RNA-protein interaction enables more stringent washes and reduces the identification of non-specifically bound proteins. Finally, the SBP domain binds streptavidin beads with high affinity and is easily eluted with free biotin.
Insertion of the MS2 loop into the RNA is advisable between the ORF and the 3' UTR to minimize translation perturbation and to ensure expression of all loops. Six to 24 MS2 loop repeats were used previously for the visualization of mRNA localization38-40. However, the insertion of a long sequence of MS2 loops may cause changes in the processing and structure of the RNA; it might destabilize the transcript or change the repertoire of RBPs bound to it. In our experience, 12 loops are sufficient to get an efficient elution of MS2-tagged RNA20. In this respect, we note that we usually observed additional shorter transcripts in our northern hybridizations. Northern analyses with different probes revealed that these shorter forms included the MS2L and part of the 3' UTR20, indicative of premature transcription termination. Such cases are likely to reduce the efficiency of the isolation of proteins associated with the 3' UTR. Therefore, the number of inserted loops and their location within the RNA should be balanced between high efficiency pull-down and transcript stability.
The RNA has to remain intact throughout the protocol to enable efficient isolation and detection of the maximal array of bound proteins. Chances for external RNase contamination can be reduced by wearing gloves, ensuring a clean work area, preparing solution with RNase-free water, and working concisely and efficiently to reduce protocol time. Nevertheless, we found that the greatest contributor to RNA degradation is the release of cellular RNases to the lysate upon cell lysis. It was proposed that cryogenic grinding of yeast cells with a mechanical mill results in improved RNA purification41. This can be tried in cases where significant degradation is observed with this protocol.
A common drawback of pull-down assays is the isolation of numerous proteins that bind nonspecifically to the column. Therefore, the use of a yeast strain with untagged RNA is an important control. This strain expresses the MS2-CP-GFP-SBP fusion protein to further exclude proteins that bind to the fusion protein and not the target RNA. We note that this does not exclude proteins that might bind the MS2 loop itself, and these need further validation with the protein pull-down assay. Figure 1B shows that we were able to identify several proteins that bind specifically to PMP1 mRNA, and some were subsequently validated by a co-IP assay20.
The RaPID methodology is currently limited to yeast systems, utilizing its well-established site-specific integration methodologies. Site-specific integration protocols are currently being developed for genes in other systems using the CRISPR-Cas system42,43. These will be of great importance in expanding the utilization of RaPID to additional systems. Another future application of RaPID is for the isolation of RNAs that are associated with an mRNA of interest. This can be easily achieved, by subjecting the isolated material to RNA-seq rather than LC-MS/MS. Considering the important roles of small RNAs in regulating mRNA expression, this application is likely to be of great importance. Finally, improvements in mass spectrometry (MS) analysis and the use of quantitative MS methods such as SILAC will result in the quantitative measure of mRNA-bound proteins under different conditions. RaPID can be used with yeast grown in media enriched with heavy isotopes and compared to cells grown under different conditions (e.g., stress) with unlabeled media44,45. Applying the RaPID method together with quantitative MS analysis will yield accurate measures of changes in the RBP repertoire that are associated with a particular mRNA.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Prof. Jeff Gerst and Boris Slobodin for their helpful advice in setting up the RaPID protocol and providing the necessary plasmids. We also thank Dr. Avigail Atir-Lande for her help in establishing this protocol and Dr. Tamar Ziv from the Smoler Proteomics Center for her help with the LC-MS/MS analysis. We thank Prof. T.G. Kinzy (Rutgers) for the YEF3 antibody. This work was supported by grant 2011013 from the Binational Science Foundation.
Materials
| Tris | sigma | T1503 | |
| SDS | bio-lab | 1981232300 | |
| DTT | sigma | D9779 | |
| Acidic Phenol (pH 4.3) | sigma | P4682 | |
| Acidic Phenol: Chloroform (5:1, pH 4.3) | sigma | P1944 | |
| Chloroform | bio-lab | 3080521 | |
| Formaldehyde | Frutarom | 5551820 | |
| Glycine | sigma | G7126 | |
| NP-40 | Calbiochem | 492016 | |
| Heparin | Sigma | H3393 | |
| Phenylmethylsulfonyl Flouride (PMSF) | Sigma | P7626 | |
| Leupeptin | Sigma | L2884 | |
| Aprotinin | Sigma | A1153 | |
| Soybean Trypsin Inhibitor | Sigma | T9003 | |
| Pepstatin | Sigma | P5318 | |
| DNase I | Promega | M610A | |
| Ribonuclease Inhibitor | Takara | 2313A | |
| Glass Beads | Sartorius | BBI-8541701 | 0.4-0.6mm diameter |
| Mini BeadBeater | BioSpec | Mini BeadBeater 16 | |
| Guanidinium | Sigma | G4505 | |
| Avidin | Sigma | A9275 | |
| Streptavidin Beads | GE Healthcare | 17-5113-01 | |
| Bovine serum albumin (BSA) | Sigma | A7906 | |
| Yeast tRNA | Sigma | R8508 | |
| Biotin | Sigma | B4501 | |
| Yeast extract | Bacto | 288620 | |
| peptone | Bacto | 211677 | |
| Glucose | Sigma | G8270 | |
| 1 x Phosphate-Buffered saline (PBS) | |||
| 0.2 M NaOH | |||
| 4 x Laemmli Sample Buffer (LSB) | 0.2 M Tris-Hcl pH 6.8, 8% SDS, 0.4 M DTT, 40% glycerol, 0.04% Bromophenol-Blue. | ||
| Hot phenol lysis buffer | 10 mM Tris pH 7.5, 10 mM EDTA, 0.5% SDS | ||
| 3 M Sodium Acetate pH 5.2 | |||
| 100% and 70% Ethanol (EtOH) | |||
| RNase-free water | |||
| RaPID lysis buffer | 20 mM Tris pH 7.5, 150 mM NaCl, 1.8 mM MgCl2, 0.5% NP-40, 5 mg/ml Heparin, 1 mM Dithiothreitol (DTT), 1 mM Phenylmethylsulfonyl Flouride (PMSF), 10 µg/ml Leupeptin, 10 µg/ml Aprotinin, 10 µg/ml Soybean Trypsin Inhibitor, 10 µg/ml Pepstatin, 20 U/ml DNase I, 100 U/ml Ribonuclease Inhibitor. | ||
| 2x Cross-linking reversal buffer | 100 mM Tris pH 7.4, 10 mM EDTA, 20 mM DTT, 2 % SDS. | ||
| RaPID wash buffer | 20 mM Tris-HCl pH 7.5, 300 mM NaCl, 0.5% NP-40 | ||
| 0.5 M EDTA pH 8 | |||
| Silver Stain Plus Kit | Bio-Rad | 161-0449 | For detecting proteins in polyacrylamide gels |
| SD selective medium | 1.7 g/l Yeast nitrogen base with out amino acids and ammonium sulfate, 5 g/l Ammonium sulfate, 2% glucose, 350 mg/l Threonine, 40 mg/l Methionine, 40 mg/l Adenine, 50 mg/l Lysine, 50 mg/l Tryptophan, 20 mg/l Histidine, 80 mg/l Leucine, 30 mg/l Tyrosine, 40 mg/l Arginine | ||
| Anti-eEF3 (EF3A,YEF3) | Gift from Kinzy TG. (UMDNJ Robert Wood Johnson Medical School) | 1:5,000 | |
| Anti GFP antibody | Santa Cruz | sc-8334 | 1:3,000 |
| Anti rabbit IgG-HRP conjugated | SIGMA | A9169 | 1:10,000 |
References
- Hogan, D. J., Riordan, D. P., Gerber, A. P., Herschlag, D., Brown, P. O. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 6, e255 (2008).
- Tsvetanova, N. G., Klass, D. M., Salzman, J., Brown, P. O. Proteome-wide search reveals unexpected RNA-binding proteins in Saccharomyces cerevisiae. PloS One. 5, (2010).
- Castello, A., et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 149, 1393-1406 (2012).
- Gerstberger, S., Hafner, M., Tuschl, T. A census of human RNA-binding proteins. Nat Rev Genet. 15, 829-845 (2014).
- Baltz, A. G., et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 46, 674-690 (2012).
- Mitchell, S. F., Parker, R. Principles and properties of eukaryotic mRNPs. Mol Cell. 54, 547-558 (2014).
- Licatalosi, D. D., Darnell, R. B. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 11, 75-87 (2010).
- Eliyahu, E., Lesnik, C., Arava, Y. The protein chaperone Ssa1 affects mRNA localization to the mitochondria. FEBS Lett. 586, 64-69 (2012).
- Ascano, M., Gerstberger, S., Tuschl, T. Multi-disciplinary methods to define RNA-protein interactions and regulatory networks. Curr Opin Genet Dev. 23, 20-28 (2013).
- Denman, R. B. mRNPs take shape by CLIPPING and PAIRING. BioEssays. 28, 1132-1143 (2006).
- McHugh, C. A., Russell, P., Guttman, M. Methods for comprehensive experimental identification of RNA-protein interactions. Genome Biol. 15, 203 (2014).
- Bernstein, D. S., Buter, N., Stumpf, C., Wickens, M. Analyzing mRNA-protein complexes using a yeast three-hybrid system. Methods. 26, 123-141 (2002).
- SenGupta, D. J., et al. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci USA. 93, 8496-8501 (1996).
- Yosefzon, Y., et al. Divergent RNA binding specificity of yeast Puf2p. RNA. 17, 1479-1488 (2011).
- Mitchell, S. F., Jain, S., She, M., Parker, R. Global analysis of yeast mRNPs. Nat Struct Mol Biol. 20, 127-133 (2013).
- Zielinski, J., et al. In vivo identification of ribonucleoprotein-RNA interactions. Proc Natl Acad Sci USA. 103, 1557-1562 (2006).
- Bell, T. J., Eberwine, J. Live Cell Genomics: RNA Exon-Specific RNA-Binding Protein Isolation. Methods Mol Biol. 1324, 457-468 (2015).
- Leppek, K., Stoecklin, G. An optimized streptavidin-binding RNA aptamer for purification of ribonucleoprotein complexes identifies novel ARE-binding proteins. Nucleic Acids Res. 42, e13 (2014).
- Slobodin, B., Gerst, J. E. A novel mRNA affinity purification technique for the identification of interacting proteins and transcripts in ribonucleoprotein complexes. RNA. 16, 2277-2290 (2010).
- Samra, N., Atir-Lande, A., Pnueli, L., Arava, Y. The elongation factor eEF3 (Yef3) interacts with mRNA in a translation independent manner. BMC Mol Biol. 16, 17 (2015).
- Loya, A., et al. The 3′-UTR mediates the cellular localization of an mRNA encoding a short plasma membrane protein. RNA. 14, 1352-1365 (2008).
- Haim-Vilmovsky, L., Gadir, N., Herbst, R. H., Gerst, J. E. A genomic integration method for the simultaneous visualization of endogenous mRNAs and their translation products in living yeast. RNA. 17, 2249-2255 (2011).
- Eldad, N., Yosefzon, Y., Arava, Y. Identification and characterization of extensive intra-molecular associations between 3′-UTRs and their ORFs. Nucleic Acids Res. 36, 6728-6738 (2008).
- Slobodin, B., Gerst, J. E. RaPID: an aptamer-based mRNA affinity purification technique for the identification of RNA and protein factors present in ribonucleoprotein complexes. Methods Mol Biol. 714, 387-406 (2011).
- Schmitt, M. E., Brown, T. A., Trumpower, B. L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18, 3091-3092 (1990).
- Eldad, N., Arava, Y. A ribosomal density-mapping procedure to explore ribosome positions along translating mRNAs. Methods Mol Biol. 419, 231-242 (2008).
- Arava, Y., Seger, R., Fbeta Walker, M. D. GRFbeta, a novel regulator of calcium signaling, is expressed in pancreatic beta cells and brain. J Biol Chem. 274, 24449-24452 (1999).
- Arava, Y., Adamsky, K., Ezerzer, C., Ablamunits, V., Walker, M. D. Specific gene expression in pancreatic beta-cells: cloning and characterization of differentially expressed genes. Diabetes. 48, 552-556 (1999).
- Bavli-Kertselli, I., Melamed, D., Bar-Ziv, L., Volf, H., Arava, Y. Overexpression of eukaryotic initiation factor 5 rescues the translational defect of tpk1w in a manner that necessitates a novel phosphorylation site. FEBS J. 282, 504-520 (2015).
- Eliyahu, E., Melamed, D., Arava, Y. Genome-wide analysis of RNA extracted from isolated mitochondria. Methods Mol Biol. 714, 287-299 (2011).
- Brunelle, J. L., Green, R. One-dimensional SDS-polyacrylamide gel electrophoresis (1D SDS-PAGE). Methods Enzymol. 541, 151-159 (2014).
- Gundry, R. L., et al. Preparation of proteins and peptides for mass spectrometry analysis in a bottom-up proteomics workflow. Current protocols in molecular biology. Chapter 10, Unit 10.25 (2009).
- Medzihradszky, K. F. In-solution digestion of proteins for mass spectrometry. Methods Enzymol. 405, 50-65 (2005).
- Bachler, M., Schroeder, R., von Ahsen, U. StreptoTag: a novel method for the isolation of RNA-binding proteins. RNA. 5, 1509-1516 (1999).
- Oeffinger, M. Two steps forward–one step back: advances in affinity purification mass spectrometry of macromolecular complexes. Proteomics. 12, 1591-1608 (2012).
- Ross, A. F., Oleynikov, Y., Kislauskis, E. H., Taneja, K. L., Singer, R. H. Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol. 17, 2158-2165 (1997).
- Deshler, J. O., Highett, M. I., Schnapp, B. J. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science. 276, 1128-1131 (1997).
- Bertrand, E., et al. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 2, 437-445 (1998).
- Aizer, A., et al. Quantifying mRNA targeting to P-bodies in living human cells reveals their dual role in mRNA decay and storage. J Cell Sci. 127, 4443-4456 (2014).
- Gadir, N., Haim-Vilmovsky, L., Kraut-Cohen, J., Gerst, J. E. Localization of mRNAs coding for mitochondrial proteins in the yeast Saccharomyces cerevisiae. RNA. 17, 1551-1565 (2011).
- Lopez de Heredia, ., M, R. P., Jansen, RNA integrity as a quality indicator during the first steps of RNP purifications : a comparison of yeast lysis methods. BMC Biochem. 5, 14 (2004).
- Lee, J. S., Kallehauge, T. B., Pedersen, L. E., Kildegaard, H. F. Site-specific integration in CHO cells mediated by CRISPR/Cas9 and homology-directed DNA repair pathway. Sci Rep. 5, 8572 (2015).
- Auer, T. O., Duroure, K., De Cian, A., Concordet, J. P., Del Bene, F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 24, 142-153 (2014).
- Oda, Y., Huang, K., Cross, F. R., Cowburn, D., Chait, B. T. Accurate quantitation of protein expression and site-specific phosphorylation. Proc Natl Acad Sci USA. 96, 6591-6596 (1999).
- de Godoy, L. M. SILAC yeast: from labeling to comprehensive proteome quantification. Methods Mol Biol. 1156, 81-109 (2014).

