Protocol for the Solid-phase Synthesis of Oligomers of RNA Containing a 2′-O-thiophenylmethyl Modification and Characterization via Circular Dichroism
Summary
This article provides a detailed procedure on the solid-phase synthesis, purification, and characterization of dodecamers of RNA modified at the C2'-O-position. UV-vis and circular dichroism photometric analyses are used to quantify and characterize structural aspects, i.e., single-strands or double-strands.
Abstract
Solid-phase synthesis has been used to obtain canonical and modified polymers of nucleic acids, specifically of DNA or RNA, which has made it a popular methodology for applications in various fields and for different research purposes. The procedure described herein focuses on the synthesis, purification, and characterization of dodecamers of RNA 5'-[CUA CGG AAU CAU]-3' containing zero, one, or two modifications located at the C2'-O-position. The probes are based on 2-thiophenylmethyl groups, incorporated into RNA nucleotides via standard organic synthesis and introduced into the corresponding oligonucleotides via their respective phosphoramidites. This report makes use of phosphoramidite chemistry via the four canonical nucleobases (Uridine (U), Cytosine (C), Guanosine (G), Adenosine (A)), as well as 2-thiophenylmethyl functionalized nucleotides modified at the 2'-O-position; however, the methodology is amenable for a large variety of modifications that have been developed over the years. The oligonucleotides were synthesized on a controlled-pore glass (CPG) support followed by cleavage from the resin and deprotection under standard conditions, i.e., a mixture of ammonia and methylamine (AMA) followed by hydrogen fluoride/triethylamine/N-methylpyrrolidinone. The corresponding oligonucleotides were purified via polyacrylamide electrophoresis (20% denaturing) followed by elution, desalting, and isolation via reversed-phase chromatography (Sep-pak, C18-column). Quantification and structural parameters were assessed via ultraviolet-visible (UV-vis) and circular dichroism (CD) photometric analysis, respectively. This report aims to serve as a resource and guide for beginner and expert researchers interested in embarking in this field. It is expected to serve as a work-in-progress as new technologies and methodologies are developed. The description of the methodologies and techniques within this document correspond to a DNA/RNA synthesizer (refurbished and purchased in 2013) that uses phosphoramidite chemistry.
Introduction
Solid-phase synthesis to obtain oligonucleotides of DNA/RNA is a powerful tool that has served several applications in various fields since the 1970s1,2,3 using phosphoramidite building blocks4. Examples of its broad influence include: its impact in labeling (via click chemistry reactions)5, structural probing6, and antisense technologies7, as well as its elucidation of biological mechanisms8,9, source as genetic material10, and the study of various natural and/or chemical modifications11,12, among many others.The modification that we use here represents the first step in our efforts to obtain RNA oligonucleotides that contain photoactive probes to enable temporal control of structure and function of this important biopolymer.
The synthesis of RNA dodecamers with sequences: 5'-[CUA CGG AAU CAU]-3'/5'-[AUG AUU CCG UAG]-3' (underlined positions represent the incorporation of a C2'-O-thiophenylmethyl modification) constitutes the focus of this study. The sequences were chosen to enable the quantification and measurement of RNA strands as single strands, or as their corresponding duplex structures (no other secondary structures were predicted as thermodynamically stable). CD was used to establish the structural parameters, i.e., duplex formation and thermal denaturation transitions.
Synthesis
The overall procedure for obtaining these oligonucleotides is illustrated in Figure 1 and follows the stepwise process: automated Solid-phase Synthesis → Deprotection → Purification → Quantification → Characterization. Figure 2 displays the monomeric units that are necessary in this procedure. The solid-phase synthesis of RNA is similar to that of DNA in that it is based on phosphoramidite chemistry (Figure 2, left) and the use of base-labile protecting groups for the nucleophilic exocyclic amines on G, A and C, e.g., acetyl, benzoyl, phenoxyacetyl, t-butyl or N,N-dimethylformamide (Figure 2, right). One more aspect to consider in RNA, due to the presence of the C2'-OH group (lacking in the deoxyoligonucleotide biopolymers), is the additional step that has to be incorporated for the protection, and subsequent deprotection, of this nucleophilic position. In this respect, silicon-based protecting groups have become an attractive strategy due to their potential as biorthogonal moieties (specifically deprotected in the presence of fluoride), with the tert-butyldimethylsilyl (TBDMS) and triisopropylsilyloxymethyl (TOM) groups as popular choices (Figure 2, bottom-left).
In this work, the automated synthesis was carried out on a DNA/RNA synthesizer that uses standard phosphoramidite chemistry. The manufacturer settings on the instrument include an automated dilution step when using the commercial versions of the phosphoramidites for DNA, or the option to dilute at volumes set by the user. However, we decided to weigh the RNA phosphoramidite and dilute manually given that: 1) the price of the canonical phosphoramidites of RNA is higher (up to 50-times more expensive in some cases); 2) the modified phosphoramidites are often obtained in small amounts; and 3) the amount of wasted material upon using an automated dilution step (set by manufacturer) is large. In addition, we used: 1) commercially available solid supports (e.g., CPG) containing a protected nucleobase to function as the 3'-end; and 2) commercial phosphoramidites (canonical nucleobases) protected with a TBDMS group at the C2'-O-position. The detailed list of the synthesis steps is provided in Figure 3 and Table 1, along with further description and comments for steps that were adjusted for the RNA synthesis. Furthermore, Figure 4 illustrates the stepwise yields that are observed for every step after selecting the 'Trityl Monitor' option, which quantitates the trityl cation released from each detritylation step.
It is worth noting that typically, in our experience, the limiting factor was obtaining the phosphoramidite containing the desired modification. That is, the development of a synthetic methodology that allows for the incorporation of modifications at select sites. In this report, we focus on the incorporation of a modified nucleotide for which we have established the corresponding synthetic methodology, the C2'-O-thiophenylmethyl group. This group is small in size and does not affect the solid-phase synthesis in any manner. Since the incorporation of this group into oligonucleotides of RNA has been reported, along with structural and thermodynamic parameters4, no aspects of the organic synthesis leading to the modified phosphoramidites will be described herein.
Deprotection, Purification, and Characterization
The deprotection of the exocyclic amines and ß-cyanoethyl groups occurs in the same step as that of the cleavage from the CPG-resin. We applied the commonly used conditions of heating the obtained resin in the presence of an aqueous solution of AMA, followed by cleavage of the C2'-O-silyl groups in the presence of fluoride ions, and then purification via gel electrophoresis. While these have become standard conditions in many cases, modifications that are labile to basic conditions or fluoride ions may require milder conditions13,14, e.g., methanol/potassium carbonate (MeOH/K2CO3), or butylamine. Thus, a different set of protecting groups on the corresponding phosphoramidites is necessary. Furthermore, we chose electrophoresis as the preferred alternative to purify the deprotected oligomers given our previous experience with this method and the lack of other instrumentation. However, HPLC can alternatively be used as an effective method15. Characterization of the purified oligonucleotides was carried out via mass spectrometry, matrix assisted laser desorption/ionization-time of flight (MALDI-TOF), using a reported procedure by our group16.
Structural characterization and thermal stability of the obtained duplexes were carried out via CD. Specifically, we make use of CD to determine the thermal denaturation transitions of modified and unmodified oligonucleotides of RNA by following the decrease in ellipticity of the band at ca. 270 nm, as well as the disappearance of the band (with negative ellipticity) with a λmax at 210 nm. A spectra comparison before and after hybridization is provided to illustrate their differences and provide validation of the employed methodology. The use of CD is widely accepted in the determination of structural motifs in nucleic acids and aminoacids17, and can therefore be employed as a tool to determine various structural and thermodynamic parameters18; however, there are not many examples where the technique is used to assess thermal denaturation transitions. Some cases include the determination of thermal stabilities on DNA containing G-quadruplexes19,20 or in duplexes and hairpins of RNA21.
This report intends to provide the non-expert reader or viewer with a set of tools that enable a smooth start to this type of research. It will serve to enhance and compare with methodologies and techniques at other research laboratories that are involved in this exciting branch of science. The content in this report adds to the existing protocols of this technology from various sources, and enriches and facilitates the experience with a visual aid for each step.
Protocol
1. Solid-phase Synthesis of RNA Oligonucleotides
- Preparation of solutions containing each phosphoramidite (Table 1).
- Count the number of nucleotides and fit in the n+1 equation (where n = number of nucleotides) corresponding to each base and fill in the table with the missing values. Volume = 0.15 mL per nucleotide at a concentration of 0.1 M, dissolved in anhydrous acetonitrile.
- Weigh each phosphoramidite into oven-dried 10 mL amber bottles (Septum Top Amber 394 Amidite 13 mm ID x 20 mm OD Top) and place immediately under vacuum using a desiccator containing a drying agent.
- Fill the desiccator with dry argon, remove the bottle and dilute immediately with the corresponding amount of acetonitrile (anhydrous) before securing onto the instrument.
NOTE: Make sure to use gas-tight syringes and keep an anhydrous atmosphere at all times. - Remove the septa from the bottle and place on the machine, via a bottle change function.
NOTE: This process requires an additional 0.15 mL (hence the excess volume in the n+1 equation above) of each solution to prime the line prior to synthesis.
- Set-up of sequence and solid phase synthesis using the software (Figure 3).
- Click on the software icon (e.g., OligoNet 1.0.1) and select Instrument name>OK. Create a new synthesis by File>New Synthesis>Order.
- Fill-in: Date; RunID; select instrument; sequence name; sequence (5'-to-3' end). Under |cycle, assign the previously created method (Figure 3). Choose the end procedure, (leaves oligonucleotide bound to the CPG resin)>manual.
- Select DMT Off >(TCA treatment in the last step to obtain a 5'-OH group at the end of the synthesis); save File>Save as and provide a name for the experiment.
- Send the file to begin synthesis Order>Send order to synthesizer and select column 1-4. Open the synthesizer window and select trityl monitor|choose function|trity>monitor every step.
- Repeat the steps as necessary depending on the number of oligonucleotides to be synthesized at once (note that all orders must have the same method, i.e., same coupling times and sequence of events).
- Begin the synthesis Synthesizer>Prepare to start. Place the columns with the desired 3'-end on the positions indicated on the instrument. On the instrument click Start>No (for ABI preparation).
- Once the synthesis is complete, remove the column from the instrument and place in a round bottom flask followed by drying under reduced pressure for about 0.5 h.
NOTE: It is recommended to check that a deep orange color is observed in random couplings (steps 1.2.4 – 1.2.7) to avoid potential errors from the trityl monitor function.
- Deprotection and purification of RNA oligonucleotides.
- Open the column by twisting the black cap and transfer all (or half, depending on the need or purpose) of the white resin into a 1.6 mL centrifuge tube. Add 0.5 mL of a methylamine (40% in water)/ammonia (40% in water) at 1:1.
- Secure the centrifuge tube cap with parafilm and, using a heat-block, heat to 60 °C for 1.5 h.
NOTE: A heavy object can be placed on top of the tube to ensure that the ammonia concentration remains constant inside the reaction tube. - Remove from heat block and cool slowly to room temperature. Briefly centrifuge the sample to spin down contents, then transfer the supernatant to a new centrifuge tube.
- Freeze the samples by submerging tubes in liquid nitrogen (or dry ice/ethanol bath) and concentrate to dryness while spinning in a centrifuge under reduced pressure.
- Re-suspend solids in 0.4 mL of a triethylamine/N-methylpyrrolidinone/triethylamine-trihydrofluoride (2:2:3 ratio) solution. Heat to 60 °C for 1.5 h followed by slow cooling to room temperature.
- Add 0.04 mL of a NaOAc solution (3M, pH 5.5 – adjusted with HCl) followed by gentle mixing with a pipette tip. Add ethanol (1 mL) and cool to ca. -70 °C (dry ice/ethanol bath) for 15 min.
- Centrifuge at 15,000 rpm and 4 °C for 10 min. Use a pipette to extract the aliquot and dry the resulting pellet under reduced pressure.
- Re-suspend the obtained solid in loading buffer (0.2 mL, 90% aq. formamide, 1 mM EDTA) and mix until mixture is homogeneous.
- Load suspension onto a previously prepared polyacrylamide gel (20% denaturing, dimensions of well: 30 cm x 1 mm)22.
- Apply a current through the gel until the bromophenol blue marker is located between 1/2-2/3 down the gel (gel dimensions: ~ 21.5 cm x 30 cm).
- Remove the gel from the stand and transfer the gel contents from the glass to plastic wrap (covered on both sides), and place over a thin layer chromatography (TLC) plate covered with silica (containing fluorescent dye at 254 nm) to visualize the bands using a UV-lamp (λmax = 254 nm).
- Use a marker to delineate the position where the upper band is located and cut it using a new razor blade. Place gel containing the RNA (without plastic wrap) into a 50-mL conical tube and crush to small pieces using a glass rod.
- Suspend the gel residues into a NaCl aq. solution (200 mM NaCl and 1 mM EDTA) and shake the suspension at 37 °C for 12 h. Centrifuge the conical tube for 10 min.
- Desalt by using a reverse-phase C18 cartridge.
- Prepare the cartridge using a 10-mL syringe by washing with:
Acetonitrile (10 mL)
H2O (twice, 10 mL)
5 mM NH4OAc aq. solution (3 mL)
Solution containing RNA, taking care not to pour any of the gel residuals. - Wash with H2O (three times, 10 mL).
- Elute from the column using a 60% aq. methanol solution (3 mL)
- Prepare the cartridge using a 10-mL syringe by washing with:
- Concentrate under reduced pressure and re-dissolve in RNase-free water (0.3 mL)
- Prepare a dilute solution (10 µL) and deposit 1 µL on the UV-vis instrument (e.g., Nanodrop) to measure the UV-vis spectrum (200-450 nm).
- Use Beer Lambert's Law to determine the concentration of the obtained solution:
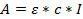
where A = obtained absorbance; ε = calculated molar extinction coefficient; c = concentration, and l = 0.1 for a path length of 1 mm. - Calculate the molar extinction coefficients for the oligonucleotides; calculated here with an on-line calculator that uses DINAMelt software (http://unafold.ra.albany.edu/?q=dinamelt)23
- Concentration determination and characterization via MALDI-TOF.
- Load samples onto a MALDI plate using a pipette tip loaded with a C18 tip to desalt, and spot each oligonucleotide.
- Wash the tip with 50% acetonitrile (10 µL x 2). Equilibrate the tip with 0.1% trifluoroacetic acid (TFA; 10 µL x 2). Load tip with sample (typically 100-150 pmol).
- Wash the tip with 0.1% TFA (10 µL x 2), and then with water (10 µL x 2).
- Elute the sample into a solution containing the desired matrix; we used a solution of: 10 µL 25 mM-2,4,6-trihydroxyacetophenone monohydrate (THAP), 10 mM ammonium citrate, and 300 mM ammonium fluoride in 50% acetonitrile.
- Spot directly onto the MALDI plate by depositing 0.9 µL (followed by air drying) and repeating the procedure as necessary.
NOTE: All spectra were obtained on the reflector positive-mode.
- Load samples onto a MALDI plate using a pipette tip loaded with a C18 tip to desalt, and spot each oligonucleotide.
2. RNA Structure Analysis via CD
- Preparation of solutions for CD.
- Prepare 0.25 mL containing the modified RNA [3 µM], NaCl [10 mM], sodium phosphate buffer [10 mM, pH 7.2], and MgCl2 [5 mM]. The sample is ready for analysis as is, particularly if it represents a unimolecular transition.
- If the goal is to analyze duplex structures or bimolecular transitions, add complement (1 molar equivalent, if applicable) at this time, or continue with the step below.
- Place the sample on a heat-block, pre-heated to 90 °C, and turn-off the heat to control slow cooling to room temperature (typically 2 – 4 h).
- Spectra acquisition
- Prepare a blank solution (NaCl 10 mM, sodium phosphate 10 mM pH 7.3, MgCl2 5 mM), transfer to a micro-cuvette (1 cm path length, 250 µL minimum volume), and place on a holder position of the sample changer within the instrument.
- Transfer the sample containing RNA into another micro-cuvette and position in sample changer. If measuring a thermal denaturation transition, carefully add a bed of oil without disrupting the aqueous layer and secure the cap of cuvette using a piece of Teflon tape.
- Open the nitrogen tank to provide a flow that moves the air floater located on the instrument to ca. 40. Turn-on the cooler.
- Turn-on the instrument and open the software Spectra Manager icon, then open the acquisition window Spectra Measurement.
- Purge the instrument with nitrogen for 5 min before acquisition.
- Acquire the spectra using the following parameters Measure>Parameters and adjust the parameters as follows:
- Under General, select: Scan 200-350 nm; Channels CD and HT: Data pitch at 0.1 nm; scanning speed at 100 nm/min; band width at 1 nm; number of accumulations at 5.
- Under cell unit, choose 20 °C. Under control, select Shutter is opened and closed automatically. Under information, choose the name, concentration, operator.
- Under data, browse to desired folder. Click OK.
- Acquire the spectra Measure>Sample measurement, identify position(s) of cuvettes in sample changer 1-6 accordingly, click OK.
- If taking a thermal denaturation transition:
- Close Acquisition software File>Exit and open a new program Variable Temperature Measurement>Measure>Parameters.
- Apply the following parameters to record a thermal transition, which can be adjusted as desired:
- Under Temperature, select start temp: 4 °C; interval: 0.2; target: 90 °C; gradient: 1 °C/min; wait 0 s. Under Start/End, select Start condition & end condition as desired.
- Under General, select 3 Channels, CD/HT/Abs; bandwidth: 1 nm; wavelength: 270 nm (or as desired). Under Control, select none. Under Information, select name, concentration, operator.
- Under data, browse to desired folder. Click OK.
- Cool samples to 4 °C and wait 5 min at this temperature before obtaining a spectrum measurement.
- Data work-up for spectra.
- Using the function on the software, subtract the blank spectrum from the target acquisition to account for background signals arising from the buffer system in use: Spectra Analysis>File>Open.
- Once a blank file and a spectra file have been opened, drag View 2 under View 1 (on left hand side of screen).
- Click on Processing>subtraction>OK or exchange data>OK.
- Extract data as an ASCII file: File>Export and select ASCII.
- Use software to plot the corresponding spectrum. Smoothening of data is optional in cases where the signal-to-noise ratio is lower than expected. This transformation can be applied by applying the smoothening data option.
- Data work-up for thermal denaturation transitions.
- Extract data from the JASCO file as an ASCII file.
- Plot the ellipticity points as a function of temperature. Typically, smoothening of the data is necessary depending on the wavelength that is being examined. In some instances, the use of the wavelength at 210 nm displayed larger variations between plots that required smoothening. However, depending on the sample and concentrations, a number of calculations were carried out using the raw data.
- To calculate the thermal denaturation transitions (Tm) value, obtain a first derivative of the curve. The maxima or minima were an indication of this parameter. As in the previous step, some cases required smoothening of the data to obtain the most accurate value.
NOTE: Experiments were typically carried out in triplicate, which resulted in the corresponding average and standard deviation for the measurement.
Representative Results
The synthesis of RNA dodecamers containing zero, one, or two 2-thiophenylmethyl modifications at the C2'-O-position is described along with its corresponding purification and characterization. Furthermore, a detailed description of the structural analysis that was carried out via CD is included.
The four strands of RNA (including a strand with a complementary sequence) were obtained via solid-phase synthesis, which was followed by a purification yield between 300 – 700 nmol of each oligonucleotide. Mass spectrometry was carried out by desalting ~150 pmol of each sample and then depositing on a plate for MALDI-TOF analysis (Figure 5). Quantification was carried out via ultraviolet photometrical analysis of each solution, while CD was used to identify the formation of duplex structures and record their corresponding Tm. No clear difference is observed between canonical and modified oligonucleotides, via UV-vis spectroscopy, whether comparing single-stranded samples or duplex structures. However, minor changes are observed upon measurement of their CD spectra (Figure 6). In addition, Tm measurements of the three duplex structures displayed a distinct value that was indicative of the destabilization induced by the incorporation of the 2'-O-thiophenylmethyl modification on one of the strands.
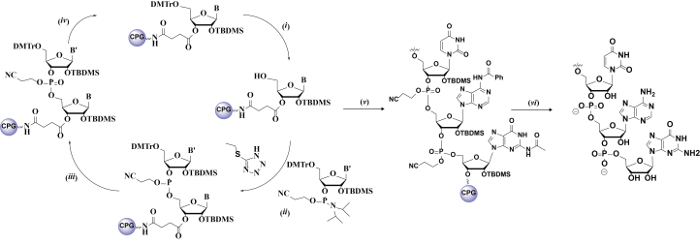
Figure 1: Procedure to Obtain RNA Strands. Solid-phase cycle using CPG as the solid support and 5-ethylthiotetrazole as the activator: (i) detritylation; (ii) coupling; (iii) oxidation; (iv) capping and onto new cycle; to yield the corresponding oligonucleotide (v) supported on the CPG resin and containing all of the protecting groups; and its subsequent deprotection in the presence of base and fluoride (vi) to yield the final RNA oligonucleotide for further analysis. Please click here to view a larger version of this figure.
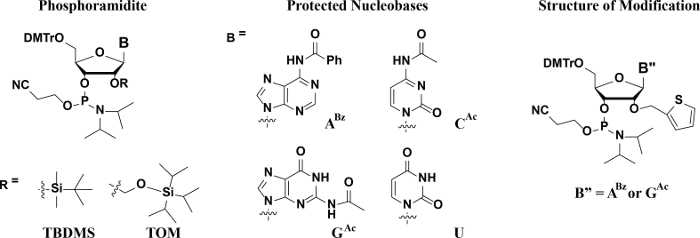
Figure 2: Structures of Phosphoramidites and Protecting Groups. The chemical structure of the phosphoramidite containing silyl based C2'-O-groups and base labile groups on the exocyclic amines of A, G, and C. The O-TBDMS group was used in this study. Please click here to view a larger version of this figure.

Figure 3: Stepwise Synthesis of the Corresponding Oligonucleotides. The cycle was edited as shown and a key for the used codes and flow rates is shown on the right. The stepwise synthesis was pasted from the software that is provided. Please click here to view a larger version of this figure.
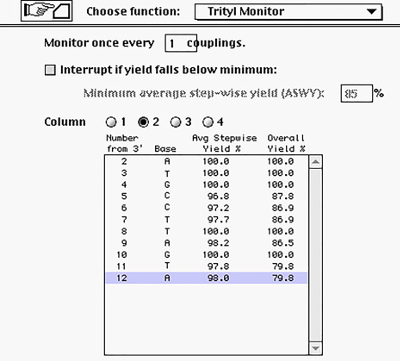
Figure 4: Representation of Obtained Yields and Tracking of Individual Couplings. The shown example has been adapted from the synthesis of the complement to display a typical oligonucleotide synthesis. Please click here to view a larger version of this figure.
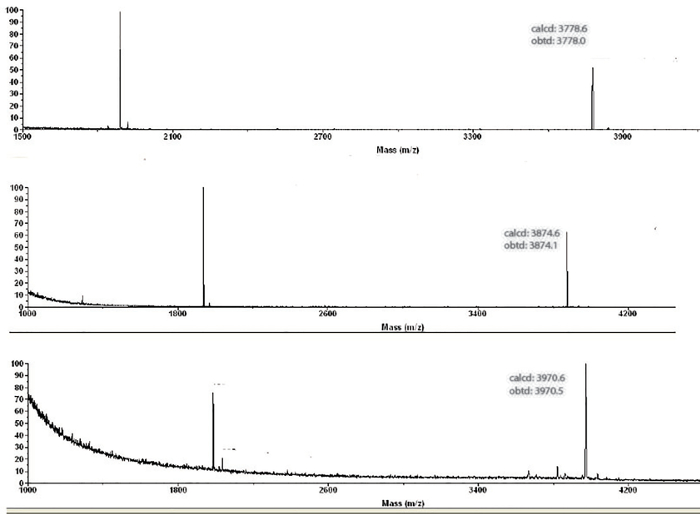
Figure 5: MS of Oligonucleotides (ON) 1-3. MALDI-TOF of ON 1–3 (top to bottom). Please click here to view a larger version of this figure.
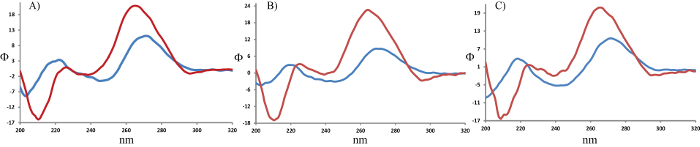
Figure 6: CD spectra of Single- and Double-stranded RNA, Canonical and Modified. CD spectra of samples containing zero (1, A), one (2, B), or two (3, C) modifications before (blue) and after (red) hybridization. Please click here to view a larger version of this figure.
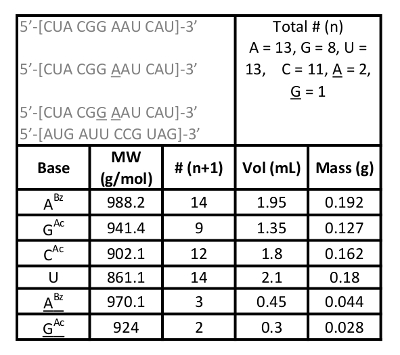
Table 1: Phosphoramidite Solutions Calculation.
Discussion
The intent of this manuscript is to serve as a guide to researchers in the field, beginner or expert, to successfully achieve or enhance the synthesis of oligonucleotides of DNA or RNA. The described methodology focuses on the use of solid-phase synthesis using an automated DNA/RNA synthesizer via standard phosphoramidite chemistry. The report describes a step-by-step depiction of the synthesis, purification, and characterization of RNA dodecamers. In addition, the use of CD is employed to identify secondary structural motifs and thermal denaturation transitions.
It is important to note that this work can be adaptable to other circumstances, i.e., various brands of equipment, modifications, protecting groups, and/or reagents in the solid-phase synthesis. Therefore, improvement may be achieved upon changing one or more of the many parameters that are involved in this process or through adjustments in the reaction conditions. In addition, the detailed structural analysis provided herein (via CD) is expected to provide an alternate approach to researchers in this field, typically carrying out analogous analyses via UV-vis.
The calculations to determine the masses of the phosphoramidites for the solid phase synthesis were based on the sequences shown below, and the four oligonucleotides were prepared and purified on the same run (underlined positions indicate the presence of a 2'-O-thiophenylmethyl modification). All calculations and values are included in Table 1
1 5'-[CUA CGG AAU CAU]-3'
2 5'-[CUA CGG AAU CAU]-3'
3 5'-[CUA CGG AAU CAU]-3'
4 5'-[AUG AUU CCG UAG]-3'
Perhaps the most important aspect in handling RNA pertains to its susceptibility toward degradation by ribonucleases, and its ease to undergo hydrolysis in aqueous solutions and in the presence of metal ions24. Thus, RNase-free conditions must be enforced at all times25 as followed here: 1) all water was autoclaved in the presence of diethyl pyrocarbonate (0.1% w/v, DEPC); 2) all glassware was autoclaved, baked in an oven (150 °C, overnight), and rinsed with DEPC treated water; 3) all tubes and pipette tips were purchased from manufacturers in their RNase-free form; 4) gloves were used at all times and work was carried out in a designated hood; and 5) all surfaces and equipment were wiped constantly with RNase decontamination solutions that are available for purchase from various manufacturers. All purified RNAs were divided into small portions and stored at -20 °C or -80 °C, depending on the frequency of usage, while uncleaved or unpurified resins were stored at 20 °C. As we have pointed out previously16, oligonucleotides obtained in the manner described herein and in sizes between 10-34 nucleotides long display a higher stability than other reports26. Therefore, the reader is referred to other storage procedures when handling longer RNAs27.
The stepwise yields (calculated via the detritylation step) in the automated synthesis were between 97 – 100% and are an indication of good coupling efficiencies, particularly of those which have been modified. Overall yields of 30 – 75% were obtained after cleavage from the resin, deprotection, and purification (via gel electrophoresis), which correspond to ~300 – 750 nmol of isolated oligonucleotides. Agreeably, the incorporation of the modifications did not affect the overall yield of the RNA strands. However, while these ranges can be considered as acceptable amounts, the synthesis of oligonucleotides larger than ~ 50 nucleotides (previous, unpublished data in our laboratory) may be significantly affected by stepwise yields below 98%. Thus, certain precautions must be taken if strands larger than 50 nucleotides are desired, e.g., change the source of the acetonitrile to higher quality, minimize the time of exposure of the phosphoramidites to the atmosphere, program the instrument to dilute the phosphoramidites to a set value while using new bottles of the canonical phosphoramidites each time, use HPLC purification in place of electrophoretic analysis, and/or use milder deprotection conditions.
Mass spectra (MALDI-TOF MS) for all the oligonucleotides were carried out on an ABI 4800 Plus MALDI-TOF/TOF mass spectrometer in the positive mode. All samples were prepared using the procedure described herein and examples of the spectra for ON 1–3 are shown in Figure 5.
All experiments are typically carried out in triplicate. All spectra and experimental setup were carried out on a spectropolarimeter equipped with a Rectangular 6-Cell Holder. All glassware was washed with an RNase removal solution and rinsed thoroughly with RNase-free water. All samples were discarded following each measurement. It is important to point out that close attention should be placed on the high-tension voltage (HT is a parameter that is measured by the instrument). This can be an indication of the saturation in the signal and thus poses a threat to the accuracy and validity of the data. This parameter is sample dependent and should be kept such that the signal does not go above 500 mV at any given time. Examples for CD spectra obtained before and after hybridization of ON 1–3 are shown on Figure 6.
In addition, increasing the concentration in the sodium salts (and other buffer systems), or using other buffer systems, e.g., HEPES, or MOPS, results in increased noise below 220 nm. Therefore, since the band at 210 nm is of particular importance to follow the formation of an A-form duplex, the maximum concentration in sodium ions was kept to ~10 mM.
We also show that the use of CD provides the same parameters as those obtained from ultraviolet spectroscopy. In conclusion, we describe and illustrate the procedure to synthesize, purify, and characterize modified and unmodified RNA oligonucleotides.
Disclosures
The authors have nothing to disclose.
Acknowledgements
Preparation of this manuscript was supported via start-up funds from the University of Colorado Denver (JMRE). AF would like to acknowledge support from a Research and Creative Activities award (RaCAS, CU Denver). Funding from the Office of Research Services, University of Colorado Denver to cover publication charges is acknowledged. We would like to thank lab members Ms. Cassandra Herbert and Mr. Yannick K. Dzowo for their contributions in the video portion.
Materials
| AbsolveTM | PerkinElmer | 6NE9711 | |
| Acetonitrile 99.9%, HPLC Grade | Fisher Scientific | 75-05-8 | |
| Acetonitrile 99.9%, anhydrous for DNA sequencing | Fisher BioReagents | 75-05-8 | |
| Acrylamide, 99+% | ACROS Organics | 164850025 | |
| Ammonium chloride 98+% | Alfa Aesar | 12125-02-9 | |
| Ammonium citrate, dibasic 98% | Sigma Aldrich | 3012-65-5 | |
| Ammonium Fluoride 98.0%, ACS grade | Alfa Aesar | 12125-01-8 | |
| Ammonium hydroxide 28-30% in water, ACS Plus | Fisher Chemical | 1336-21-6 | |
| Ammonium persulfate | ACROS Organics | 1444 | |
| Argon-ultra high purity | Airgas | 7440-37-1 | |
| Bis-acrylamide Ultra pure | VWR-Amresco | 172 | |
| Boric Acid | Fisher Scientific | A73-1 | |
| Diethyl pyrocarbonate, 97% | ACROS Organics | A0368487 | |
| Ethanol, anhydrous, histological grade | Fisher Chemical | 64-17-5 | |
| Ethylenediamine tetraacetic acid, disodium salt dehydrate 100.2% | Fisher Chemical | 6381-92-6 | |
| Formamide | Thermo Scientific | 75-12-7 | |
| Hydrochloric acid, 36.5-38.0%, Certified ACS Plus | Fisher Chemical | 7647-01-0 | |
| Magnesium chloride hexahydrate, 99% | Fisher Scientific | 7786-30-3 | |
| Methanol, 99.9%, HPLC Grade | Fisher Chemical | 67-56-1 | |
| Methylamine 40% in water | Sigma Aldrich | 74-89-5 | |
| 1-Methyl-2-pyrrolidinone, andhydrous, 99.5% | Aldrich | 872-50-4 | |
| Opti-TOFTM 96 Well Insert (123 x 81 mm) | MDS SCIEX | 1020157 | |
| RNase Away | Molecular BioProducts | 7005-11 | |
| Sodium acetate, anhydrous 99.2%, Certified ACS | Fisher Chemical | 127-09-3 | |
| Sodium chloride, 100.5%, Certified ACS | Fisher Chemical | 7647-14-5 | |
| Sodium phosphate monobasic dihydrate 99.0% | Sigma | 13472-35-0 | |
| 2’,4’ Triethylamine, 99+% | Alfa Aesar | 121-44-8 | |
| TEMED | Amresco | 761 | |
| Triethylamine trihydrofluoride, 98% | Aldrich | 73602-61-6 | |
| Trifluoroacetic acid, 99% | Alfa Aesar | 76-05-1 | |
| 6’-Trihydroxyacetophenone monohydrate 98% | Sigma Aldrich | 480-66-0 | |
| Tris Base | Fisher Scientific | BP154-3 | |
| Urea | Fisher Scientific | U15-3 | |
| Reagents for the RNA synthesis: | |||
| Deblocking mix, 3% trichloroacetic acid in dichloromethane | Glen Research | 40-4140-57 | |
| Cap Mix A, THF/Pyridine/Acetic anhydride | Glen Research | 40-4110-52 | |
| Cap Mix B, 10% 1-methylimidazole in THF | Glen Research | 40-4120-52 | |
| Activator, 0.25 M 5-ethylthio-1H-tetrazole in anhydrous acetonitrile | Glen Research | 30-3140-52 | |
| Oxidizing Solution, 0.02 M iodine in THF/Pyridine/Water | Glen Research | 40-4330-52 | |
| U-RNA-CPG | Glen Research | 20-3330-xx | |
| Ac-G-RNA-CPG | Glen Research | 20-3324-xx | |
| Ac-G-CE Phosphoramidite | Glen Research | 10-3025-xx | |
| U-CE Phosphoramidite | Glen Research | 10-3030-xx | |
| Ac-C-CE Phosphoramidite | Glen Research | 10-3015-xx | |
| Bz-A-CE Phosphoramidite | Glen Research | 10-3003-xx |
References
- Matteucci, M. D., Caruthers, M. H. The synthesis of oligodeoxypyrimidines on a polymer support. Tetrahedron Lett. 21 (8), 719-722 (1980).
- Beaucage, S. L., Caruthers, M. H. Deoxynucleoside phosphoramidites-A new class of key intermediates for deoxypolynucleotides. Tetrahedron Lett. 22 (20), 1859-1862 (1981).
- Caruthers, M. H. Gene synthesis machines: DNA Chemistry and its uses. Science. 230 (4723), 281-285 (1985).
- Nguyen, J. C., et al. Synthesis, Thermal Stability, Biophysical Properties, and Molecular Modeling of Oligonucleotides of RNA Containing 2′-O-2-Thiophenylmethyl Groups. J Org Chem. 81 (19), 8947-8958 (2016).
- El-Sagheer, A. H., Brown, T. Click chemistry with DNA. Chem Soc Rev. 39, 1388-1405 (2010).
- Puffer, B., et al. 5-Fluoro pyrimidines: labels to probe DNA and RNA secondary structures by 1D 19F NMR spectroscopy. Nucleic Acids Res. 37 (22), 7728-7740 (2009).
- Wan, W. B., Seth, P. P. The medicinal chemistry of therapeutic oligonucleotides. J Med Chem. 59 (21), 9645-9667 (2016).
- Zhou, C., Greenberg, M. M. DNA Damage by histone radicals in nucleosome core particles. J Am Chem Soc. 136 (18), 6562-6565 (2014).
- Zhang, Y., et al. UV-Induced proton-coupled electron transfer in cyclic DNA miniduplexes. J Am Chem Soc. 138 (23), 7395-7401 (2016).
- Anosova, I., et al. The structural diversity of artificial genetic polymers. Nucleic Acids Res. 44 (3), 1007-1021 (2016).
- Riml, C., Micura, R. Synthesis of 5-Hydroxymethylcytidine- and 5-Hydroxymethyl-uridine-Modified RNA. Synthesis. 48, 1108-1116 (2016).
- Anderson, B. A., Hrdlicka, P. J. Merging Two Strategies for Mixed-Sequence Recognition of Double-Stranded DNA: Pseudocomplementary Invader Probes. J Org Chem. 81 (8), 3335-3346 (2016).
- Beaucage, S. L., Iyer, R. P. Synthesis of oligonucleotides by the phosphoramidite approach. Tetrahedron. 48 (12), 2223-2311 (1992).
- Gillet, L. C. J., Alzeer, J., Schärer, O. D. Site-specific incorporation of N-(deoxyguanosin-8-yl)-2-acetylaminofluorene (dG-AAF) into oligonucleotides using modified ‘ultra-mild’ DNA synthesis. Nucleic Acids Res. 33 (6), 1961-1969 (2005).
- Shiba, Y., et al. Chemical synthesis of a very long oligoribonucleotide with 2-cyanoethoxymethyl (CEM) as the 2′-O.-protecting group: structural identification and biological activity of a synthetic 110mer precursor-microRNA candidate. Nucleic Acids Res. 35 (10), 3287-3296 (2007).
- Choi, Y. J., Gibala, K. S., Ayele, T., Deventer, K. D., Resendiz, M. J. E. Biophysical properties, thermal stability and functional impact of 8-oxo-7,8-dihydroguanine on oligonucleotides of RNA – a study of duplex, hairpins and the aptamer for preQ1 as models. Nucleic Acids Res. 45 (4), 2099-2111 (2017).
- Ranjbar, B., Gill, P. Circular dichroism techniques: Biomolecular and nanostructural analyses – A review. Chem Biol Drug Des. 74 (2), 101-120 (2009).
- Mergny, J. -. L., Lacroix, L. Analysis of thermal melting curves. Oligonucleotides. 13 (6), 515-537 (2003).
- Jin, R., Breslauer, K. J., Jones, R. A., Gaffney, B. L. Tetraplex formation of a guanine-containing nonameric DNA fragment. Science. 250 (4980), 543-546 (1990).
- Virgilio, A., et al. 5-Hydroxymethyl-2′-deoxyuridine residues in the thrombin binding aptamer: Investigating anticoagulant activity by making a tiny chemical modification. CHEMBIOCHEM. 15 (16), 2427-2434 (2014).
- Chauca-Diaz, A. M., Choi, Y. J., Resendiz, M. J. E. Biophysical properties and thermal stability of oligonucleotides of RNA containing 7,8-dihydro-8-hydroxyadenosine. Biopolymers. 103 (3), 167-174 (2015).
- Maniatis, T., Fritsch, E. F., Sambrook, J. . Molecular cloning: a laboratory manual. 14, 173-185 (1987).
- Markham, N. R., Zuker, M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol. 453, 3-31 (2008).
- Breslow, R., Huang, D. -. L. Effects of metal ions including Mg2+ and lanthanides on the cleavage of ribonucleotides and RNA model compounds. Proc Natl Acad Sci U S A. 88 (10), 4080-4083 (1991).
- Blumberg, D. D. Creating a ribonuclease-free environment. Methods Enzymol. 152 (2), 20-24 (1987).
- AbouHaidar, M. G., Ivanov, I. G. Non-enzymatic RNA promoted by the combined catalytic activity of buffers and magnesium ions. Z Naturforsch C. 54 (7-8), 542-548 (1999).
- Farrell, R. E. . RNA Methodologies (4th Ed): A laboratory guide for isolation and characterization. 2, 45-80 (2010).

