Methods to Study Lipid Alterations in Neutrophils and the Subsequent Formation of Neutrophil Extracellular Traps
Summary
Lipids are known to play an important role in cellular functions. Here, we describe a method to determine the lipid composition of neutrophils, with emphasis on the cholesterol level, by using both HPTLC and HPLC to gain a better understanding of the underlying mechanisms of neutrophil extracellular trap formation.
Abstract
Lipid analysis performed by high performance thin layer chromatography (HPTLC) is a relatively simple, cost-effective method of analyzing a broad range of lipids. The function of lipids (e.g., in host-pathogen interactions or host entry) has been reported to play a crucial role in cellular processes. Here, we show a method to determine lipid composition, with a focus on the cholesterol level of primary blood-derived neutrophils, by HPTLC in comparison to high performance liquid chromatography (HPLC). The aim was to investigate the role of lipid/cholesterol alterations in the formation of neutrophil extracellular traps (NETs). NET release is known as a host defense mechanism to prevent pathogens from spreading within the host. Therefore, blood-derived human neutrophils were treated with methyl-β-cyclodextrin (MβCD) to induce lipid alterations in the cells. Using HPTLC and HPLC, we have shown that MβCD treatment of the cells leads to lipid alterations associated with a significant reduction in the cholesterol content of the cell. At the same time, MβCD treatment of the neutrophils led to the formation of NETs, as shown by immunofluorescence microscopy. In summary, here we present a detailed method to study lipid alterations in neutrophils and the formation of NETs.
Introduction
Lipids have been shown to play important roles in cell homeostasis, cell death, host-pathogen interactions, and cytokine release1. Over time, interest for and knowledge on the impact of lipids in host-pathogen interactions or inflammation have increased, and several publications confirm the central role of certain lipids, especially the steroid cholesterol, in cellular responses. Pharmacological treatment with statins, which are used as inhibitors of cholesterol biosynthesis by blocking 3-hydroxy-3-methylglutaryl-coenzym-A-reductase (HMG-CoA-reductase), can act as anti-inflammatory agents by lowering the serum levels of interleukin 6 and C-reactive protein2. Cholesterol- and glycosphingolipid-enriched structures can be used by several pathogens, such as bacteria and viruses, as a gateway into the host3,4,5,6. Sphingolipids (e.g., sphingomyelin) have been shown to be used by pathogens to promote their pathogenicity7. In macrophages, mycobacteria use cholesterol-enriched domains for entering cells; a depletion of cholesterol inhibits mycobacterial uptake8. Furthermore, infection of macrophages with Francisella tularensis, a zoonotic agent responsible for tularemia (also known as rabbit fever)9, led to an infection that was abolished when cholesterol was depleted from the membranes10. Similarly, the invasion of host cells by Escherichia coli via lipid-rich structures was demonstrated to be cholesterol-dependent4. Moreover, Salmonella typhimurium infection experiments of epithelial cells demonstrated that cholesterol is essential for pathogen entry into the cells11. Cholesterol depletion inhibited the uptake of Salmonella11. Furthermore, a recent study by Gilk et al. demonstrated that cholesterol plays an important role in the uptake of Coxiella burnetti12. Additionally, Tuong et al. found that 25-hydroxycholesterol plays a crucial role in phagocytosis by lipopolysaccharide (LPS)-stimulated macrophages13. Phagocytosis was reduced when macrophages were pharmacologically treated to deplete cholesterol14. Thus, cholesterol and other lipids seem to play an important role in infection and inflammation, since their depletion can reduce the risk of invasion from several pathogens10,11,12.
Recently, we were able to show that lipid alterations, especially the depletion of cholesterol from the cell, induce the formation of neutrophil extracellular traps (NETs) in human blood-derived neutrophils15. Since the discovery of NETs in 2004, they have been shown to play critical roles in bacterial entrapment, and thus in hindering the spread of infection16,17. NETs consist of a DNA backbone associated with histones, proteases, and antimicrobial peptides16. The release of the NETs by neutrophils can be induced by invading pathogens18,19 and chemical substances such as phorbol-myristate-acetate (PMA) or statins16,20. However, the detailed cellular mechanisms, and especially the role of lipids in this process, are still not entirely clear. The analysis of lipids can lead to a better understanding of the mechanisms involved in a wide variety of cellular processes and interactions, such as the release of NETs. Cholesterol and sphingomyelin are vital constituents of the cell membrane and lipid microdomains, where they add stability and facilitate the clustering of the proteins involved in protein trafficking and signaling events21. To investigate the mechanistic role of certain lipids, amphiphilic pharmacological agents, such as the cyclic oligosaccharide methyl-β-cyclodextrin (MβCD), can be used to alter the lipid composition of a cell and to reduce cholesterol in vitro15. Here, we present a method to use HPTLC to analyze the lipid composition of neutrophils in response to MβCD. HPLC was used to confirm the level of cholesterol in the neutrophil population. Furthermore, we describe a method to visualize the formation of NETs by immunofluorescence microscopy in human blood-derived neutrophils in response to MβCD.
Protocol
The collection of the peripheral blood in this protocol was approved by the local human research ethics commission. All human subjects provided their written informed consent.
1. Isolation of Human Blood-derived Neutrophils by Density Gradient Centrifugation
- Isolation of human blood-derived neutrophils
- Layer ~20 mL of blood onto 20 mL of sodium diatrizoate/dextran solution near a flame and without mixing.
- Centrifuge for 30 min at 470 x g without brake.
- Remove the mononuclear cells and the yellowish plasma layer. Transfer the polymorphonuclear cell (PMN) phase (second phase with accumulating cells; Figures 1 and 2) into a new 50 mL tube and fill it up to 50 mL with 1x phosphate-buffered saline (PBS).
- Centrifuge for 10 min at 470 x g with brake.
- Remove the supernatant and resuspend the pellet in 5 mL of sterile water for 5 s to lyse the erythrocytes.
- Immediately fill up to 50 mL with 1x PBS and centrifuge for 10 min at 470 x g.
- Remove the supernatant. The pellet should be white. If it is still red, repeat steps 1.1.5 and 1.1.6.
- Resuspend the pellet in 1,000 µL of Roswell Park Memorial Institute (RPMI) medium. Count the cell number using trypan blue staining in a hemocytometer under a light microscope.
- Prepare a cell suspension in RPMI at a concentration of 2 x 106/mL. Approximately 2.5 x 107 neutrophils can be harvested from 20 mL of blood.
- Pharmacological treatment of neutrophils for lipid analysis
- Use 5 x 107 cells from step 1.1.9. Adjust the cell number in pure Hank´s Balanced Salt Solution (HBSS) medium in the presence or absence of 10 mM MβCD or 25 nM PMA in a total volume of 300 µL in a 1.5 mL reaction tube.
- Incubate the samples in the reaction tubes for 2 h at 37 °C and 5% CO2.
- Centrifuge for 10 min at 470 x g with brake.
- Remove the supernatant and resuspend the cell pellet in 300 µL of HBSS.
- Centrifuge for 10 min at 470 x g with brake.
- Resuspend the cell pellet in 1 mL of chloroform/methanol (1:1), homogenize it with a 26-G cannula by sucking the sample in and out into a 1 mL syringe 10 times, and store the samples at -20 °C.
- Pharmacological treatment of neutrophils for NET quantification by immunofluorescence
- Prepare poly-L-Lysine-coated 48-well plates with glass coverslips.
- Place one 8-mm glass coverslip in each well, add 55 µL of sterile 0.01% Poly-L-Lysine, and incubate for ~20 min at room temperature (RT).
- Wash twice with 200 µL of 1x PBS. Store the plates with 200 µL of 1x PBS per well in a refrigerator until required.
- Carefully add 100 µL of cell suspension (2 x 105/100 µL) from step 1.1.9 per well in the center of each coverslip.
- Add 100 µL per well of either the final 10 mM MβCD or the final 25 nM PMA.
- Centrifuge for 5 min at 370 x g with brake.
- Incubate for 2 h at 37 °C and 5% CO2.
- Centrifuge for 5 min at 370 x g with brake.
- Fix the cells with 155 µL of 4% PFA, wrap them in plastic paraffin film, and store them overnight at 4 °C or for 10 min at RT.
NOTE: For data on NET formation induced by MβCD, see Neumann et al.15.
- Prepare poly-L-Lysine-coated 48-well plates with glass coverslips.
2. Lipid Isolation and Analysis of Human Blood-derived Neutrophils
- Isolate the lipids from the human peripheral blood-derived neutrophils based on Bligh and Dyer22 and Brogden et al.23
- Take neutrophils from step 1.2.6 and place them on ice.
- On ice, pipette the neutrophils (present in methanol and chloroform solution) to a 15 mL screw-cap glass tube with a polytetrafluoroethylene (PTFE) seal and homogenize them by shaking for 1 min. Use glass tubes to prevent the lipids from binding to plastic surfaces. Use PTFE caps to prevent contamination from rubber/plastic.
- Add 2 mL of methanol followed 1 min later by 1 mL of chloroform. Shake again for 1 min.
- Rotate the glass tubes at RT and 50 rpm for 30 min.
- Pellet the protein fraction by centrifuging the solution at 7 °C and 1,952 x g for 10 min.
- Carefully decant the supernatant into a new 15 mL glass screw-cap tube, leaving the protein-containing pellet behind. Store the pellet at -20 °C for future quantification.
- Add 1 mL of chloroform, wait 1 min, add 1 mL of double-distilled water, and invert the glass screw-cap tube with the sample for 30 s.
- Centrifuge at 7 °C and 1,952 x g for 10 min and discard the upper phase, down to, but not including, the cloudy layer.
- If required, perform an optional further purification step by repeating step 2.1.8.
- Dry the samples in a vacuum concentrator at 60 °C and store them at -20 °C until required.
- High performance thin layer chromatography (HPTLC) to semi-quantify the amount of several lipids, including cholesterol, phospholipids, and sphingomyelin
- Prepare the following four running solutions and staining solution.
- Solution 1: Mix ethyl acetate (26.6%), 1-propanol (26.6%), chloroform (26.6%), methanol (26.6%), and potassium chloride (9.6%). Prepare KCl by dissolving 0.25 g of KCl in 100 mL of HPLC-quality water.
- Solution 2: Mix n-hexane (73%), diethyl ether (23%), and citric acid (2%).
- Solution 3: 100% n-hexane.
- Prepare the staining solution by mixing distilled water (90 mL) with 7.5 g of copper sulphate, and then add 10 mL of phosphoric acid.
- Fill up to 5 mm of each solution in a separate glass chamber. Add any kind of filter paper to increase the running speed in each chamber.
- Pre-incubate a 20 x 10 cm HPTLC silica gel 60 glass plate in the first running solution until the running solution reaches the top of the plate. Then dry it for 10 min at 110 °C.
NOTE: These plates can be stored for future use in aluminum foil and dried for 10 min at 110 °C prior to use. - Dissolve the lipid pellet obtained from step 2.1.10 in 200 µL of chloroform/methanol (1:1) solution and incubate for 15 min at 37 °C to dissolve.
- Use a ruler and a soft pencil to mark the loading spots for the desired number of samples, plus at least one standard. Mark the running distance at approximately 4 cm and 6 cm for the first and second running solution, respectively.
- To load the samples, wash the 10 µL syringe 3 times in chloroform/methanol (1:1) prior to loading each new sample. Load 10 µL of each sample drop-wise, trying to concentrate the sample on as small an area as possible. Samples should be loaded at least in duplicates.
- Place the plate vertically into the first chamber with running solution 1 (step 2.2.1.1). Ensure that the plate is parallel to the wall of the glass chamber to achieve a uniform migration speed.
- Once the solvent line has reached the first mark, remove the plate, dry it, and place it in the second solution. Repeat similar steps for the second and for the third running solutions; leave the plate in the solution until the solvent front reaches the top of the plate, then remove and dry it at RT for 1 min.
- Place plate in copper sulphate solution for 7 s. Remove the plate, dry it thoroughly, and bake it in an oven for 7 min at 170 °C. Wait for the plate to cool.
- Using a thin layer chromatography and gel analysis software, as well as an image processing software, scan and analyze as described previously by Brogden et al.23.
- Prepare the following four running solutions and staining solution.
- High performance liquid chromatography (HPLC) to quantify the amount of cholesterol
- Attach a 100 x 4.6 mm column to a 5 µm/4.6 mm guard cartridge and heat it to 32 °C. Use methanol as the mobile phase at a flow rate of 1 mL/min at 65 bar and a UV detector measuring at 202 nm to quantify the amount of cholesterol in each sample.
- Wash the HPLC machine thoroughly prior to analyzing the samples, performing the following steps: cleaning the pump, washing the needle, rinsing the pot, purging the syringe, and washing the pump purge. Wash all with water. Lastly, perform a system purge with methanol at a flow rate of 5 mL/min for 5 min.
- To establish a cholesterol standard curve, prepare at least 4 concentrations ranging from 0.05 mg/mL to 2 mg/mL of cholesterol in chloroform/methanol (1:1)24.
- Resuspend the samples obtained from step 2.1.10 in 500 µL of chloroform/methanol (1:1) in amber-colored 1.5 mL glass bottles with screw-top red PTFE/white silicone lids.
- Quantify the cholesterol concentrations using the standard prepared in step 2.3.3.
NOTE: Fill in the sampling protocol, starting with at least one standard and one negative control, followed by the samples. - Express the results as the area under the curve and compare between appropriate samples using any statistical software.
NOTE: Results can also be quantified against a standard curve and can be shown as the total cholesterol amount per mL, g, or number of cells. The standard curve equation should be calculated by using the values obtained in step 2.3.3.
3. Visualization and Quantification of NETs
- Visualization of NETs
NOTE: The visualization of NETs is based on previously-published work by Neumann et al.15.- Wash the fixed samples from step 1.3.8 3 times with 200 µL of 1x PBS.
- Block and permeabilize with 100 µL of 2% BSA, 0.2% Triton X-100 in PBS per well for 45 min at RT.
- Add 100 µL of primary antibodies: mouse monoclonal anti DNA-histone 1-antibody (dilute the stock with 2.2 mg/mL 1:5,000 in PBS containing 2% BSA and 0.2% Triton X-100) or polyclonal antibody against myeloperoxidase (MPO; rabbit anti-MPO; dilute 1:300 in PBS containing 2% BSA and 0.2% Triton X-100). Incubate overnight at 4 °C.
- Wash 3 times with 200 µL of 1x PBS.
- Add 100 µL of the secondary antibody (fluorescence-labelled goat anti-mouse; 1:1,000 in BSA-PBS-Triton X-100 and goat anti-rabbit; 1:1,000 BSA-PBS-Triton X-100) for 1 h at RT in the dark.
- Wash 3 times with 200 µL of 1x PBS.
- Remove coverslips using tweezers and place them face down with 3 µL of mounting medium containing DAPI on glass slides.
- Examine the samples using a confocal inverted-base fluorescence microscope equipped with a HCX PL APO 40X 0.75-1.25 oil immersion objective15.
- Quantification of the NET-releasing nuclei
- Open an image processing software (e.g., ImageJ) and drag and drop picture of interest into the tool bar.
- Click on "Plugins," "Analyze," and "Cell counter."
- Press the "Initialize" button in the counter window and choose a counter type (e.g., click on 7 in red for NET-releasing cells and 8 in yellow for non-releasing cells, as displayed in Figure 6B-i and ii).
NOTE: The criteria for NET-positive cells: Positively stained green nucleus + a less dense nucleus (loss of lobulation) or a loss of the round shape of the nucleus + an increased size of the nucleus, or an occurrence of a distinct extracellular off-shoot. - Click every cell adhering to the above criteria and write the counted cell number into a data sheet of an analysis software.
- Calculate the percentage of NET-releasing cells using the counted cell numbers of NET-releasing and non-releasing cells.
Representative Results
Human blood-derived neutrophils were isolated by density gradient centrifugation (Figure 2). To investigate the effect of lipid alterations on neutrophils, the cells were treated with 10 mM MβCD, which depletes cholesterol from the cell. Subsequently, the lipids were isolated from the samples by Bligh and Dyer (Figure 1, left panel), as described by Brogden et al.23. The prepared lipid samples were loaded onto silica gel HPTLC plates and run using a three-solution protocol, which has been optimized to separate and visualize a broad range of lipids, including cholesterol, cholesterol esters, sphingomyelin, phospholipids, and triacylglycerides, free fatty acids, monoacylglycerol, phosphatidylethanolamine, cardiolipin, phosphatidylserine, and phosphatidylcholine (Figure 3A). Oxygenated derivatives of cholesterol (oxysterols) are not detectable with this method. An additional quantitative analysis of cholesterol was performed using HPLC (Figure 3B). The regression value for cholesterol was markedly better when using HPLC (Figure 4A) compared to HPTLC (Figure 4B). To visualize the NETs, the cells were stimulated with the above-mentioned stimuli, fixed, and stained for DNA/histone 1 (green), MPO (red), and DNA (blue) as typical NET markers (Figure 5A). As displayed in Figure 5A, a clear occurrence of MPO, DNA/histone 1, and DNA occurs in the NETs when the cells were treated with 10 mM MβCD for 2 h. Subsequently, the effect of cholesterol reduction was microscopically analyzed. Therefore, cells were stained for DNA (blue) and DNA/histone 1 (green), and NET-releasing nuclei were counted using the cell counter plugin from the image processing software ImageJ (Figure 5B). Untreated neutrophils served as a control for spontaneous NET formation (Figure 5B-i). In the displayed figure, the NET release in untreated neutrophils after 2 h of incubation was 3.89%, whereas the treatment of the cells with 10 mM MβCD resulted in 35.17% NET formation (Figure 5B).
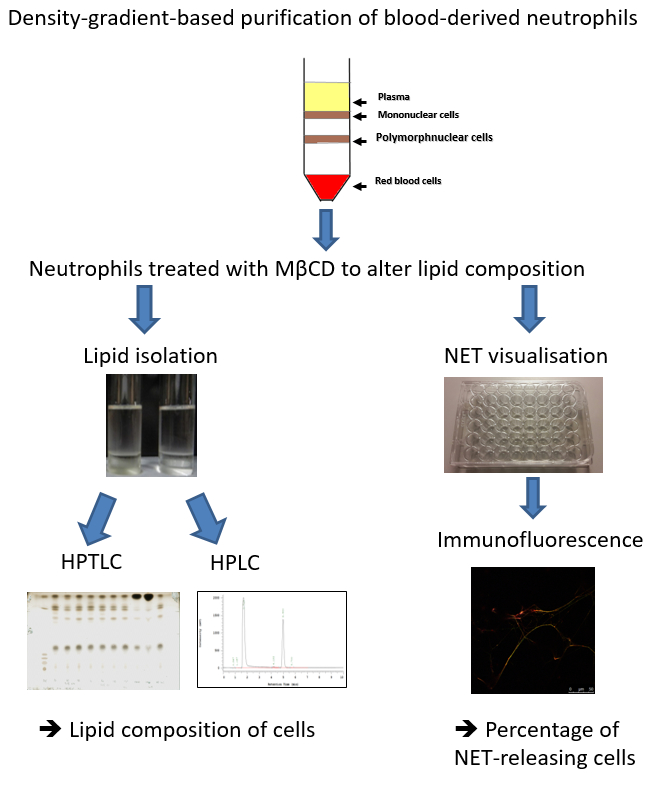
Figure 1: Schematic showing the steps involved in determining the lipid composition and extracellular trap release from human blood-derived neutrophils. Neutrophils are isolated using a density gradient and treated with Methyl-β-cyclodextrin (MβCD) to induce lipid alterations and NET formation. Thereafter, lipids are isolated and analyzed by using either HPTLC or HPLC, and NETs are visualized and quantified by immunofluorescence.
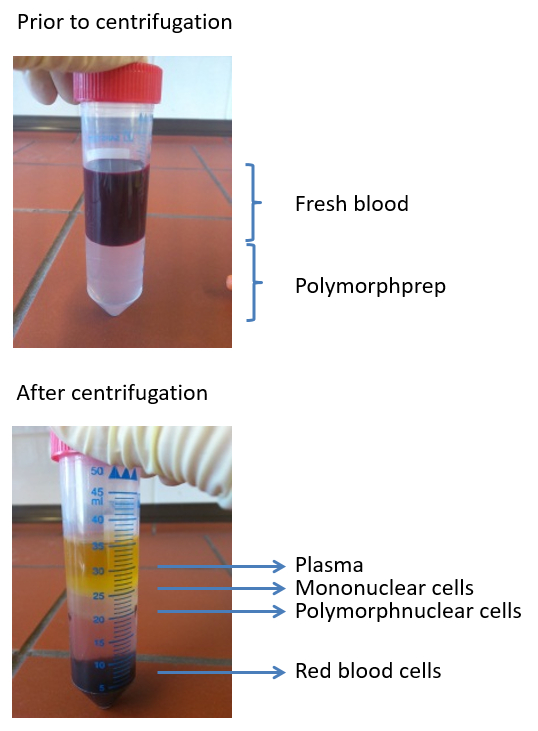
Figure 2: Images depicting a typical density gradient used to isolate polymorphonuclear cells from freshly-isolated blood. Four layers are visible post centrifugation: plasma, mononuclear cells, polymorphonuclear cells, and erythrocytes.
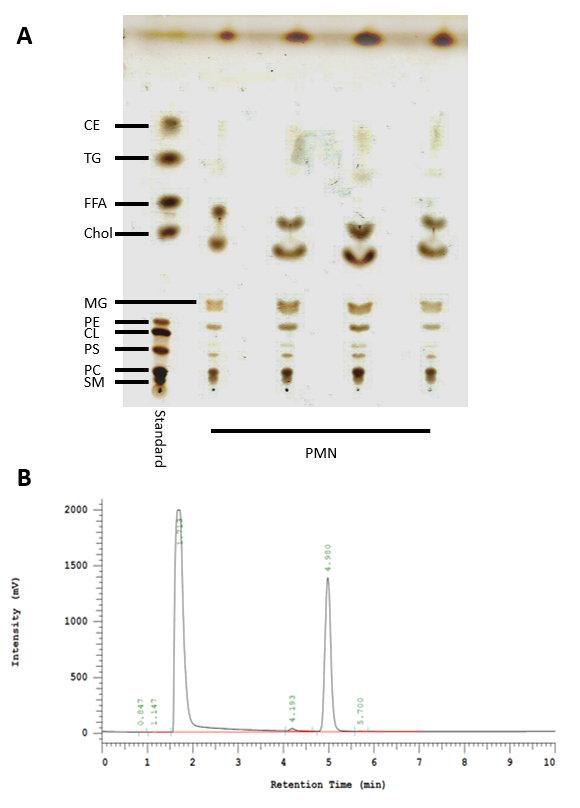
Figure 3: HPTLC and HPLC analysis of lipids isolated from neutrophils. (A) Standard used to identify the lipids present in neutrophils via HPTLC (left lane). CE: Cholesterol esters, TG: Triacylglycerides, FFA: Free fatty acids, Chol: Cholesterol, MG: Monoacylglycerol, PE: Phosphatidylethanolamine, CL: Cardiolipin, PS: Phosphatidylserine, PC: Phosphatidylcholine, and SM: Sphingomyelin. (B) Representative result showing a cholesterol-specific peak for 2 mg/mL at 4.980 min using the HPLC protocol described here.
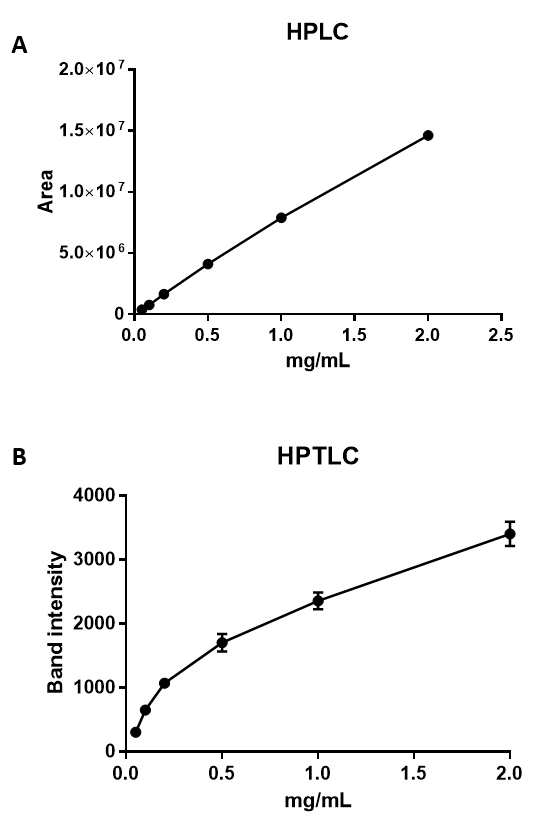
Figure 4: HPLC and HPTLC analyses of cholesterol. Graphs showing the relationship between cholesterol concentration and area, as measured by HPLC (A), and band intensity, as measured by HPTLC (B). The minimum detection limit for HPLC was 0.0016 mg/mL, with a regression value for the standard curve of 0.998 N = 3, SEM (A). Cholesterol ranging from 2 mg/mL down to 0.05 mg/mL can be semi-quantified using HPTLC (B), with a minimum detection limit of 0.05 mg/mL N = 3, SEM. The regression value for the HPTLC standard curve is 0.918.
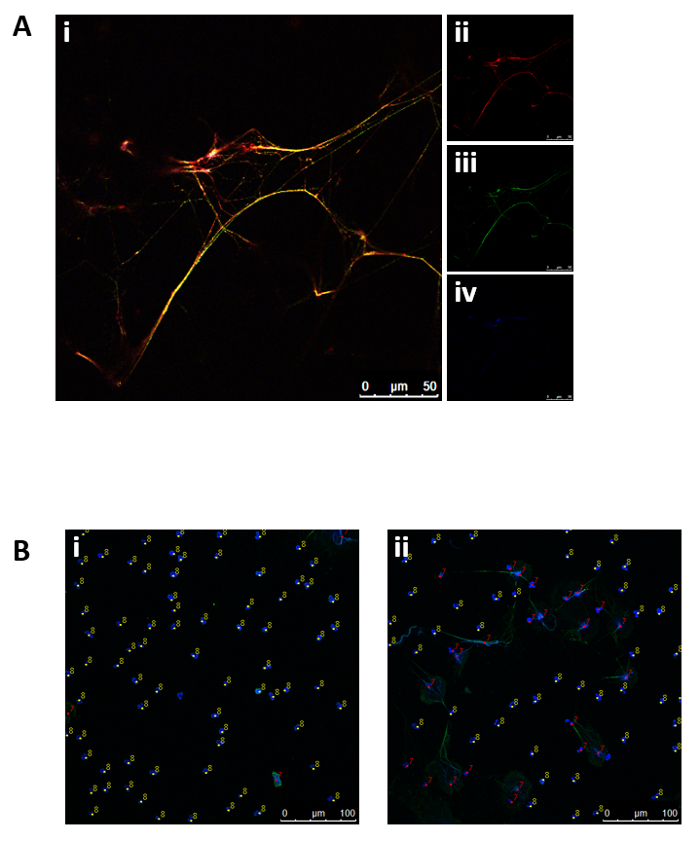
Figure 5: Representative fluorescence micrographs displaying NET structures and subsequent NET quantification. (A) Neutrophils stimulated with 10 mM MβCD for 2 h were stained for (ii) MPO (red), (iii) DNA/histone 1 (green), and (iv) DNA (blue). (i) Overlay of (ii), (iii), and (iv). (B) The cells were incubated for 2 h with (i) HBSS medium or (ii) 10 mM MβCD, and released NETs were stained for nuclei (blue) and DNA/histone 1 (green). NET-negative nuclei were marked with counter 8 (yellow) and NET-positive nuclei with counter 7 (red).
Discussion
The methods described here can be used to analyze specific lipids, such as cholesterol, by HPTLC or HPLC and to investigate the effects of pharmacological lipid alterations on the formation of NETs (see Neumann et al.15).
HPTLC is a relatively cost-effective and simple method to analyze a broad range of lipids in a large number of samples. This method has been used in many research areas, including antibiotic quantification25, lipid storage in lysosomal storage diseases26, and the determination of cholesterol and cholesterylglucoside levels in epithelial cells27. The method described here was modified and optimized to isolate lipids from a purified neutrophil population; however, a slightly modified version can also be used to isolate lipids from tissue samples (see Brogden et al.23). Using this method, lipids can also be effectively separated, identified, and semi-quantified against a known standard. A critical step in this method is the usage of the respective named buffer, since the usage of any other buffer or medium may lead to lipid contamination and unspecific lipid bands in the samples. Since sensitivity is limited with HPTLC, HPLC should be used as a more accurate method for absolute quantification.
HPLC facilitates the quantification and identification of cholesterol and its various forms or derivatives by performing a comparison to a known lipid. By using the protocol described above, it is possible to reliably quantify down to 0.0016 mg/mL cholesterol (Figure 4A), with a linear ratio up to 10 mg/mL (R = 0.990). The HPTLC method enables cholesterol detection down to 0.05 mg/mL, but with a sigmoid curve and a lower correlation coefficient of R = 0.906 (Figure 4B). The higher sensitivity and higher correlation coefficient thus makes HPLC a much superior method for the exact quantification of lipids, which subsequently enables smaller differences between the samples to be detected. However, both methods can be used in combination; HPTLC can be used to gain an overview into which lipids may be altered, and HPLC may be used in a more targeted approach to quantify the difference of a specific lipid in a given sample. When comparing our measurements with the values obtained by others28,29, less cholesterol was quantified in total neutrophils. This might be explained by the methodology used. Other studies used a fluorimetric detection, which is based on an enzyme-coupled reaction that detects both free cholesterol and cholesterylesters.
Here, HPTLC and HPLC are used in combination to verify that MβCD leads to a significant reduction of cholesterol as also previously shown by Gorudko et al.28. They demonstrated a 60% reduction of cholesterol in neutrophils by 10 mM MβCD. Additionally, a slight reduction of sphingomyelin level but not cholesterol esters was detectable in the neutrophils when treated with MβCD (see Neumann et al.15). At the same time, the depletion of cholesterol leads to the formation of NETs (see Neumann et al.15). These finding correlate well with the phenomenon that treatment of neutrophils with statins, inhibitors of the 3-hydroxy 3-methylglutaryl coenzyme A (HMG-CoA) reductase, the rate-limiting enzyme in cholesterol biosynthesis, boosts formation of NETs. However, since statin treatment of neutrophils exhibits stronger NET induction compared to MβCD treatment (see Neumann et al.15 and Chow et al.20), additional effects of statins e.g. on prenylated membrane-anchored effector proteins might also be involved in formation of NETs.
The formation of NETs is a relatively novel described host defense mechanism. The extracellular DNA fibers have now been described not only in mammals30,31, but also in chicken, fish, shrimps, and plants32,33,34,35. Upon stimulation of the neutrophils with pathogens or other stimuli, NETs are released into the extracellular space, where they entrap and possibly kill invading pathogens36. Studying the lipid composition of an activated neutrophil may help to understand the cellular mechanisms mediating its antimicrobial activity. Since we measured only the total lipid level of the neutrophils, it remains to be determined how the lipid content differs and is affected by MβCD in whole cell versus plasma membranes and among different membrane micro domains. However, this requires specific membrane separation procedures as previously described (Xu et al.).37
Besides chromatin, human NETs contain several neutrophil proteins, such as MPO; elastase, the antimicrobial peptide LL-37; or calgranulin17,36. Recent research has focused heavily on the role of those specific proteins and morphological changes, which initiate and facilitate the formation of NETs15,38,39. However, so far, there is only limited knowledge about the importance of certain lipids during this process15,20. Therefore, this is a particularly interesting area to be explored in more detail in the future. Finally, knowledge on lipid membrane modifications could serve as a basis for therapeutic approaches to boost the immune system against infections. As an example, it was shown that the depletion of cholesterol might help in fighting antibiotic-resistant pathogens such as H. pylori, since it was demonstrated that cholesterol enhances the resistance of those bugs against antibiotics and antimicrobial peptides40. Similar studies might be helpful for understanding host-pathogen interactions with different bacterial species.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by a fellowship of the Akademie für Tiergesundheit (AfT) and a fellowship from the PhD program, “Animal and Zoonotic Infections,” of the University of Veterinary Medicine, Hannover, Germany, provided to Ariane Neumann.
Materials
| Neutrophil isolation, NET staining and quantification | |||
| Alexa Flour 633 goat anti-rabbit IgG | Invitrogen | A-21070 | |
| Anti-MPOα antibody | Dako | A0398 | |
| BSA | Sigma-Aldrich | 3912-100G | |
| Marienfeld-Neubauer improved counting chamber | Celeromics | MF-0640010 | |
| Confocal microscope TCS SP5 AOBS with tandem scanner | Leica | DMI6000CS | |
| Dulbecco´s PBS 10X | Sigma-Aldrich | P5493-1L | Dilute 1:10 in water for 1X working solution |
| Dy Light 488 conjugated highly cross-absorbed | Thermo Fisher Scientific | 35503 | |
| Excel | Microsoft | 2010 | |
| DNA/Histone 1 antibody | Millipore | MAB3864 | |
| Image J | NIH | 1.8 | http://imagej.nih.gov/ij/ |
| Light microscope | VWR | 630-1554 | |
| Methyl-β-cyclodextrin | Sigma-Aldrich | C4555-1G | |
| PFA | Carl Roth | 0335.3 | dissolve in water, heat up to 65 °C and add 1N NaOH to clear solution |
| PMA | Sigma-Aldrich | P8139-1MG | Stock 16 µM, dissolved in 1X PBS |
| Poly-L-lysine | Sigma-Aldrich | P4707 | |
| Polymorphprep | AXIS-SHIELD | AN1114683 | |
| ProLong Gold antifade reagent with DAPI | Invitrogen | P7481 | |
| Quant-iT PicoGreen dsDNA Reagent | Invitrogen | P7581 | |
| RPMI1640 | PAA | E 15-848 | |
| HBSS with CaCl and Mg | Sigma | H6648 | |
| Triton X-100 | Sigma-Aldrich | T8787-50ml | |
| Trypanblue | Invitrogen | 15250-061 | 0.4% solution |
| Water | Carl Roth | 3255.1 | endotoxin-free |
| Name | Company | Catalog Number | Comments |
| Lipid isolation and analysis | |||
| 1-propanol | Sigma-Aldrich | 33538 | |
| 10 µl syringe | Hamilton | 701 NR 10 µl | |
| Diethyl ether | Sigma-Aldrich | 346136 | |
| Ethyl acetate | Carl Roth | 7336.2 | |
| Canullla 26G | Braun | 4657683 | |
| Copper(II)sulphatepentahydrate | Merck | 1027805000 | |
| Chloroform | Carl Roth | 7331.1 | |
| CP ATLAS software | Lazarsoftware | 2.0 | |
| Chromolith HighResolution RP-18 endcapped 100-4.6 mm column | Merck | 152022 | |
| High Performance Liquid Chromatograph Chromaster | Hitachi | HITA 892-0080-30Y | Paramaters are dependent on individual HPLC machine |
| HPLC UV Detector | Hitachi | 5410 | |
| HPLC Column Oven | Hitachi | 5310 | |
| HPLC Auto Sampler | Hitachi | 5260 | |
| HPLC Pump | Hitachi | 5160 | |
| Methanol | Carl Roth | 7342.1 | |
| n-Hexane | Carl Roth | 7339.1 | |
| Phosphoric acid | Sigma-Aldrich | 30417 | |
| Potassium chloride | Merck | 49,361,000 | |
| Potters | LAT Garbsen | 5 ml | |
| SDS | Carl Roth | CN30.3 | |
| HPTLC silica gel 60 | Merck | 105553 | |
| Vacufuge plus basic device | Eppendorf | 22820001 | |
| Corning Costar cell culture 48-well plate, flat bottom | Sigma | CLS3548 | |
| Coverslip | Thermo Fisher Scientific | 1198882 | |
| Glass slide | Carl Roth | 1879 | |
| BD Tuberculin Syringe Only 1 ml | BD Bioscience | 309659 |
References
- Riethmuller, J., Riehle, A., Grassme, H., Gulbins, E. Membrane rafts in host-pathogen interactions. Biochim Biophys Acta. 1758 (12), 2139-2147 (2006).
- Shahbazian, H., Atrian, A., Yazdanpanah, L., Lashkarara, G. R., Zafar Mohtashami, A. Anti-inflammatory effect of simvastatin in hemodialysis patients. Jundishapur J Nat Pharm Prod. 10, e17962 (2015).
- Bavari, S., et al. Lipid raft microdomains: A gateway for compartmentalized trafficking of Ebola and Marburg viruses. J Exp Med. 195, 593-602 (2002).
- Zaas, D. W., Duncan, M., Rae Wright, ., J, S. N., Abraham, The role of lipid rafts in the pathogenesis of bacterial infections. Biochim Biophys Acta. 1746 (3), 305-313 (2005).
- Rohde, M., Muller, E., Chhatwal, G. S., Talay, S. R. Host cell caveolae act as an entry-port for group A streptococci. Cell Microbiol. 5 (5), 323-342 (2003).
- Grassme, H., et al. Host defense against Pseudomonas aeruginosa requires ceramide-rich membrane rafts. Nat Med. 9 (3), 322-330 (2003).
- Heung, L. J., Luberto, C., Del Poeta, M. Role of sphingolipids in microbial pathogenesis. Infect Immun. 74 (1), 28-39 (2006).
- Gatfield, J., Pieters, J. Essential role for cholesterol in entry of mycobacteria into macrophages. Science. 288 (5471), 1647-1650 (2000).
- Rapini, R. P., Bolognia, J. L., Jorizzo, J. L. . Dermatology 2-Volume Set. , (2007).
- Tamilselvam, B., Daefler, S. Francisella targets cholesterol-rich host cell membrane domains for entry into macrophages. J Immunol. 180 (12), 8262-8271 (2008).
- Garner, M. J., Hayward, R. D., Koronakis, V. The Salmonella pathogenicity island 1 secretion system directs cellular cholesterol redistribution during mammalian cell entry and intracellular trafficking. Cell Microbiol. 4 (3), 153-165 (2002).
- Gilk, S. D., et al. Bacterial colonization of host cells in the absence of cholesterol. PLoS Pathog. 9 (1), e1003107 (2013).
- Tuong, Z. K., et al. Disruption of Rorα1 and cholesterol 25-hydroxylase expression attenuates phagocytosis in male Roralphasg/sg mice. Endocrinology. 154 (1), 140-149 (2013).
- Bryan, A. M., Farnoud, A. M., Mor, V., Del Poeta, M. Macrophage cholesterol depletion and its effect on the phagocytosis of Cryptococcus neoformans. J Vis Exp. (94), (2014).
- Neumann, A., et al. Lipid alterations in human blood-derived neutrophils lead to formation of neutrophil extracellular traps. Eur J Cell Biol. 93 (8-9), 347-354 (2014).
- Brinkmann, V., et al. Neutrophil extracellular traps kill bacteria. Science. 303 (5663), 1532-1535 (2004).
- von Köckritz-Blickwede, M., Nizet, V. Innate immunity turned inside-out: antimicrobial defense by phagocyte extracellular traps. J Mol Med. 87 (8), 775-783 (2009).
- Fuchs, T. A., et al. Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 176 (2), 231-241 (2007).
- Lauth, X., et al. M1 Protein Allows Group A Streptococcal Survival in Phagocyte Extracellular Traps through Cathelicidin Inhibition. J Innate Immun. 1 (3), 202-214 (2009).
- Chow, O. A., et al. Statins Enhance Formation of Phagocyte Extracellular Traps. Cell Host Microbe. 8 (5), 445-454 (2010).
- Simons, K., Vaz, W. L. Model systems, lipid rafts, and cell membranes. Annu Rev Biophys Biomol Struct. 33, 269-295 (2004).
- Bligh, E. G., Dyer, W. J. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 37, 911-917 (1959).
- Brogden, G., Propsting, M., Adamek, M., Naim, H. Y., Steinhagen, D. Isolation and analysis of membrane lipids and lipid rafts in common carp (Cyprinus carpio L). Comp Biochem Physiol B Biochem Mol Biol. 169, 9-15 (2014).
- Saldanha, T., Sawaya, A. C., Eberlin, M. N., Bragagnolo, N. HPLC separation and determination of 12 cholesterol oxidation products in fish: comparative study of RI, UV, and APCI-MS detectors. J Agric Food Chem. 54 (12), 4107-4113 (2006).
- Hubicka, U., Krzek, J., Woltynska, H., Stachacz, B. Simultaneous identification and quantitative determination of selected aminoglycoside antibiotics by thin-layer chromatography and densitometry. J AOAC Int. 92 (4), 1068-1075 (2009).
- Maalouf, K., et al. A modified lipid composition in Fabry disease leads to an intracellular block of the detergent-resistant membrane-associated dipeptidyl peptidase IV. J Inherit Metab Dis. 33 (4), 445-449 (2010).
- Correia, M., et al. Helicobacter pylori’s cholesterol uptake impacts resistance to docosahexaenoic acid. Int J Med Microbiol. 304 (3-4), 314-320 (2014).
- Gorudko, I. V., et al. Lectin-induced activation of plasma membrane NADPH oxidase in cholesterol-depleted human neutrophils. Arch Biochem Biophys. 516 (2), 173-181 (2011).
- Masoud, R., Bizouarn, T., Houée-Levin, C. Cholesterol: A modulator of the phagocyte NADPH oxidase activity – A cell-free study. Redox Biol. 3, 16-24 (2014).
- Knight, J. S., et al. Peptidylarginine deiminase inhibition is immunomodulatory and vasculoprotective in murine lupus. J Clin Invest. 123 (7), 2981-2993 (2013).
- Reichel, M., et al. Harbour seal (Phoca vitulina) PMN and monocytes release extracellular traps to capture the apicomplexan parasite Toxoplasma gondii. Dev Comp Immunol. 50 (2), 106-115 (2015).
- Chuammitri, P., et al. Chicken heterophil extracellular traps (HETs): novel defense mechanism of chicken heterophils. Vet Immunol Immunopathol. 129, 126-131 (2009).
- Palic, D., Ostojic, J., Andreasen, C. B., Roth, J. A. Fish cast NETs: Neutrophil extracellular traps are released from fish neutrophils. Dev Comp Immunol. 31 (8), 805-816 (2007).
- Ng, T. H., Chang, S. H., Wu, M. H., Wang, H. C. Shrimp hemocytes release extracellular traps that kill bacteria. Dev Comp Immunol. 41 (4), 644-651 (2013).
- Hawes, M. C., et al. Extracellular DNA: the tip of root defenses?. Plant Sci. 180 (6), 741-745 (2011).
- Brinkmann, V., Zychlinsky, A. Neutrophil extracellular traps: is immunity the second function of chromatin?. J Cell Biol. 198 (5), 773-783 (2012).
- Xu, T., et al. Lipid raft-associated β-adducin is required for PSGL-1-mediated neutrophil rolling on P-selectin. J Leukoc Biol. 97 (2), 297-306 (2015).
- Ermert, D., et al. Mouse neutrophil extracellular traps in microbial infections. J Innate Immun. 1 (3), 181-193 (2009).
- Metzler, K. D., Goosmann, C., Lubojemska, A., Zychlinsky, A., Papayannopoulos, V. A myeloperoxidase-containing complex regulates neutrophil elastase release and actin dynamics during NETosis. Cell Rep. 8 (3), 883-896 (2014).
- McGee, D. J., et al. Cholesterol enhances Helicobacter pylori resistance to antibiotics and LL-37. Antimicrob Agents Chemother. 55 (6), 2897-2904 (2011).

