Antibody Labeling with Fluorescent Dyes Using Magnetic Protein A and Protein G Beads
Summary
The on-bead method for labeling antibodies with small molecules enables labeling of a small amount of antibodies directly from cell media. This method is compatible with amine and thiol chemistry, and can handle multiple samples in parallel, manually or using automated platforms.
Abstract
Antibodies labeled with small molecules like fluorescent dyes, cytotoxic drugs, and radioactive tracers are essential tools in biomedical research, immunodiagnostics and more recently as therapeutic agents. Traditional methods for labeling antibodies with small molecules require purified antibodies at relatively high concentration, involve multiple dialysis steps and have limited throughput. However, several applications, including the field of Antibody Drug Conjugates (ADCs), will benefit from new methods that will allow labeling of antibodies directly from cell media. Such methods may allow antibodies to be screened in biologically relevant assays, for example, the receptor-mediated antibody internalization assay in the case of ADCs. Here, we describe a method (on-bead method) that enables labeling of small amounts of antibodies directly from cell media. This approach utilizes high capacity magnetic Protein A and Protein G affinity beads to capture antibodies from the cell media followed by labeling with small molecules using either amine or thiol chemistry and subsequent elution of the labeled antibodies. Taking fluorescent dyes as surrogates for small molecules, we demonstrate the on-bead labeling of three different mouse antibodies directly from cell media using both amine and thiol labeling chemistry. The high binding affinity of antibodies to Protein A and Protein G ensures high recoveries as well as high purity of the labeled antibodies. In addition, use of magnetic beads allows multiple samples to be handled manually, thereby significantly improving labeling throughput.
Introduction
Antibodies labeled with small molecules are perhaps the most commonly used reagents in biology1,2. Antibodies labeled with fluorescent dyes and biotin are extensively used in imaging, immunoassays, flow cytometry, western blots, and immunoprecipitation among other applications3-6. Radiolabeled antibodies3,7 find extensive use in imaging and therapy, antibodies labeled with cytotoxic drugs (ADCs) are offering new options for the treatment of cancers, and two ADCs have already been approved for therapeutic use8. In spite of their extensive use, the methods for labeling antibodies have remained surprisingly unchanged and typically involve multiple reactions and desalting steps9-12. Solution methods work very well in cases where only a few antibodies need to be labeled and are available in highly purified form at high concentration and in sufficient volumes. However, for newer applications like ADCs, there is a need to label antibodies at the early hybridoma stage so that they can be screened for biologically relevant properties, for example, receptor-mediated antibody internalization13-16. At the hybridoma stage, sample volumes are limited, antibodies are expressed at low concentrations and a number of samples are large, hence solution based labeling methods are not suitable.
To simplify and improve the throughput of the traditional antibody labeling methods, a few alternative approaches have been proposed17,18. One approach is to use non-magnetic Protein A affinity beads packed in small columns to capture antibodies followed by the labeling reaction and elution of labeled and purified protein. This method can be used to label antibodies directly from cell media, however, the use of columns can be laborious. A magnetic bead based method has recently been reported19 that eliminates the use of columns and improves throughput but due to the limited antibody binding capacity of the beads, only nanogram to low microgram quantities of the antibodies could be labeled.
We recently developed and used high capacity magnetic Protein A and Protein G beads (>20 mg of Human IgG/ml of settled beads) to label antibodies present in cell media with small molecules20. The high capacity of the beads allows tens to hundreds of micrograms of antibody to be labeled conveniently and the rapid magnetic response of the beads simplifies handling and processing of a large number of samples in parallel. Using fluorescent dyes as surrogates for small molecules, we show that the method is compatible with amine and thiol labeling chemistry and offers high recoveries of labeled and very pure antibodies.
This protocol and the accompanying video describe on-bead labeling of mouse antibodies present in the cell media using Magnetic Protein A and Protein G beads. The protocol is divided into four sections: Section 1 describes the capture of antibodies onto the bead from biological samples. Following capture, the labeling of antibodies with fluorescent dye using amine chemistry or using thiol chemistry is described in sections 2 and section 3, respectively. Finally, section 4 describes the method for the calculations of the antibody concentration and the dye to antibody ratio.
Protocol
1. Antibody Capture onto High Capacity Magnetic Protein A or Magnetic Protein G Beads
- Uniformly re-suspend magnetic beads by gentle shaking. Keep the suspension uniform when aliquoting beads.
- Add 50 µl of bead slurry to a 1.5 ml microcentrifuge tube. Place in the magnetic stand for 10 sec. Carefully remove the storage buffer.
- Add 250 µl of antibody binding buffer.
- Mix and place in the magnetic stand for 10 sec. Carefully remove the binding buffer.
- Add 1.0 ml of the sample containing 50-100 µg of antibody to the beads. Samples can be purified antibodies or antibodies in cell media.
- Mix sample for 60 min at RT using a vortex mixer or end-over-end mixer.
- Place tube in the magnetic stand for 10 sec and remove the supernatant.
- Add 250 µl of antibody binding/washing buffer and mix. Place in the magnetic stand for 10 sec and remove binding/washing buffer. Repeat this step for a total of two washes.
- Proceed to Section 2 to label antibodies using amine chemistry or to Section 3 to label antibodies using thiol chemistry.
2. Antibody Labeling Using Amine Chemistry
- Conjugate Antibody
- Add 100 µl of amine conjugation buffer to the beads.
- Dissolve the amine-reactive fluorescent dyes (AlexaFluor 532-SE) at 10 mg/ml by adding 100 µl of Dimethyl sulfoxide (DMSO) to 1.0 mg of dye. Mix by vortexing. Make this solution just before use.
- Add 2.5 µl of amine-reactive dye for 100 µg of antibody.
Note: Typically a 5-20 molar excess of reactive dye is recommended. However, amount of reactive dye added to the reaction needs to be empirically optimized depending on desired dye to antibody ratio and intrinsic antibody properties. - Mix sample for 60 min at RT using a vortex mixer or end-over-end mixer. Make sure that the beads remain in suspension.
- Place tube in the magnetic stand for 10 sec and remove the supernatant.
- Add 250 µl of antibody binding/washing buffer and mix. Place in the magnetic stand for 10 sec. Remove and discard binding/washing buffer. Repeat this step for a total of two washes.
- Antibody Recovery
- Add 50 µl of elution buffer to the beads.
- Mix for 5 min at RT using a vortex mixer or end-over-end mixer. Make sure that the beads remain in suspension.
- Place tube in the magnetic stand for 10 sec. Remove eluted sample and transfer to a new micro-centrifuge tube containing 5 µl of neutralization buffer.
- Repeat the process one more time and pool the eluted samples.
- Quantitate the antibody concentration and dye-to-antibody ratio as described in Section 4.
3. Antibody Labeling Using Thiol Chemistry
- Antibody Reduction
- Add 250 µl of thiol conjugation buffer and mix. Place the tube in the magnetic stand for 10 sec. Remove and discard the buffer. Repeat this step twice.
- Add 100 µl of thiol conjugation buffer.
- Add Dithiothreitol (DTT) to a final concentration of 2.5 mM.
- Mix sample for 60 min at RT using a vortex mixer or end-over-end mixer. Make sure that the beads remain in suspension.
- Place the tube in the magnetic stand for 10 sec and discard the buffer.
- Add 250 µl of thiol conjugation buffer and mix. Place the tube in the magnetic stand for 10 sec. Remove and discard the buffer. Repeat this step for a total of two washes.
- Add 100 µl of thiol conjugation buffer.
- Conjugate Antibody
- Dissolve the thiol-reactive dye at 10 mg/ml by adding 100 µl of DMSO to 1.0 mg of dye. Mix by vortexing. Make this solution just before use.
- Add 2.5 µl of thiol-reactive dye for 100 µg of antibody.
Note: Typically a 5-20 molar excess of reactive dye is recommended. However, amount of reactive dye added to the reaction needs to be empirically optimized depending on desired dye to antibody ratio and intrinsic antibody properties. - Mix sample for 60 min at RT using a vortex mixer or end-over-end mixer. Make sure that the beads remain in suspension.
- Place the tube in the magnetic stand for 10 sec and remove supernatant.
- Add 250 µl of thiol conjugation buffer and mix. Place in the magnetic stand for 10 sec and remove the buffer. Repeat this step for a total of two washes.
- Elute Antibody
- Add 50 µl of elution buffer to the beads.
- Mix for 5 min at RT using a vortex mixer or end-over-end mixer. Make sure that the beads remain in suspension.
- Place tube in the magnetic stand for 10 sec. Remove eluted sample and transfer to a new micro-centrifuge tube containing 5 µl of neutralization buffer.
- Repeat the process one more time and pool the eluted samples.
- Quantitate the antibody concentration and dye-to-antibody ratio as described in Section 4.
4. Calculate Dye-to-Antibody Ratio
- Measure the absorbance of the antibody-dye conjugate at 280 nm (A280) and at the λmax for the dye (Amax).
- Calculate the antibody concentration:
Antibody Concentration (mg/ml) = A280 – (Amax × CF) / 1.4
where CF = Correction factor of the dye (provided by the manufacturer). - Calculate the dye-to-antibody ratio:
Dye-to-Antibody Ratio (DAR) = (Amax × 150,000)/Ab Concentration (mg/ml) × εdye
where, εdye = the extinction coefficient of the dye at its absorbance maximum and molecular weight of antibody = 150,000 Da.
Representative Results
A schematic for labeling antibodies with small molecules using high capacity magnetic Protein A and Protein G beads is shown in Figure 1. Antibodies captured on the magnetic Protein G beads can be labeled with small molecules, for example fluorescent dyes, using either amine chemistry which labels primary amines of the lysine amino acids or using thiol chemistry which labels at the reduced thiols in the hinge region of the antibodies. Affinity between antibodies and Protein G is strong21 and results in minimal loss of antibody during the labeling reaction. This is reflected in the efficient recovery (50-90%) of three different mouse antibody isotypes (Table 1) after labeling with the fluorescent dyes when compared to simple purification. Efficient recovery (similar to that reported before19) is also evident in the Coomassie gels (Figure 2) where band intensities of the heavy and light chains from the purified antibodies and labeled antibodies reflect the result reported in Table 1. Magnetic Protein G beads have low non-specific binding properties resulting in highly purified antibodies as seen in the Coomassie gel (Figure 2) and both the heavy and light chains are fluorescently labeled (Figure 2). On-bead labeling can also be done on Magnetic Protein A beads and is compatible with multiple fluorescent dyes resulting in high recovery and good dye to antibody ratios (Table 2).
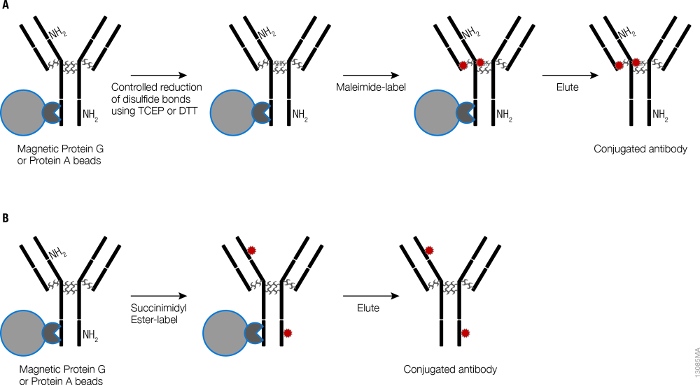
Figure 1. Schematic of on-bead labeling of antibodies with fluorescent molecules. (A) Antibody labeling using thiol chemistry. Antibodies are captured on high capacity magnetic Protein A or magnetic Protein G beads followed by reduction of inter-chain disulfide bonds using reducing agents like DTT or tris(2-carboxyethyl)phosphine (TCEP). Thiol-reactive maleimide containing fluorescent dyes are added to label antibodies. Labeled antibodies are recovered by low pH elution and neutralized immediately. (B) Antibody labeling using amine chemistry. Antibodies captured on high capacity magnetic Protein A and Protein G Beads are reacted with amine-reactive fluorescent dyes to label antibodies at the primary amines of the lysines. Labeled antibodies are eluted by low pH elution and neutralized immediately. Adapted from Nath, N. et al.20 Please click here to view a larger version of this figure.
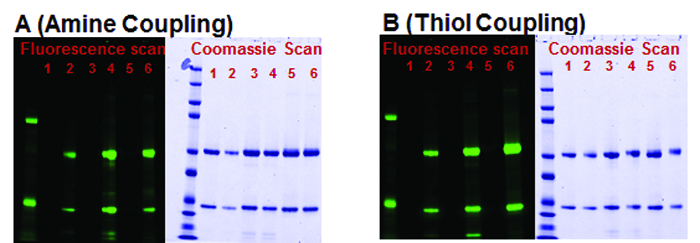
Figure 2. Denaturing SDS-PAGE gel images of three mouse IgG isotypes purified and labeled with AlexaFluor 532, using on-bead method. (A) Amine reaction (B) Thiol reaction. Gel images of antibodies labeled with thiol-reactive dye. Lanes 1-6 corresponds to the samples shown in Table 1. Gels were first imaged using fluorescent scanner and show only AlexaFluor 532 labeled antibody heavy and light chains. Samples where antibodies were only purified are not visible. Gels were subsequently stained with Coomassie stain to show heavy and light chains from antibodies that were purified as well as antibodies that were labeled. Adapted from Nath, N. et al.20 Please click here to view a larger version of this figure.
| Amine Reaction | Thiol Reaction | |||||
| Well | Isotype | Antibody Recovery (µg) | Dye to Antibody Ratio | Antibody Recovery (µg) | Dye to Antibody Ratio | |
| 1 | mouse IgG1 | Purification | 54.8 ± 1.8 | 0 | 49.2 ± 2.0 | 0 |
| 2 | Conjugation | 27.4 ± 3.2 | 4.2 ± 0.2 | 46.2 ± 3.5 | 1.6 ± 0.2 | |
| 3 | mouse IgG2A | Purification | 85.9 ± 6.4 | 0 | 75.3 ± 8.0 | 0 |
| 4 | Conjugation | 60.7 ± 3.2 | 4.4 ± 0.1 | 61.5 ± 7.4 | 5.5 ± 0.4 | |
| 5 | mouse IgG2B | Purification | 79.9 ± 6.9 | 0 | 73.3 ± 1.4 | 0 |
| 6 | Conjugation | 66.7 ± 0.5 | 5.6 ± 0.1 | 55.7 ± 0.9 | 5.2 ± 0.5 | |
Table 1: On-bead labeling of three mouse IgG isotypes directly from cell media using amine and thiol chemistries. For each sample, one aliquot of 1.0 ml cell media was used just for purification of antibodies using Magnetic Protein G beads (50 µl slurry) and second aliquot was used for on-bead labeling with amine-reactive AlexaFluor 532 dye. Antibody recoveries in both cases were calculated (as described in text) and compared to determine antibody losses during labeling reaction. Dye to antibody ratio was also calculated for all three antibodies. All the reactions were done in triplicate. Adapted from Nath, N. et al.20
| Magnetic Protein G Beads | Magnetic Protein A Beads | ||||
| Antibody Recovery (µg) | Dye to antibody Ratio | Antibody Recovery (µg) | Dye to antibody Ratio | ||
| 1 | Purification | 263.7 ± 10.4 | 0 | 269.4 ± 5.1 | 0 |
| 2 | AlexaFluor532 | 182.9 ± 15.3 | 5.3 ± 0.04 | 189.1 ± 6.9 | 6.9 ± 0.2 |
| 3 | AlexaFluor647 | 192.5 ± 2.9 | 3.3 ± 0.1 | 112.3 ± 11.4 | 3.6 ± 0.2 |
| 4 | Fluorescein | 179.5 ± 5.3 | 6.8 ± 0.1 | 201.5 ± 6.5 | 6.8 ± 0.1 |
Table 2: On-bead labeling of mouse IgG2A with three different thiol-reactive fluorescent dyes. Concentrated cell media samples were used in this experiment to show that method can also be adopted for different amount of antibody. In addition, conjugation reactions were performed using magnetic Protein G as well as magnetic Protein A. One aliquot of 1.0 ml concentrated cell media was used for antibody purification using Magnetic Protein G beads (100 µl slurry). Three other 1.0 ml aliquots were labeled and purified with thiol-reactive AlexaFluor 532, AlexaFluor 647 and Fluorescein. Antibody recoveries were calculated (as described in text) and compared for various labeling reactions. Dye to antibody ratio was also calculated for all three fluorescent dyes. Similar purification and labeling was done using Magnetic Protein A beads. All the reactions were done in triplicate. Adapted from Nath, N. et al.'20
| Buffer | Comments/Description |
| Amine conjugation buffer (10 mM sodium bicarbonate buffer, pH 8.5) | 1. 0.084 g sodium bicarbonate 2. Dissolve in deionized water. Adjust to pH 8.5. 3. Adjust the final volume to 100 ml with deionized water. |
| Thiol conjugation buffer (10 mM phosphate buffer with 1 mM EDTA, pH 7.0) | 1. 0.0378 g sodium phosphate, monobasic, monohydrate 2. 0.195 g sodium phosphate, dibasic, heptahydrate 3. Dissolve in deionized water. 4. Add EDTA to a final concentration of 1.0 mM. 5. Adjust to pH 7. 6. Adjust the final volume to 100 ml with deionized water. |
| Antibody binding/washing buffer (10 mM phosphate buffer, pH 7.0) | 1. 0.0378 g sodium phosphate, monobasic, monohydrate 2. 0.195 g sodium phosphate, dibasic, heptahydrate 3. Dissolve in deionized water. Adjust to pH 7. 4. Adjust the final volume to 100 ml with deionized water. |
| Elution buffer (50 mM Glycine Buffer, pH 2.7) | 1. 0.188 g glycine 2. Dissolve in deionized water. Adjust pH to 2.7 with HCl. 3. Adjust the final volume to 50 ml with deionized water. |
| Neutralization buffer (2 M Tris Buffer, pH 7.5) | 1. 0.472 g trizma base 2. 2.54 g trizma hydrochloride 3. Dissolve in deionized water. Adjust to pH 7.5. 4. Adjust the final volume to 10 ml with deionized water. |
Table 3: Buffer compositions.
Discussion
The goal of this study was to develop a method to label antibodies, present in the cell media at low concentrations, with a variety of small molecules. Such a method will allow a large number of antibodies, during the early stages of antibody discovery, to be labeled with small molecules and screened using a biologically relevant assay. One such assay is the receptor-mediated antibody internalization assay where internalization may vary between antibodies even with similar binding affinities. Hence, it is important to screen antibodies specifically for their internalization properties in addition to traditional Enzyme Linked Immunosorbent Assay (ELISA) based screening. The key requirements for the method to label antibodies at the early antibody development stage are: a) ability to label antibodies directly from cell media; b) ability to label antibodies present at low concentrations, for example when the antibody concentration in cell media is 50 µg/ml; c) purity of the labeled antibody and antibody concentration should be compatible with downstream applications; d) the method should be compatible with both amine and thiol based conjugation chemistry; e) the method should be compatible with processing multiple samples simultaneously.
Considering these requirements, we developed an on-bead antibody labeling and purification method using high capacity magnetic Protein A and Protein G beads. These beads are based on hydrophilic cellulose with low non-specific binding properties and result in highly pure antibody preparation as shown in Figure 2. The high capacity of the beads means that a small amount of beads can efficiently capture antibodies from the media and antibodies can be eluted in a small volume hence concentrating the antibodies. In Table 1, only 10 µl settled beads were used to capture antibodies from 1.0 ml samples (expected concentrations 50-100 µg/ml) and labeled antibodies were eluted in a 120 µl volume at concentrations between 200-500 µg/ml. These concentrations are compatible with cell based internalization experiments where typical antibody concentrations range from 1.0 µg/ml to 10 ng/ml15,16. The on-bead labeling method is compatible with both amine and thiol chemistries and a wide range of dye-to-antibody ratios can be achieved along with efficient recovery of the labeled antibodies (Table 1). Experiments were done in triplicates and up to 12 samples were handled in parallel manually, significantly reducing processing time. The throughput of the labeling could be further increased by using robotic platforms as it has been shown in other applications using magnetic beads19,22.
Samples with higher antibody amount can be easily accommodated by increasing the amount of beads used for capture while maintaining the antibody recovery and labeling efficiency (Table 2). The method is also compatible with multiple dyes so that various dye antibody conjugates can be made, however, dye-to-antibody ratios will need to be optimized. Dye to antibody ratio of 2-4 has been determined to be optimum11,12 and in the case of approved ADCs an average of 3.5 drugs per antibody has been reported23. With on-bead conjugation chemistry, it is relatively easy to tune an optimum number of small molecule per antibody depending on the requirements of the downstream application. Moreover, both the thiol and amine chemistry for labeling can be tested to determine the best chemistry option which is critical for ADCs as it has been shown that antigen and receptor binding, in vivo stability, and therapeutic activity of antibody depends on the conjugation chemistry24,25.
There are some key factors to achieve good antibody labeling and recovery. Properties of the small molecule are the critical driver for both the antibody recovery and dye to antibody ratio. For a hydrophobic small molecule, high dye to antibody ratio will cause non-specific sticking of the labeled antibody to the bead and significantly reduced antibody recovery. Sometimes, depending on the dye and antibody characteristics, we found that one bead may work better than another hence, testing of both the beads during optimization may be useful. Protein A and Protein G have different affinities21 for antibodies from different species and of different subtypes, which should be considered when selecting the appropriate beads. For example, Protein A has low affinity for mouse IgG1 hence Protein G beads are strongly recommended. For thiol chemistry, DTT and TCEP reducing agents work equally well. DTT will however interfere with the maleimide reaction hence should be completely removed by washing. Low pH of 2.7 is recommended for elution, but lower pH of 2.2 may be required for efficient recovery in some cases. For labeled antibodies that may denature at low pH a balance between efficient recovery and antibody functionality will have to be empirically determined. It is well known that labeling chemistry may impact the antibody-antigen binding activity11,23, hence, it is absolutely necessary to test for antibody functionality after labeling using desired downstream application.
In summary, we describe a method to label antibodies with fluorescent dyes directly from the cell media without any prior purification steps. This method leverages high capacity magnetic Protein A and Protein G beads and is a significant improvement over solution based labeling method requiring highly purified antibodies at high concentrations10,11. The use of magnetic beads enables manual as well as automated sample handling, which is an advantage over non-magnetic bead based labeling techniques17,18. Finally, the use of high capacity beads allow scale up of antibody labeling reaction from low microgram levels to hundreds of micrograms using small amounts of beads and differentiates this method from other magnetic bead based labeling methods19. The protocol presented here describes labeling of antibodies with fluorescent dyes but the method can be extended to other labels including biotin, cytotoxic drugs and possibly enzymes.
Disclosures
The authors have nothing to disclose.
Acknowledgements
None.
Materials
| Magne Protein A Beads | Promega Corporation | G8781 | |
| Magne Protein G Beads | Promega Corporation | G7471 | |
| AlexaFluor 532-SE (Succinimidyl Ester) | Life Technologies | A20001 | |
| AlexaFluor 532-ME (Malemide) | Life Technologies | A10255 | |
| AlexaFluor 647-ME (Maleimide) | Life Technologies | A20347 | |
| Fluorescein-ME (Maleimide) | Life Technologies | F-150 | |
| Magnetic Stand | Promega | Z5332 |
References
- Colwill, K., Graslund, S. A roadmap to generate renewable protein binders to the human proteome. Nat Methods. 8 (7), 551-558 (2011).
- Silverstein, A. M. Labeled antigens and antibodies: the evolution of magic markers and magic bullets. Nat Immunol. 5 (12), 1211-1217 (2004).
- Day, J. J., et al. Chemically modified antibodies as diagnostic imaging agents. Curr Opin Chem Biol. 14 (6), 803-809 (2010).
- Cunningham, R. E. Overview of flow cytometry and fluorescent probes for flow cytometry. Methods Mol Biol. 588, 319-326 (2010).
- Eaton, S. L., et al. A guide to modern quantitative fluorescent western blotting with troubleshooting strategies. J Vis Exp. (93), e52099 (2014).
- Dundas, C. M., Demonte, D., Park, S. Streptavidin-biotin technology: improvements and innovations in chemical and biological applications. Appl Microbiol Biotechnol. 97 (21), 9343-9353 (2013).
- Kraeber-Bodere, F., et al. Radioimmunoconjugates for the treatment of cancer. Semin Oncol. 41 (5), 613-622 (2014).
- Leal, M., et al. Antibody-drug conjugates: an emerging modality for the treatment of cancer. Ann N Y Acad Sci. 1321, 41-54 (2014).
- Drachman, J. G., Senter, P. D. Antibody-drug conjugates: the chemistry behind empowering antibodies to fight cancer. Hematology Am Soc Hematol Educ Program. 2013, 306-310 (2013).
- Hermanson, G. T. . Bioconjugate Techniques. , 380-382 (1996).
- Shrestha, D., Bagosi, A., Szollosi, J., Jenei, A. Comparative study of the three different fluorophore antibody conjugation strategies. Anal Bioanal Chem. 404 (5), 1449-1463 (2012).
- Vira, S., Mekhedov, E., Humphrey, G., Blank, P. S. Fluorescent-labeled antibodies: Balancing functionality and degree of labeling. Anal Biochem. 402 (2), 146-150 (2010).
- Isa, M., et al. High-throughput screening system to identify small molecules that induce internalization and degradation of HER2. ACS Chem Biol. 9 (10), 2237-2241 (2014).
- Liao-Chan, S., et al. Quantitative assessment of antibody internalization with novel monoclonal antibodies against Alexa fluorophores. PLoS One. 10 (4), e0124708 (2015).
- Riedl, T., van Boxtel, E., Bosch, M., Parren, P. W., Gerritsen, A. F. High-Throughput Screening for Internalizing Antibodies by Homogeneous Fluorescence Imaging of a pH-Activated Probe. J Biomol Screen. 21 (1), 12-23 (2016).
- Nath, N., et al. Homogeneous plate based antibody internalization assay using pH sensor fluorescent dye. J Immunol Methods. 431, 11-21 (2016).
- Lundberg, E., Sundberg, M., Graslund, T., Uhlen, M., Svahn, H. A. A novel method for reproducible fluorescent labeling of small amounts of antibodies on solid phase. J Immunol Methods. 322 (1-2), 40-49 (2007).
- Strachan, E., et al. Solid-phase biotinylation of antibodies. J Mol Recognit. 17 (3), 268-276 (2004).
- Dezfouli, M., et al. Magnetic bead assisted labeling of antibodies at nanogram scale. Proteomics. 14 (1), 14-18 (2014).
- Nath, N., Godat, B., Benink, H., Urh, M. On-bead antibody-small molecule conjugation using high-capacity magnetic beads. J Immunol Methods. 426, 95-103 (2015).
- Lund, L. N., et al. Exploring variation in binding of Protein A and Protein G to immunoglobulin type G by isothermal titration calorimetry. J Mol Recognit. 24 (6), 945-952 (2011).
- Safarik, I., Safarikova, M. Magnetic techniques for the isolation and purification of proteins and peptides. Biomagn Res Technol. 2 (1), 7 (2004).
- Flygare, J. A., Pillow, T. H., Aristoff, P. Antibody-drug conjugates for the treatment of cancer. Chem Biol Drug Des. 81 (1), 113-121 (2013).
- Shen, B. Q., et al. Conjugation site modulates the in vivo stability and therapeutic activity of antibody-drug conjugates. Nat Biotechnol. 30 (2), 184-189 (2012).
- Acchione, M., Kwon, H., Jochheim, C. M., Atkins, W. M. Impact of linker and conjugation chemistry on antigen binding, Fc receptor binding and thermal stability of model antibody-drug conjugates. MAbs. 4 (3), 362-372 (2012).

