In Vitro Polymerization of F-actin on Early Endosomes
Summary
Early endosome functions depend on F-actin polymerization. Here, we describe a microscopy-based in vitro assay that reconstitutes the nucleation and polymerization of F-actin on early endosomal membranes in test tubes, thus rendering this complex series of reactions amenable to biochemical and genetic manipulations.
Abstract
Many early endosome functions, particularly cargo protein sorting and membrane deformation, depend on patches of short F-actin filaments nucleated onto the endosomal membrane. We have established a microscopy-based in vitro assay that reconstitutes the nucleation and polymerization of F-actin on early endosomal membranes in test tubes, thus rendering this complex series of reactions amenable to genetic and biochemical manipulations. Endosomal fractions are prepared by floatation in sucrose gradients from cells expressing the early endosomal protein GFP-RAB5. Cytosolic fractions are prepared from separate batches of cells. Both endosomal and cytosolic fractions can be stored frozen in liquid nitrogen, if needed. In the assay, the endosomal and cytosolic fractions are mixed, and the mixture is incubated at 37 °C under appropriate conditions (e.g., ionic strength, reducing environment). At the desired time, the reaction mixture is fixed, and the F-actin is revealed with phalloidin. Actin nucleation and polymerization are then analyzed by fluorescence microscopy. Here, we report that this assay can be used to investigate the role of factors that are involved either in actin nucleation on the membrane, or in the subsequent elongation, branching, or crosslinking of F-actin filaments.
Introduction
In higher eukaryotic cells, proteins and lipids are internalized into early endosomes where sorting occurs. Some proteins and lipids, which are destined to be reutilized, are incorporated into tubular regions of the early endosomes and then transported to the plasma membrane or to the trans-Golgi network (TGN)1,2. By contrast, other proteins and lipids are selectively packaged into regions of the early endosomes that exhibit a multivesicular appearance. These regions expand and, upon detachment from early endosomal membranes, eventually mature into free endosomal carrier vesicles or multivesicular bodies (ECV/MVBs), which are responsible for cargo transport towards late endosomes1,2.
Actin plays a crucial role in the membrane remodeling process associated with endosomal sorting capacity and endosome biogenesis. Protein sorting along the recycling pathways to the plasma membrane or to the TGN depends on the retromer complex and the associated proteins. This sorting machinery seems to be coupled to the formation of the recycling tubules via interactions of the retromer complex, with the WASP and Scar homologue (WASH) complex and branched actin3,4,5. In contrast, molecules destined for degradation, particularly activated signaling receptors, are sorted into intraluminal vesicles (ILVs) by the endosomal sorting complexes required for transport (ESCRT)2,6,7. While the possible role of actin in the ESCRT-dependent sorting process is not known, F-actin plays an important role in the biogenesis of ECV/MVBs and in transport beyond early endosomes. In particular, we found that annexin A2 binds cholesterol-enriched regions of early endosome, and together with spire1, nucleates F-actin polymerization. The formation of the branched actin network observed on endosomes requires the branching activity of the actin-related protein (ARP) 2/3 complex, as well as the ERM protein moesin and the actin-binding protein cortactin8,9.
Here, we describe a microscopy-based in vitro assay that reconstitutes the nucleation and polymerization of F-actin on early endosomal membranes in test tubes. This assay has been used previously to investigate the role of annexin A2 in F-actin nucleation and of moesin and cortactin in the formation of endosomal actin networks8,9. With this in vitro protocol, the complex series of reactions that occur on endosomes during actin polymerization become amenable to the biochemical and molecular analysis of the sequential steps of the process, including actin nucleation, linear polymerization, branching, and crosslinking.
Protocol
1. Solutions and Preparations
NOTE: All buffers and solutions should be prepared in double-distilled (dd) H2O. Because the hydration state of sucrose varies, the final concentration of all sucrose solutions must be determined using a refractometer.
- Prepare phosphate-buffered saline without divalent cations (PBS-): 137 mM NaCI, 2.7 mM KCl, 1.5 mM KH2PO4, and 6.5 mM Na2HPO4. Adjust the pH to 7.4. Sterilize by autoclaving (1 cycle at 121 °C for 15 min) and store at room temperature.
- Prepare stock solution of 300 mM imidazole: 0.204 g of imidazole in 10 mL of ddH2O.Adjust the pH to 7.4 at 4 °C using HCl. Filter-sterilize using a 0.22 μm pore size filter and store at 4 °C.
- Prepare homogenization buffer (HB; prepare approximately 100 mL): 250 mM sucrose in 3 mM imidazole. Adjust the pH to 7.4 at 4 °C using a pH-meter. Filter-sterilize using a 0.22 μm pore size filter and store at 4 °C. Complement the HB just before use with a cocktail of protease inhibitors at final concentrations corresponding to 10 mM leupeptin, 1 mM pepstatin A, and 10 ng/mL aprotinin.
- For the step floatation gradients, prepare 62%, 39%, and 35% sucrose solutions in 3 mM imidazole, pH 7.4, as described below. Precisely determine the final concentration of all sucrose solutions using a refractometer.
- Prepare 100 mL of 62% sucrose solution in 3 mM imidazole, pH 7.4. Dissolve by stirring 80.4 g of sucrose in ddH2O pre-warmed to 35 °C. Add 1 mL of 300 mM imidazole stock solution and fill to 100 mL with ddH2O. Adjust the pH to 7.4 at 4 °C. Filter-sterilize using a 0.22 μm pore size filter and store at 4 °C.
- Prepare 100 mL of 35% sucrose solution in 3 mM imidazole, pH 7.4. Dissolve by stirring 40.6 g of sucrose in ddH2O. Add 1 mL of 300 mM imidazole stock solution and fill to 100 mL with ddH2O. Adjust the pH to 7.4 at 4 °C. Filter-sterilize using a 0.22 μm pore size filter and store at 4 °C.
- Prepare 50 mL of 39% sucrose in 3 mM imidazole, pH 7.4. Dilute the 62% sucrose solution with the 35% sucrose solution. Store at 4 °C.
- Dissolve 3 g of paraformaldehyde (PFA) by stirring in 100 mL of PBS- pre-warmed to 60 °C while adding 1 M NaOH dropwise; adjust the final pH to 7.4 using NaOH. Filter-sterilize using a 0.22-μm pore size filter and store at -20 °C.
CAUTION: 3% PFA is a toxic reagent. - Prepare 1 M KCl stock solution. Dissolve 3.73 g of KCl by stirring in 50 mL of ddH2O. Filter-sterilize using a 0.22-μm pore size filter and store at room temperature.
- Prepare 50X concentrated stock solution containing 0.625 M Hepes at pH 7.0 and 75 mM MgOAc2in ddH2O. Aliquot and store at -20 °C.
- Prepare mounting medium. Mix by stirring 2.4 g of poly(vinyl alcohol) Mw ~31,000 (PVA) with 6 g of glycerol. Add 6 mL of ddH2O and leave for at least 2 h at room temperature. Add 12 mL of 0.2 M Tris-Cl (pH 8.5) and heat to approximately 53 °C with occasional stirring until the PVA has dissolved.
- Clarify the solution by centrifugation at 5,000 x g for 20 min at room temperature. Collect the supernatant and add 2.5% (w/v) 1,4-diazabicyclo-[2.2.2]octane (DABCO) to prevent photobleaching during fluorescence detection. Vortex for about 30 s to dissolve. Aliquot into 1.5-mL plastic tubes and store at -20 °C.
2. Cell Culture
- Culture HeLa cells in Dulbecco's Modified Eagle Medium supplemented with 10% fetal calf serum, 10% non-essential amino acids, 10% L-glutamine, and 1% penicillin-streptomycin. Maintain them at 37 °C in a 5% CO2 incubator.
- Transfect the cells 24 h before the experiment with GFP-RAB510, using commercial transfection reagent according to the manufacturer's instructions.
- When needed, prepare endosomal fractions and cytosol from cells depleted of a protein of interest (i.e., using RNAi or CRISPR/Cas9) and/or overexpressing a wildtype or mutant form of the protein of interest.
3. Endosomal Fraction Preparation
NOTE: This protocol describes the straightforward preparation of subcellular fractions containing endosomes and other light membranes. If needed, purified endosome fractions can also be used11. Prepare 2 Petri dishes (10-cm outer diameter; 57 cm2) of confluent cells expressing GFP-RAB5 as starting material. The total number of confluent HeLa cells in 2 Petri dishes corresponds roughly to 2.5 x 107 cells. When needed, the endosomes can also be prepared from cells depleted of a protein of interest (i.e., using RNAi or CRISPR/Cas9) and/or overexpressing a wildtype or mutant form of the protein of interest, always expressing GFP-RAB5.
CAUTION: All steps of the fractionation protocol should be performed on ice.
- Place the Petri dishes onto a wet metal plate in a flat ice bucket and immediately wash the cells twice with 5 mL of ice-cold PBS-.
- Remove all PBS- from the last wash and add 3 mL of ice-cold PBS- per dish. Cells should not dry during handling: if necessary, place the ice bucket on a rocking platform to adequately cover cells with fluid.
- Mechanically remove the cells in PBS from the Petri dishes using a flexible rubber policeman (i.e., homemade cell scraper). Scrape the cells off the dishes first in a quick, circular motion around the outside of the dish, followed by a downward motion in the middle of the dish. Scrape gently to obtain "sheets" of attached cells. Gently transfer the cells using a plastic Pasteur pipette with a wide opening to a 15-mL conical polypropylene tube on ice.
- Centrifuge for 5 min at 175 x g and 4 °C.
- Gently remove the supernatant (SN) and add 1 – 3 mL of HB to the pellet. Using a plastic Pasteur pipette with a wide opening, gently pipette up and down once to resuspend the cells.
- Centrifuge for 7 – 10 min at 1,355 x g and 4 °C. Gently remove the SN.
- Perform cell homogenization.
NOTE: Conditions should be gentle so that the plasma membranes of the cells are broken but the endosomes remain intact.- Add a known volume of HB with protease inhibitors onto each cell pellet (approximately 100 μL per cell pellet). Using a 1,000-µL micropipette, gently pipette up and down until the cells are resuspended. Do not introduce air bubbles.
- Prepare a 1-mL tuberculin syringe with a 22G needle pre-rinsed with HB and free of air or bubbles.
- Fill the syringe with the cell suspension relatively slowly (to avoid creating air bubbles), place the beveled tip of the needle against the wall of the tube, and expel the cell suspension rapidly to shear off the plasma membranes of the attached cells.
- Take a small aliquot of the homogenate (approximately 3 µL) and dilute it into 50 µL of HB on a glass slide. Mix and cover with a glass coverslip. Inspect the homogenate under a phase-contrast microscope equipped with a 20X or 40X objective.
NOTE: Under optimal conditions, most cells are broken, but nuclei, which appear as dark grey and round structures, are not (Figure 1). - Repeat step 3.7.3 and 3.7.4 until most cells are broken; usually, 3 – 6 up-and-down strokes through the needle are necessary. Keep a small aliquot (10 – 30 µL) of the homogenate for protein determination.
- Centrifuge homogenate for 7 min at 1,355 x g and 4 °C.
NOTE: After centrifugation, the pellet (defined as the nuclear pellet; NP) contains the nuclei and the supernatant (defined as the postnuclear supernatant; PNS) contains the cytosol and the organelles released upon homogenization and in free suspension. - Carefully collect the PNS. Keep small aliquots (10 – 30 µL) of the PNS and NP for protein determination and discard the NP.
- Adjust the PNS to 40.6% sucrose by diluting the 62% sucrose solution with PNS using a 1:1.1 ratio of PNS:62% sucrose. Mix gently but thoroughly, without creating air bubbles. Check the sucrose concentration using the refractometer.
- Place the PNS in 40.6% sucrose at the bottom of an ultracentrifugation tube. Overlay carefully with 2.5 mL of the 35% sucrose solution and fill the centrifuge tube with HB.
NOTE: The sucrose solutions should be layered so that interfaces are clearly visible. - Ultracentrifuge the gradients for 1 h at ̴165,000 x g and 4 °C.
- Carefully remove the white layer of lipids on top of the gradient. Next, collect the interface containing endosomes, between the HB and 35% sucrose, using a 200-μL micropipette with a cut tip.
NOTE: The endosomal fractions can be used directly in the in vitro actin polymerization assay or can be aliquoted on ice, flash-frozen in liquid nitrogen, and stored at -80 °C. - Determine the protein concentration of the PNS and endosomal fractions.
NOTE: This concentration should be at least 100 µg/mL. Approximately 2.5 x 107 cells grown in 2 Petri dishes are needed to obtain a PNS with at least 100 µg/mL (see the Note above).
4. Cytosol Preparation
NOTE: Prepare 2 Petri dishes (10 cm diameter; 57 cm2) of confluent HeLa cells as starting material.When needed, the cytosol can also be prepared from cells depleted of a protein of interest (i.e., using RNAi or CRISPR/Cas9) and/or overexpressing a wildtype or mutant form of the protein of interest. As for the endosome fractionation protocol, all steps should be performed on ice.
- Repeat steps 3.1 – 3.8.
- Place the PNS in a centrifuge tube and ultracentrifuge for 45 min at ̴250,000 x g and 4 °C.
- Carefully remove the white layer on top and collect the SN (cytosol fraction) without disturbing the microsome pellet. Keep an aliquot of the cytosol fraction for protein determination.
NOTE: The cytosol can be used directly in the in vitro actin polymerization assay or can be aliquoted on ice, flash-frozen in liquid nitrogen, and stored at -80 °C. - Determine the protein concentration of the cytosol.
NOTE: It should be at least 3 mg/mL.
5. Assay Measuring Endosome-dependent Actin Polymerization In Vitro
NOTE: Endosome-dependent actin polymerization can be performed using two alternative approaches. In the first approach, the materials are mixed in a test tube, fixed, and transferred to a coverslip and analyzed. In the second approach, described here, the assay can be carried out directly in the imaging chamber and can be fixed and analyzed without mechanical perturbation (Figure 2C). In this second approach, the assay mixture is not transferred from a test tube to a coverslip and thus remains unperturbed, decreasing the danger of F-actin networks being physically perturbed during the transfer. In addition, this second approach is compatible with a time-lapse analysis of F-actin polymerization. It should be noted that no difference in the analysis of F-actin polymerization was observed with either approach.
NOTE: Use cut tips throughout the protocol.
- In a test tube
- Gently mix ice-cold purified GFP-RAB5 endosomes (step 3) with cytosol (step 4) at a ratio of 1:10 (protein concentration) in an ice-cold 1.5-mL conical test tube.
NOTE: A typical experiment is carried out with approximately 40 – 50 µL. - Adjust the ice-cold reaction mixture to final concentrations of 125 mM KCl (using the 1 M KCl stock solution), 12.5 mM Hepes, and 1.5 mM MgOAc2 (using the 50x stock solution). Then, adjust the mixture with protease inhibitors to final concentrations of 10 mM leupeptin, 1 mM pepstatin A, and 10 ng/mL aprotinin. Gently mix all components.
- Place the test tube containing the reaction mixture at 37 °C, without shaking, and incubate for the desired time.
NOTE: Typically, it will take approximately 3 min to detect actin polymerization on endosomes and 30 min for the formation of an extensive network. - At the desired time (i.e., 3 min or 30 min), stop the reaction by placing the test tube on ice and add 1/10 (vol:vol) of the precooled 3% PFA solution.
- Add 0.3:10 (vol:vol) of 200 units/mL (~6.6 μM) phalloidin conjugated to red-orange dye to stain the polymerized actin.
- Place 12 μL of the mixture onto 12 μL of PVA mounting medium on a micro glass slide. Put an 18 x 18 mm2 glass coverslipon top.
- Analyze the sample with confocal microscopy.
- Gently mix ice-cold purified GFP-RAB5 endosomes (step 3) with cytosol (step 4) at a ratio of 1:10 (protein concentration) in an ice-cold 1.5-mL conical test tube.
- In the imaging chamber
- To make microscopic imaging chambers, use plasma cleaner for 1 min to clean a round 18-mm diameter coverslip and a round 35 mm diameter Petri dish equipped with a 20 mm glass bottom (0.16-0.19 mm).
- Incubate the cleaned surfaces with 1% β-casein for 20 min to minimize protein binding to the glass. Wash twice with 3 mM imidazole.
- Add endosome and cytosol, in the same assay mixture as in steps 5.1.1 and 5.1.2, to a precooled 1.5 mL test tube and complement the assay mixture with 0.1 μg/μL rhodamine-actin.
- Adjust the final refractive index of the mixture to 1.375 (26.5% sucrose) using 39% sucrose solution.
NOTE: Typically, the same volume is added; however, this has to be empirically re-adjusted depending upon the sucrose concentration of the collected fraction, determined using a refractomer, since the sucrose concentration may slightly change from experiment to experiment. - Place the mixture on the pretreated 35-mm dish (step 5.2.1) and cover it with the pretreated 18-mm coverslip (step 5.2.1).
- Place the chamber at 37 °C, without shaking.
- Analyze the sample using fluorescence confocal microscopy.
6. Image Acquisition and Analysis of Actin Network
- Image acquisition
- Acquire the images on an inverted scanning confocal microscope.
NOTE: Image acquisition settings were optimized for an inverted scanning confocal microscope with a 63X 1.25 NA oil objective. Adjust the laser power (usually around 50%) to achieve a good signal-to-noise ratio.
- Acquire the images on an inverted scanning confocal microscope.
- Quantification of the actin network
- Analyze the fluorescent micrographs. Use the opensource software CellProfiler (v2.1.1)12 to measure the amount of actin per endosome (alternative software can be used).
- First, identify endosomes as the primary object using the GFP-RAB5 signal. Then, quantify the F-actin associated with individual endosomes using the propagation of the actin signal (red-orange dye conjugated Phalloidin) as a secondary object.
- Average and normalize the number of associated material for each object to the control condition.
- Analyze the fluorescent micrographs. Use the opensource software CellProfiler (v2.1.1)12 to measure the amount of actin per endosome (alternative software can be used).
Representative Results
To gain insights into the formation of F-actin patches on early endosome membranes, we followed the protocol outlined in Figure 2. Briefly, cells were transfected with GFP-RAB5 and then early endosomes were prepared by subcellular fractionation. These purified early endosomes were incubated with cytosol in order to provide actin itself as well as other factors possibly involved in the reaction. At the end of the incubation period, the reaction was stopped by fixation of the mixture. A sample was then collected and deposited on a microscopic slide. Finally, F-actin was revealed with phalloidin (Figure 3A). The nucleation of F-actin onto early endosomal membranes as well as the subsequent polymerization process occurred rapidly. Indeed, F-actin could already be visualized on endosomes within 1min after the beginning of the reaction (Figure 3A). These actin structures, which were initially short, rapidly became longer, branched and linked to one another leading to the formation of an interwoven network of actin filaments (Figure 3A), presumably reflecting unbalanced actin dynamics in vitro. Similar nucleation and polymerization rates were observed when the assay was carried out with purified rhodamine-labeled actin in addition to cell cytosol. This was done in glass-bottomed dishes so that structures could be imaged directly, in the absence of any perturbation (Figure 3B). In the assay described here, early endosomes nucleate de novo actin polymerization selectively. Indeed, actin polymerization did not occur when the in vitro reaction was carried out in the absence of either endosomes or cytosol. It can thus be concluded that early endosomes possess the fundamental ability to support the nucleation and subsequent polymerization of actin filaments9,11.
To analyze the amount of actin polymerized from endosomes, images acquired at different times were analyzed using CellProfiler (Figure 4A). The amount of actin polymerized per endosome was measured as the area of actin surrounding a single early endosome. The actin polymerization rate was higher at the beginning of the process and decreased with time (Figure 4B). The actin network could be analyzed in more detail at early time-points after the beginning of the reaction. At these early time-points, individual actin-containing structures could be still be differentiated from each other. In order to evaluate the complex organization of actin networks, the number of actin filaments emanating from each endosome was counted (i.e., primary filaments). Similarly, the number of actin filaments originating from primary filaments (i.e., secondary filaments) were counted, as well as the number of all other actin filaments that could be identified in the networks and that did not originate from endosomes (i.e., other filaments) (Figure 4C-E. Once the actin filaments were traced, other parameters, such as length of individual actin filaments and number of endosomes connected to the same actin filaments, could be easily calculated (Figure 4F).
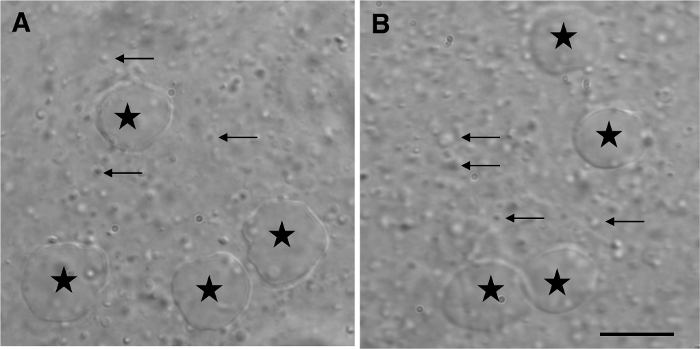
Figure 1: Phase Contrast Micrograph of a HeLa Cell Homogenate. After homogenization, the extract was mounted between a microscopic slide and a coverslip. For publication, micrographs were captured with a 100X objective after oil immersion. For routine inspection, however, the cell extract was visualized under a 20X or a 40X objective without oil immersion. Two examples of high-magnification views of the homogenate are shown (A-B). Nuclei (star) appear as round structures dark gray in color, without attached cell remnants. Arrows point to the characteristic granules of varying size, shape, and translucence, which are released upon cell homogenization and correspond to intracellular materials (i.e., organelles). Scale bar = 10 µm. Please click here to view a larger version of this figure.
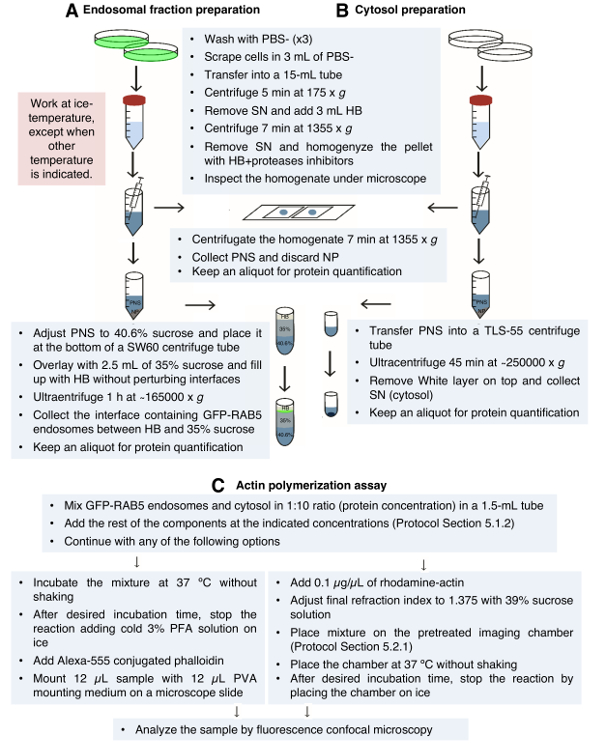
Figure 2: Schematic Representation of the Protocol. Schematic description of the protocols used to prepare the endosomal extracts (A) and cytosol extracts (B) and to monitor actin polymerization from endosomes in vitro (C). Supernatant (SN), homogenization buffer (HB), postnuclear supernatant (PNS), nuclear pellet (NP), and paraformaldehyde (PFA). Please click here to view a larger version of this figure.
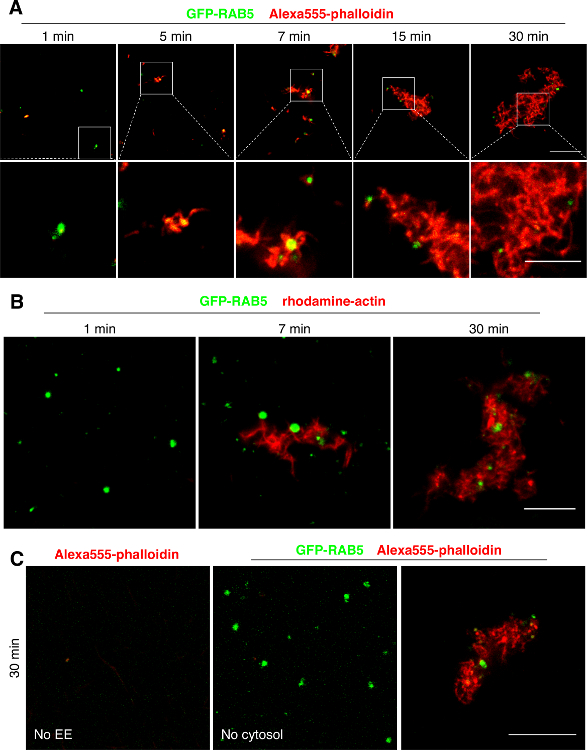
Figure 3: Nucleation and Polymerization of F-actin on Early Endosomes In Vitro. (A) Endosomes purified from Hela cells expressing GFP-Rab5 (green) and HeLa cytosol were prepared separately. In the assay, early endosomes (EEs) were mixed with cytosol, and the mixture was incubated for the indicated times. The mixture was then fixed, labeled with phalloidin (red) to label F-actin, and analyzed by fluorescence confocal microscopy.Bars: 10 µm (top panel) and 5 µm (bottom panel). (B) EEs purified from Hela cells expressing GFP-Rab5 (green) and HeLa cytosol were prepared separately. These cell extracts were incubated with purified rhodamine-actin (red) for the indicated times in microscope chambers and were analyzed by confocal microscopy. Scale bar = 10 µm. (C) The experiment was performed as in A. HeLa cytosol (without EEs) and HeLa endosomes (green) labeled with GFP-Rab5 (without cytosol) were incubated separately (left and middle panels) or together (right panel) for 30 min, fixed, labeled with phalloidin (red) to label F-actin, and analyzed by fluorescence confocal microscopy. Scale bar = 10 µm. Please click here to view a larger version of this figure.
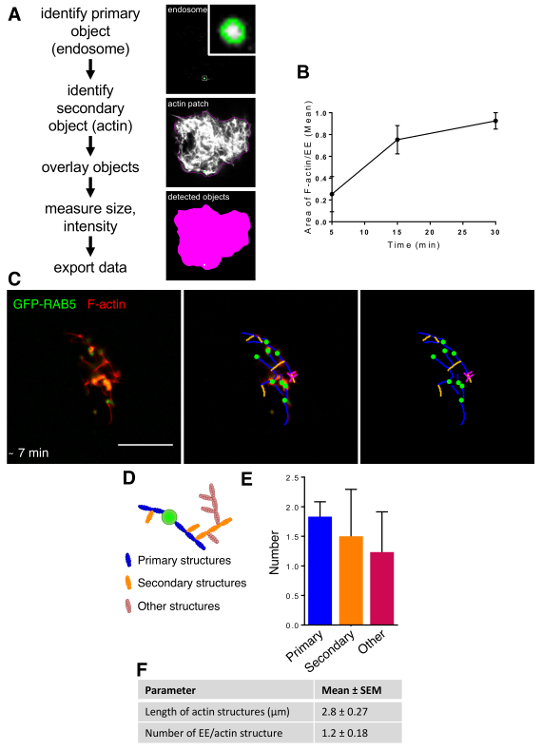
Figure 4: Analysis of the Actin Network. (A) Workflow of CellProfiler analysis. (B) Number of polymerized actin were quantified relative to the number of endosomes using the CellProfiler software. Data are median ± s.e.m. (n= 3). Data are means ± s.d. (n = 3). 5 min versus 15 min (ns P= 0.0736), 5 min versus 30 min (* P = 0.0195), and 15 min versus 30 min (ns P = 0.3129). (C) Early endosomes (EEs) purified from Hela cells expressing GFP-Rab5 and HeLa cytosol were prepared separately, and the assay was carried out as in Figure 2A. Samples were fixed after 7 min. The middle panel shows the same micrograph, with the overlap of the traces of F-actin structures drawn using the NeuronJ plugin13 of the ImageJ software. The right panel shows the traces only. Scale bar = 10 µm. (D)Representation of the F-actin structure numbering sequence used in the quantification with ImageJ software. (E) Quantification of the number of actin filaments, as defined in C-D. Data are means ± s.d. (n = 3). Primary branches versus secondary branches (ns P = 0.5262), secondary branches versus other branches (ns P = 0.6815), and primary branches versus other branches (ns P = 0.2254). (F) The length of F-actin structures and the number of EEs per actin structure are shown as examples of other parameters than can be analyzed with the quantification defined in C-D. Please click here to view a larger version of this figure.
Discussion
Actin plays a crucial role in endosome membrane dynamics4,14. We previously reported that actin nucleation and polymerization occur on early endosomes, forming small F-actin patches or networks. These F-actin networks are absolutely required for membrane transport beyond early endosomes along the degradation pathway. Interfering at any step of this nucleation and polymerization process prevents endosome maturation and thus downstream transport towards late endosomes andlysosomes9,11,15. In particular, we used the assay described above to analyze the sequential steps of F-actin polymerization on early endosomes and to characterize the process at the molecular level.
In these experiments, early endosomes are labeled with GFP-RAB5 in vivo, purified by subcellular fractionation, and incubated in a test tube in the presence of cytosol to allow for F-actin polymerization in vitro. At the desired time, the reaction is stopped and the reaction mixture is transferred onto a microscopic slide for analysis by fluorescence microscopy, using phalloidin to label the F-actin. Using this experimental strategy, we showed that annexin A2 is necessary for the nucleation of F-actin on early endosomes, together with SPIRE1, while ARP2/3 mediates F-actin branching, as expected. We also showed that moesin and cortactin are required for the formation of the F-actin networks that mediate ECV/MVB biogenesis and maturation along the degradation pathway of mammalian cells9,11.
It is important to keep in mind that, although low levels of ectopically expressed RAB5 do not alter endosomes to any significant extent10, this small GTPase is a key regulator of early endosome membrane dynamics16,17. In some experiments, it may therefore be appropriate to label endosomes in the assay using alternative protocols. For example, early endosomes can be conveniently labeled using endocytosed fluorescent tracers, such as epidermal growth factor or transferrin4,14.
It should also be noted that it is technically difficult to image individual actin filaments with conventional confocal microscopy. Therefore, we refer to F-actin structures throughout the text to describe actin networks on endosomes. As a consequence, we feel that the formation of an F-actin network can be quantified more accurately by measuring the fluorescence surface area of the networks rather than their intensity.
It is not easy to completely rule out the possibility that the actin nucleation activity observed in vitro is mediated by some other structure or organelle that remains bound to RAB5 endosomes during purification. However, such contamination is highly unlikely. While some organelles co-purify on gradients because of similar physical properties, they do not tend to remain quantitatively bound to each other after purification. Moreover, the actin nucleation activity strictly depends on annexin A24,14, which is present on early endosomes containing RAB514. Finally, we find that F-actin structures are associated selectively with RAB5 endosomes in vivo and are not found on other endosomes or on other cellular membranes4,14. In conclusion, actin nucleation activity is most likely associated with RAB5 endosomes, since this activity co-purifies and co-localizes with RAB5 endosomes.
In this assay, much like in other assays that reconstitute the complex biochemical reactions that control the dynamics of endosomes16 or other membranes18 and that are performed in a test tube, it is important to ensure that the preparation of the cellular extracts is optimal. Cells should be homogenized under gentle conditions to limit damage to endosomes and other organelles, particularly nuclei and lysosomes. Also, endosome fractions should be kept on ice under conditions that minimize osmotic and ionic stress, as well as spontaneous actin polymerization. In additional, relatively high concentrations of endosome fraction (≥0.1 mg/mL) and cytosol (≥3 mg/mL) are required for adequate F-actin polymerization on endosomes and detection, as indicated in steps 3.13 and 4.4, respectively. Similarly, it is important to carry out the assay with a 1:10 ratio of endosome to cytosol (protein:protein), as indicated in step 5.1.1. Finally, pipetting after actin polymerization (i.e., to add PFA and phalloidin) should be done very gently to avoid collapsing or breaking the actin network.
If necessary, the in vitro actin polymerization assay can also be carried out directly in a microscope imaging chamber, after adding purified rhodamine-actin to the assay mixture, to limit mechanical perturbations of the actin networks polymerized onto endosomes (see step 5.2.3 and Figure 2). In this case, it is best to adjust the extract to 26.5% (0.85 M) sucrose (refractive index of 1.375), limiting diffusion and Brownian-like motion (step 5.2.3), and to remove possible aggregates of purified rhodamine-actin by ultracentrifugation before use. Our observations that moesin controls the formation of F-actin networks on endosomes were fully recapitulated using this alternative version of our assay4,14.
Either version of our protocol, in the test tube or in the imaging chamber, is optimal for a global analysis of F-actin nucleation and polymerization on endosomes. However, it is not easy to follow the actin polymerization process through time using this approach, in part because endosomes are not immobile. Consequently, it is not easy to monitor the growth of individual filaments and thus to determine the history of the actin network. For example, it can be difficult to discriminate F-actin branching from crosslinking. However, we are confident that conditions to monitor F-actin polymerization in real time can be improved in the imaging chamber by optimizing the conditions to immobilize endosomes. Similarly, it will be interesting to monitor F-actin polymerization from artificial membranes using purified components. In fact, we previously showed that F-actin nucleation and polymerization can occur on liposomes after binding the nucleation factor annexin A2 onto the liposome bilayer11. This strategy will be useful to further dissect the role of specific lipids and proteins in the F-actin nucleation and polymerization process and to characterize the minimal components necessary to form an F-actin network on endosomal membranes. Finally, we believe that this strategy will be very useful to study other F-actin-dependent processes that occur on endosome membranes, particularly the recruitment and mobilization of the retromer/WASH machinery.
Disclosures
The authors have nothing to disclose.
Acknowledgements
Support was received from the Swiss National Science Foundation; the Swiss Sinergia program; the Polish-Swiss Research Programme (PSPB-094/2010); the NCCR in Chemical Biology; and LipidX from the Swiss SystemsX.ch initiative, evaluated by the Swiss National Science Foundation (to J. G.). O. M. was supported by an EMBO long-term fellowship (ALTF-516-2012).
Materials
| NaCl | Sigma-Aldrich | 71380 | |
| KCl | Acros Organics | 196770010 | |
| KH2PO4 | AppliChem | A1042 | |
| Na2HPO4 | Acros Organics | 424370025 | |
| Hepes | AppliChem | A3724 | |
| Magnesiun acetate tetrahydrate | Fluka | 63047 | |
| Dithiothreitol (DTT) | AppliChem | A2948 | |
| Imidazole | Sigma-Aldrich | 10125 | |
| NaOH | Fluka | 71690 | |
| Sucrose | Merck Millipore | 107687 | |
| Leupeptin | Roche | 11017101001 | |
| Pepstatin | Roche | 10253286001 | |
| Aprotinin | Roche | 10236624001 | |
| Paraformaldehide | Polysciences. Inc | 380 | |
| Alexa Fluor 555 phalloidin | Molecular Probes | A34055 | |
| Actin rhodamine | Cytoskeleton. Inc | APHR-A | |
| Mowiol 4-88 | Sigma-Aldrich | 81381 | poly(vinyl alcohol), Mw ~31 000 |
| DABCO | Sigma-Aldrich | D-2522 | |
| Tris-HCl | AppliChem | A1086 | |
| β-casein | Sigma-Aldrich | C6905 | |
| Filter 0.22um | Millex | SL6V033RS | |
| Round 10cm dishes for cell culture | Thermo Fisher Scientific | 150350 | |
| Plastic Pasteur pipette | Assistent | 569/3 40569003 | |
| 15-ml polypropylen tube | TPP | 91015 | |
| Hypodermic Needle 22G Black 30mm | BD Microlance | 300900 | |
| Sterile Luer-slip 1ml Syringes without needle | BD Plastipak | 300013 | |
| Micro glass slides | Assistent | 2406 | |
| 18X18-mm glass coverslip | Assistent | 1000/1818 | |
| SW60 centrifuge tube | Beckman coulter | 344062 | |
| TLS-55 centrifuge tube | Beckman coulter | 343778 | |
| 200-μl yellow tip | Starlab | S1111-0706 | |
| 1000-μl Blue Graduated Tip | Starlab | S1111-6801 | |
| 1.5-ml test tube | Axygen | MCT-175-C 311-04-051 | |
| 18-mm diameter round coverslip | Assistent | 1001/18 | |
| 35-mm diameter round dish with a 20-mm glass bottom (0.16-0.19 mm) | In vitro Scientific | D35-20-1.5-N | |
| Refractometer | Carl Zeiss | 79729 | |
| Plasma cleaner | Harrick Plasma | PDC-32G | |
| Sorvall WX80 Ultracentrifuge | Thermo Fisher Scientific | 46900 | |
| Tabletop ultracentrifuge | Beckman coulter | TL-100 | |
| SW60 rotor | Beckman coulter | 335649 | |
| TLS-55 rotor | Beckman coulter | 346936 | |
| Confocal microscopy | Carl Zeiss | LSM-780 | |
| Fugene HD transfection reagent | Promega | E2311 | |
| Protein assay reagent A | Bio-Rad | 500-0113 | |
| Protein assay reagent B | Bio-Rad | 500-0114 | |
| Protein assay reagent S | Bio-Rad | 500-0115 | |
| Cell scraper | Homemade | Silicone rubber piece of about 2 cm, cut at a very sharp angle and attached to a metal bar or held with forceps | |
| Refractometer | Carl Zeiss | ||
| Minimum Essential Medium Eagle (MEM) | Sigma-Aldrich | M0643 | |
| FCS | Thermo Fisher Scientific | 10270-106 | |
| MEM Non-Essential Amino Acids (NEAA) | Thermo Fisher Scientific | 11140-035 | |
| L-Glutamine | Thermo Fisher Scientific | 25030-024 | |
| Penicillin-Streptomycin | Thermo Fisher Scientific | 15140-122 | |
| pH Meter 691 | Metrohm | ||
| ImageJ software | NIH, Bethesda MD |
References
- Huotari, J., Helenius, A. Endosome maturation. EMBO J. 30 (17), 3481-3500 (2011).
- Scott, C. C., Vacca, F., Gruenberg, J. Endosome Maturation, Transport and Functions. Semin. Cell Dev Biol. 31, 2-10 (2014).
- Puthenveedu, M. A., et al. Sequence-dependent sorting of recycling proteins by actin-stabilized endosomal microdomains. Cell. 143 (5), 761-773 (2010).
- Seaman, M. N., Gautreau, A., Billadeau, D. D. Retromer-mediated endosomal protein sorting: all WASHed up!. Trends Cell Biol. 23 (11), 522-528 (2013).
- Burd, C., Cullen, P. J. Retromer: a master conductor of endosome sorting. Cold Spring Harb Perspect Biol. 6 (2), a016774 (2014).
- Woodman, P. G., Futter, C. E. Multivesicular bodies: co-ordinated progression to maturity. Curr Opin Cell Biol. 20 (4), 408-414 (2008).
- Henne, W. M., Buchkovich, N. J., Emr, S. D. The ESCRT pathway. Dev Cell. 21 (1), 77-91 (2011).
- Morel, E., Gruenberg, J. Annexin A2 binding to endosomes and functions in endosomal transport are regulated by tyrosine 23 phosphorylation. J Biol Chem. 284 (3), 1604-1611 (2009).
- Muriel, O., Tomas, A., Scott, C. C., Gruenberg, J. Moesin and cortactin control actin-dependent multivesicular endosome biogenesis. Mol Biol Cell. 27 (21), 3305-3316 (2016).
- Sonnichsen, B., De Renzis, S., Nielsen, E., Rietdorf, J., Zerial, M. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J Cell Biol. 149 (4), 901-914 (2000).
- Morel, E., Parton, R. G., Gruenberg, J. Annexin A2-dependent polymerization of actin mediates endosome biogenesis. Dev Cell. 16 (3), 445-457 (2009).
- Kamentsky, L., et al. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics. 27 (8), 1179-1180 (2011).
- Meijering, E., et al. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 58 (2), 167-176 (2004).
- Granger, E., McNee, G., Allan, V., Woodman, P. The role of the cytoskeleton and molecular motors in endosomal dynamics. Semin Cell Dev Biol. 31, 20-29 (2014).
- Mayran, N., Parton, R. G., Gruenberg, J. Annexin II regulates multivesicular endosome biogenesis in the degradation pathway of animal cells. EMBO J. 22 (13), 3242-3253 (2003).
- Gorvel, J. P., Chavrier, P., Zerial, M., Gruenberg, J. rab5 controls early endosome fusion in vitro. Cell. 64 (5), 915-925 (1991).
- Wandinger-Ness, A., Zerial, M. Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harb Perspect Biol. 6 (11), a022616 (2014).
- Balch, W. E., Glick, B. S., Rothman, J. E. Sequential intermediates in the pathway of intercompartmental transport in a cell-free system. Cell. 39 (3 Pt 2), 525-536 (1984).

