CRISPR-mediated Loss of Function Analysis in Cerebellar Granule Cells Using In Utero Electroporation-based Gene Transfer
Summary
Conventional loss-of-function studies of genes using knockout animals have often been costly and time-consuming. Electroporation-based CRISPR-mediated somatic mutagenesis is a powerful tool to understand gene functions in vivo. Here, we report a method to analyze knockout phenotypes in proliferating cells of the cerebellum.
Abstract
Brain malformation is often caused by genetic mutations. Deciphering the mutations in patient-derived tissues has identified potential causative factors of the diseases. To validate the contribution of a dysfunction of the mutated genes to disease development, the generation of animal models carrying the mutations is one obvious approach. While germline genetically engineered mouse models (GEMMs) are popular biological tools and exhibit reproducible results, it is restricted by time and costs. Meanwhile, non-germline GEMMs often enable exploring gene function in a more feasible manner. Since some brain diseases (e.g., brain tumors) appear to result from somatic but not germline mutations, non-germline chimeric mouse models, in which normal and abnormal cells coexist, could be helpful for disease-relevant analysis. In this study, we report a method for the induction of CRISPR-mediated somatic mutations in the cerebellum. Specifically, we utilized conditional knock-in mice, in which Cas9 and GFP are chronically activated by the CAG (CMV enhancer/chicken ß-actin) promoter after Cre-mediated recombination of the genome. The self-designed single-guide RNAs (sgRNAs) and the Cre recombinase sequence, both encoded in a single plasmid construct, were delivered into cerebellar stem/progenitor cells at an embryonic stage using in utero electroporation. Consequently, transfected cells and their daughter cells were labeled with green fluorescent protein (GFP), thus facilitating further phenotypic analyses. Hence, this method is not only showing electroporation-based gene delivery into embryonic cerebellar cells but also proposing a novel quantitative approach to assess CRISPR-mediated loss-of-function phenotypes.
Introduction
Brain diseases are one of the most dreadful mortal diseases. They often result from genetic mutations and subsequent dysregulation. To understand molecular mechanisms of brain diseases, ever-lasting efforts to decipher the genomes of human patients have discovered a number of potential causative genes. So far, germline genetically engineered animal models have been utilized for in vivo gain-of-function (GOF) and loss-of-function (LOF) analyses of such candidate genes. Due to the accelerated development of functional validation studies, a more feasible and flexible in vivo gene assay system for studying gene function is desirable.
The application of an in vivo electroporation-based gene transfer system to the developing mouse brain is suitable for this purpose. In fact, several studies using in utero electroporation have shown their potential to conduct functional analyses in the developing brain1,2,3. Actually, several regions of the mouse brain, such as the cerebral cortex4, retina5, diencephalon6, hindbrain7, cerebellum8, and spinal cord9 have been targeted by somatic gene delivery approaches, so far.
Indeed, transient gene expression by in vivo electroporation on embryonic mouse brains has long been used for GOF analysis. Recent transposon-based genomic integration technologies further enabled long-term and/or conditional expression of genes of interest10,11, which is advantageous to dissect gene function in a spatial and temporal manner during development. In contrast to GOF analysis, LOF analysis has been more challenging. While transient transfection of siRNAs and shRNA-carrying plasmids was performed, long-term effects of LOF of genes are not guaranteed due to eventual degradation of exogenously introduced nucleic acids, such as plasmids and dsRNAs. However, the CRISPR/Cas technology provides a break-through in LOF analyses. Genes encoding fluorescent proteins (e.g., GFP) or bioluminescent proteins (e.g., firefly luciferase) have been co-transfected with CRISPR-Cas9 and sgRNAs to label the cells exposed to CRISPR-Cas9-mediated somatic mutations. Nevertheless, this approach might have limitations in functional studies on proliferating cells, since exogenous marker genes are diluted and degraded after long-term proliferation. While the transfected cells and their daughter cells undergo CRISPR-induced mutations in their genomes, their footprints might get lost over time. Thus, genetic labeling approaches would be suitable to overcome this issue.
We recently developed a CRISPR-based LOF method in cerebellar granule cells that undergo long-term proliferation during their differentiation12. To genetically label the transfected cells, we constructed a plasmid carrying a sgRNA together with Cre and introduced the plasmid into the cerebella of Rosa26-CAG-LSL-Cas9-P2A-EGFP mice13 using in utero electroporation. Unlike regular plasmid vectors encoding EGFP, this approach successfully labeled transfected granule neuron precursors (GNPs) and their daughter cells. This method provides great support in understanding in vivo function of genes of interest in proliferating cells in normal brain development and a tumor-prone background.
Protocol
All animal experiments were conducted according to animal welfare regulations and have been approved by the responsible authorities (Regierungspräsidium Karlsruhe, approval numbers: G90/13, G176/13, G32/14, G48/14, and G133/14).
1. Generate pU6-sgRNA-Cbh-Cre Plasmids
- Design the sgRNA to target a gene-of-interest according to the previously published protocol14.
NOTE: In this experiment, two sgRNAs are designed to target the mouse Top2b gene and a non-targeted control sgRNA (Table 1). To knockout gene(s) using the CRISPR technology, sgRNAs need to be designed to target either the coding sequence of the 5' region or the essential functional domain of the protein. One convenient starting point is to test publicly available pre-designed sgRNA sequences, like the GeCKO v2 mouse knockout pool library15 from the Zhang lab. Alternatively, sgRNAs can be designed using public available software such as the following website: crispr.mit.edu. - Clone the sgRNAs into the pU6-sgRNA-Cbh-Cre vector.
NOTE: The pU6-sgRNA-Cbh-Cre vector12 used in this study is derived from the original pX330 plasmid16 by replacing the coding sequence of SpCas9 with the Cre recombinase.- Synthesize two DNA oligos as follows. Sense: 5' – CACCGNNNNNNNNNNNNNNNNNNNN – 3'; Antisense: 3' – CNNNNNNNNNNNNNNNNNNNNCAAA – 5'.
NOTE: The 20 N's shown above stand for the sgRNA sequence. - Digest pU6-sgRNA-Cbh-Cre with BbsI for 30 min at 37 °C using the following reagents, load the digested sample to a 1% agarose gel, and purify them using a gel-extraction kit. Use 3 µg Plasmid; 3 µL BbsI; 1 µL FastAP; 5 µL 10X digestion buffer; add ddH2O to bring the total volume to 50 µL.
- Phosphorylate and anneal each pair of sgRNA oligos using the following reagents (10 µL total volume): 1 µL Customized sense oligo (100 mM); 1 µL Customized antisense oligo (100 mM); 1 µL 10X T4 Ligation Buffer; 6.5 µL ddH2O; 0.5 µL T4 PNK. Incubate in a thermo-cycler for 30 min at 37 °C, and 5 min at 95 °C, and then ramp down to 25 °C at 5 °C/min.
- Set up the following ligation reaction, and incubate for 10 min at room temperature (10 µL total volume): X µL BbsI digested plasmid (50 ng); 1 µL phosphorylated and annealed, oligo duplex (1:200 dilution); 5 µL 2X ligation buffer; X µL ddH2O; 1 µL quick ligase.
- Add 2 µL of the ligation product into 50 µL of chemically competent DH5α Escherichia coli cells. After 30 min of incubation on ice, heat shock the sample for 90 s at 42 °C, and incubate for 2 min on ice. Add 100 µL of LB medium and plate it onto a LB plate containing 100 µg/mL ampicillin.
NOTE: The integration of the sgRNA sequence into the pU6-sgRNA-Cbh-Cre plasmid is performed according to the cloning steps for the pX330 plasmid14. - Inoculate two colonies, each in 2 mL of LB medium at 37 °C for a minimum of 6 h, and perform a mini-prep DNA isolation using a kit.
- Sanger sequence the mini-prep DNA using the hU6-Forward primer, and identify colonies with the correct insertion of sgRNAs by aligning with the sequence shown in step 1.2.1. The primer sequence is shown in Table 1. Plasmids used in this study were named pU6-sgTop2b-1-Cbh-Cre, pU6-sgTop2b-2-Cbh-Cre, and pU6-sgControl-Cbh-Cre.
- Inoculate the correctly-identified colonies and purify the endotoxin-free plasmid DNA for in utero electroporation using a kit, e.g., from Qiagen. Elute the DNA with endotoxin-free TE-Buffer diluted in ddH2O at 1:10 (10% TE buffer). The DNA concentration of the plasmid needs to be higher than 2 mg/mL.
NOTE: Do not use a regular maxi-prep kit for DNA preparation. Store plasmid-solutions at 4 °C for regular short-term use (up to 4 weeks), or aliquot an appropriate volume of about 25 µL and keep at -20 °C for long-term storage.
- Synthesize two DNA oligos as follows. Sense: 5' – CACCGNNNNNNNNNNNNNNNNNNNN – 3'; Antisense: 3' – CNNNNNNNNNNNNNNNNNNNNCAAA – 5'.
2. Test the Efficiency of the sgRNAs Using the EGxxFP Plasmid System
NOTE: The efficiency of the sgRNAs is normally tested by Surveyor or T7E1 (T7 endonuclease I) assays. In this protocol, an easy and efficient alternative approach is used. The key of this approach is to use the pCAG-EGxxFP plasmid which contains overlapping EGFP fragments separated by a DNA sequence containing the sgRNA targeting site17. Upon the expression of pCAG-EGxxFP together with the sgRNA and Cas9 in the transfected cells, the Cas9-mediated double strand break (DSB) in the target sequence is repaired by endogenous homology-dependent mechanisms, which reconstitutes the EGFP expression cassette.
- Design primers to amplify the genomic region centered with the sgRNA targeting sequences. The PCR amplicon is approximately 500 bp. In this experiment, a DNA assembly was applied to clone a PCR-amplified fragment into the pCAG-EGxxFP plasmid.
NOTE: Alternatively, appropriate restriction sites can be added to the 5' end of the PCR primers. Available restriction sites in the pCAG-EGxxFP vector are BamHI, NheI, PstI, SalI, EcoRI, and EcoRV. - Amplify the genomic DNA with designed primers using the form and PCR parameters below. Load the sample (19.7 µL H2O; 0.3 µL of 10 ng/µL mouse genomic DNA; 2.5 µL 10 µM EGxxFP-Top2b-F; 2.5 µL 10 µM EGxxFP-Top2b-R; 25 µL 2X PCR master mix; 50 µL total volume) onto a 1% agarose gel and gel-purify the PCR fragments using a gel-extraction kit. Use the following reaction conditions:95 °C for 3 min; then 35x cycles of 98 °C for 20 s, 60 °C for 15 s, 72 °C for 20 s, and 72 °C for 1 min; 4 °C, indefinitely.
NOTE: In this experiment, one region in Top2b exon 1 was amplified; find the primer sequences in Table 1. - Digest the pCAG-EGxxFP vector with restriction enzymes BamHI and EcoRI for 2 h at 37 °C, then load the sample onto a 1% agarose gel and gel-purify the vector backbone using a gel-extraction kit. Use the following reagents (50 µL total volume): X µL Plasmid (3 µg); 2 µL BamHI; 2 µL EcoRI; 5 µL 10X Buffer; X µL ddH2O.
- Set up the DNA assembly reaction18 according to the manufacturer's instructions, and transform into DH5α competent E. coli.
- Inoculate two colonies, each in 2 mL of LB medium at 37 °C for a minimum of 6 h, perform a mini-prep DNA isolation using a kit, e.g., from Qiagen, and identify colonies with the correct insertion (hereafter referred to as pCAG-EG-Top2b-FP) using Sanger sequencing with the EGxxFP-Top2b-F primer.
- Transfect HEK293T cells.
- Seed the HEK293T cells into a 24-well plate 24 h before transfection.
NOTE: Cells were cultured in a 37 °C cell incubator with 5% CO2, in DMEM with glutamine supplement containing 10% FBS and 1% Penicillin/Streptomycin. Ensure that the cells are 60% confluent on the day of transfection. In this experiment, we used self-made HEK293T cells stably expressing SpCas9 to test pU6-sgRNA-Cbh-Cre. However, wildtype HEK293T cells can be used if an SpCas9 expression vector is provided. - Transfect the HEK293T cells using the polyethylenimine (PEI) reagent. Transfect the cells with 120 ng/well of pCAG-EG-Top2b-FP plasmid12 and 120 ng/well of pU6-sgTop2b-1-Cbh-Cre or pU6-sgTop2b-2-Cbh-Cre in a 24-well plate. Mix the plasmid DNAs and the 1.8 µL of PEI reagent in 80 µL of unsupplemented DMEM medium with glutamine supplement and incubate for 15 min at room temperature after vortexing.
- 6 h after the transfection, remove the medium and replace with fresh medium.
- Monitor the presence of live GFP+ cells 48 h after transfection using a fluorescence microscope at 20x magnification.
NOTE: The number of GFP+ cells indicates the targeting efficiency of the sgRNA. Results of effective and non-effective sgRNAs (sgTop2b-1 and sgTop2b-2, respectively) are shown in Figure 2A.
- Seed the HEK293T cells into a 24-well plate 24 h before transfection.
3. Perform In Utero Electroporation
- Breed 6 to 8-week-old mice and prepare the tools for in utero electroporation. Cross male homozygous Rosa26-CAG-LSL-Cas9-P2A-EGFP mice with female CD-1 mice. Time the pregnancy of the CD-1 mice from the day the vaginal plug is detected (E0.5). Embryos are electroporated at day E13.5.
- Pull borosilicate glass capillaries (inside diameter: 0.6-0.8 mm) with a micropipette puller. Use the following settings: P = 500, Heat = 560, Pull = 150, Vel = 75, Time = 250. Label the capillary tip with black ink for better visualization during the injection. Trim the capillary taper under a stereomicroscope using a ruler and sharp needle tweezers and trim the tip at a length of about 6 mm.
- Maintain the unity of the capillaries to keep the experiment reproducible. Create a one-sided beveled edge during the trimming. The tip should be as sharp as possible for easy penetration of the uterine wall and to avoid tissue damage of the embryo.
NOTE: Alternatively, use a micro grinder to adjust a 35° angle19. The capillaries can be stored at room temperature in a 10 cm Petri dish under pathogen-free conditions. - Autoclave the surgical instruments.
- Mix the sgRNA-plasmids in equal parts with pT2K-IRES-Luc to a final concentration of at least 1 µg/µL for each plasmid and color the plasmid-solution with fast green (final concentration of 0.05%). Prepare 1% fast green stock solution with endotoxin-free distilled water and filter through a 0.22 µm filter to remove dye particles from the solution.
NOTE: Co-transfection of pT2K IRES-Luc is optional, but allows for identification of viable electroporated animals after birth using bioluminescence imaging. Please see step 3.5. Prepare a sufficient amount of plasmid-mix for the total number of pregnant mice to be operated. Use about 15-20 µL mix per mouse (~ 12 embryos). Proceed immediately with the surgery.
- Prepare mice for surgery.
- Anesthetize pregnant CD-1 mice with isoflurane. Induce anesthesia in a box with 3-4 vol-% isoflurane (oxygen flow rate: 1.5 L/min) and maintain it with 2 vol-% during surgery via nose cone.
NOTE: The complete surgery should not take longer than 30 min. Reduce the number of injected and electroporated embryos, if necessary. Use sterile gloves for surgery. - Place the mouse on its back on a heating pad and adjust anesthesia.
- Inject analgesic (Metamizol, 800 mg/kg body weight, subcutaneously (s.c.), 24 h depot).
NOTE: You may use other authorized analgesics than Metamizol. - Apply eye ointment to prevent eye-dryness during surgery.
- Fix limbs with tape and sterilize the abdomen with a disinfectant. Cover the mouse with gauze, leaving only the surgical area exposed. Spread 70% ethanol over surgical area and gauze.
- Make a skin incision of about 2 cm in length and widen the gap gently with straight surgical scissors. Locate the linea alba and make a slightly smaller second incision through the peritoneum using tissue scissors with blunt tip.
- Moisten the opened abdominal cavity with pre-warmed sterile 1x PBS.
- Carefully extract the uterine horns from the abdominal cavity using ring forceps. Pull gently by grabbing the uterine wall solely at the gap between the yolk sacs of two neighboring embryos.Avoid any pressure on the embryo while pulling it through the incision. Enlarge the incision if necessary and take extra care to position the uterine horns onto the abdomen while not compressing blood flow through the uterine artery.
NOTE: In some cases, it is advised to extract one uterine horn at a time and replace it before proceeding with the second uterine horn.
- Anesthetize pregnant CD-1 mice with isoflurane. Induce anesthesia in a box with 3-4 vol-% isoflurane (oxygen flow rate: 1.5 L/min) and maintain it with 2 vol-% during surgery via nose cone.
- Injection and electroporation of plasmid DNAs
- Aspirate approximately 15 µL of colored plasmid-solution with the prepared glass capillary. Avoid any air-bubbles during this process.
NOTE: The plasmid-solution can clog the tip easily if its concentration is high. Test the capillary by releasing some DNA right before the injection. - Gently hold the embryo with ring forceps and locate the neck area.
NOTE: If the embryo is in a position difficult to inject, skip it and go for the next embryo. Increased repositioning of the embryo may increase chances of damage to them. - Slowly pierce with the tip of the previously prepared glass capillary into the fourth ventricle by penetrating through the dorsal hindbrain and subsequently inject about 1 µL of the plasmid-mix. Confirm that the dyed DNA is not leaked from the brain and is visible as a diamond shaped structure.
NOTE: As an option, use 2.5-fold magnifying glasses for better visualization of the fourth ventricle and surrounding blood vessels. Deliver electric pulses immediately after injection to prevent diffusion of the plasmid-solution to the other ventricles. - Apply electric square pulses (5 pulses, 32 mV, 50 ms-on, 950 ms-off) with forceps-like platinum tweezers (see Table of Materials). Place the electrodes laterally with the negative pole covering the ear and the positive pole positioned at the cerebellar primordium over the uterine wall. Use 5 mm-diameter electrode plates for effective electroporation of E13.5 embryos.
NOTE: Increase the survival rate of pups by manipulating less than 2/3rd of the total number of embryos per dam. Avoid embryos too close to the cervix. If manipulated, they might cause complications during labor and jeopardize the entire litter. If electrodes are too close to the embryo's heart, this may cause cardiac arrest.
- Aspirate approximately 15 µL of colored plasmid-solution with the prepared glass capillary. Avoid any air-bubbles during this process.
- Post-electroporation and post-surgery care
- Carefully place uterine horns back into the abdominal cavity and fill with pre-warmed sterile 1x PBS. Let the uterine horns slide into a natural position.
- Close the peritoneum and skin separately with a simple continuous suture.
NOTE: Take extra care not to pierce through the uterine wall when suturing the peritoneum. - Shut-down the anesthesia and place the animal belly-down into a new cage.
- Monitor the animal during the waking phase and again 24 h after the surgery. Place the cage under a heating lamp, if the trembling doesn't improve within a 15 min time-frame and the animal doesn't start grooming.
- Confirm successful electroporation by luciferase imaging.
- Make sure that the operated dams drop embryos on time (at E19.5).
NOTE: The birthdate may be delayed to the next day probably because of the operation. - Anesthetize the electroporated pups (P5-P7) with 2.5% isoflurane and inject (intraperitoneal (i.p.)) with D-Luciferin (3 mg/kg body weight) 5 min prior to imaging.
- Detect luciferase signals in the electroporated pups using the bioluminescence imager.
NOTE: A 1 min exposure time is sufficient to detect the luciferase signal. Radiance (p/s/cm2/sr) is approximately >1 x 106.
- Make sure that the operated dams drop embryos on time (at E19.5).
4. Prepare Cryosections from the Electroporated Cerebella
- Euthanize the electroporated P7 pups and dissect out the cerebellum.
NOTE: The GFP signal can be observed using an epifluorescent stereoscope. - Fix the cerebella overnight in 5 mL per brain of 4% PFA in PBS at 4 °C.
- Transfer the fixed cerebella overnight in 10 mL per brain of 30% sucrose in PBS at 4 °C.
NOTE: The tissue should reach the bottom of a 15-mL tube after incubation in the sucrose solution. - After soaking up the remaining sucrose solution with a piece of cellulose filter paper, immerse the cerebellum in an optimal cutting temperature (OCT) compound in a disposable plastic mold and freeze it on dry ice. The cryogenic block can be stored at -80 °C until use.
NOTE: The protocol can be paused here. - Cut the cryogenic block into sections with 10 µm in thickness using a cryostat and keep the sections at -80 °C until use.
NOTE: One can confirm GFP expression in the sections, after washing out the OCT compound with PBS.
5. Immunostaining of the Cryosections
- Dry the slides at room temperature for 30 min and wash twice for 10 min in PBS.
- Circle the sections with a liquid blocker pen and incubate the sections in a blocking solution (10% normal donkey serum in PBS containing 0.1% Triton-X100) for 1 h at room temperature.
- Add primary antibodies against the molecules of interest (e.g., GFP and topoisomerase II beta (Top2B) in this study) in the blocking solution and incubate overnight at 4 °C.
NOTE: The names and concentrations of antibodies used are listed in the Table of Materials. In order to enhance GFP signals, an anti-GFP antibody can be optionally used together with the other antibodies. - Wash the slides with 0.1% Triton-containing PBST 3X for 10 min. Incubate the sections for 1 h at room temperature with a fluorophore-conjugated secondary antibody diluted in the blocking solution containing DAPI.
- Wash the slides in PBS 3X for 10 min. Mount the slides and keep at 4 °C until imaging.
NOTE: The protocol can be paused here.
6. Imaging and Analysis
- Image the sections using a confocal microscope at 20x magnification.
- Count the number of GFP positive cells losing the expression of the target protein. A representative image is shown in Figure 2B.
- Check the molecular phenotype upon ablation of the target genes in GNPs and their progenies by immuostaining12.
Representative Results
For in vivo functional analysis, it is critical to identify the cells into which exogenous gene(s) have been introduced. While the expression of a marker, such as GFP in non-proliferating cells can be followed-up for a long period of time, the signal gets sequentially lost in proliferating cells. An illustration of this effect is demonstrated in Figure 1. To circumvent losing the footprint of transfected cells in LOF analyses, we developed a novel approach by combining electroporation-based gene delivery with the CRISPR/Cas9 technology.
Representative results are shown in Figure 2. Functionality of the plasmid constructs is tested by transient transfection of pU6-sgTop2b-Cbh-Cre and pCAG-EG-Top2b-FP17, carrying the sgTop2b target sequence, into HEK293T cells stably expressing SpCas9 (Figure 2A). The sgRNA-guided Cas9 endonuclease activity induces DSB-mediated homology-directed repair of the EGFP expression cassette. Hence, the function of the sgRNA was directly analyzed by detection of GFP expression. According to Mashiko et al., a transfection efficiency of more than 30% determines an sgRNA as effective17.
GNPs originate from the rhombic lip (RL), a region bordering the roof of the fourth ventricle at embryonic stages E12.5 until E16.5. These cells have been known to undergo massive proliferation after birth in the outer external granule layer (oEGL) and shift to the inner EGL (iEGL) after exiting cell cycle20. Thus, GNPs are an appropriate example to test the advantages of our genetic labeling approach.
By following this protocol, in utero electroporation of GNPs allows tracing of edited cells with GFP. We introduced the pU6-sgRNA-Cbh-Cre plasmid expressing sgControl into the cerebellar primordium of Rosa26-CAG-LSL-Cas9-P2A-EGFP mice at embryonic stage E13.5. Mice were sacrificed at postnatal day P7, and sagittal sections of cerebellar tissue sections were stained by immunohistochemistry. Outer and inner EGL were defined by Ki67 expression (oEGL) and p27 expression (iEGL), respectively. Despite undergoing proliferation and maturation, mature cerebellar granule neurons (CGNs) still retained strong GFP expression (Figure 2B).
Emphasizing the utility of this protocol, we furthermore used the sgRNA targeting DNA Top2b as a representative example. Experimental procedures were performed as described above (see Figure 2B). Most of the GFP-positive cells transfected with sgRNAs for Top2b exhibited a loss of Top2b expression in the internal granule layer (IGL), while the introduction of control sgRNAs did not result in clear reduction of its expression (Figure 2C). A quantification of this result is shown in Figure 2D. These data present a valuable tool for tracing edited cells for a long period of time during development or tumor formation.
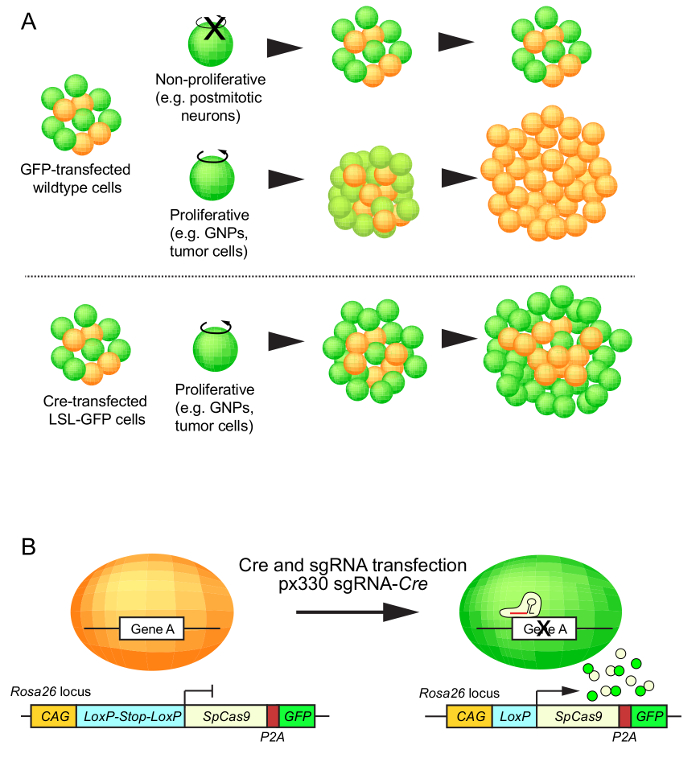
Figure 1: Schematic representation of GFP expression in proliferating and non-proliferating cells. (A) When an exogenous gene encoding GFP (e.g., pCAG-EGFP) is transfected in non-proliferating cells, the cells may express GFP for a long time (upper lane). Meanwhile, GFP expression could disappear in proliferating cells due to dilution of exogenous GFP (middle lane). In contrast, the cells that carry the LoxP-Stop-LoxP-GFP (LSL-GFP) transgene can keep the GFP expression during proliferation after transfection with Cre recombinase (lower lane). (B) The principle of SpCas9-mediated gene silencing and GFP expression after Cre and sgRNA transfection in a Rosa26-CAG-LSL-Cas9-P2A-EGFP mouse strain. Please click here to view a larger version of this figure.
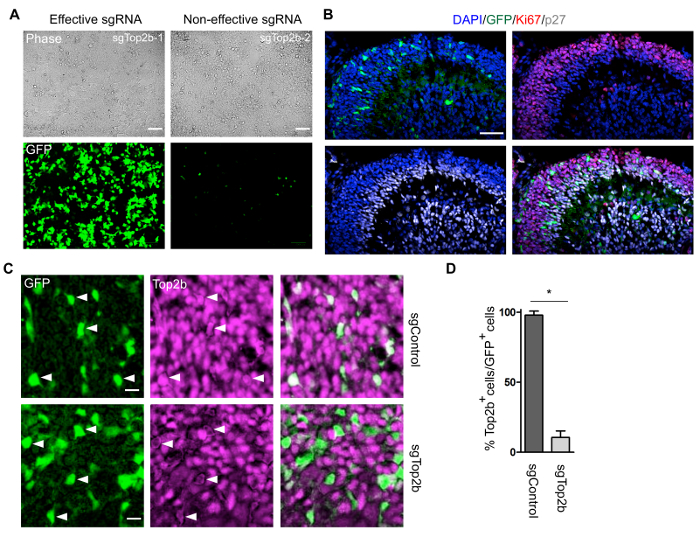
Figure 2: Representative images of results from this protocol. (A) HEK293T cells stably expressing SpCas9 were transfected with pU6-sgTop2b-Cbh-Cre and pCAG-EG-Top2b-FP plasmids. GFP expression in live cells was monitored using a fluorescent cell imager (see Table of Materials) 48 h after transfection. Left and right panels show an effective and non-effective sgRNA sequence based on GFP expression, respectively. Scale bars: 100 µm. (B) Immunostaining of GFP, Ki67, and p27 on the P7 cerebella from a Rosa26-CAG-LSL-Cas9-P2A-EGFP mouse subjected to electroporation at E13.5 with pU6-sgRNA-Cbh-Cre plasmid constructs expressing control sgRNA. The section is counterstained with DAPI (blue). Scale bars: 50 µm; 20x magnification. Note GFP-expressing cells in the oEGL marked by Ki67. (C) Immunostaining of GFP (green) and Top2b (magenta) on the P7 cerebella from a Rosa26-CAG-LSL-Cas9-P2A-EGFP mouse that was electroporated at E13.5 with pU6-sgRNA-Cbh-Cre plasmid constructs expressing sgRNA against Top2b (sgTop2b) and a control sequence (sgControl). Arrows indicate Top2b expression in GFP+ cells in the same field. Scale bars: 20 µm; 20x magnification. (D) Quantification of Top2b in electroporated (GFP-positive) cells in sgControl and sgTop2b experiments. Two and three pups were investigated for sgControl and sgTop2b, respectively, and 25 cells from each brain were analyzed. Error bars represent ± standard error of the mean (SEM) and p-values were calculated by unpaired t-test, *p = 0.0002. Please click here to view a larger version of this figure.
| Name | Sequence | Application | |
| sgTop2b-1 | CTTCGTCCTGATACATACAT | sgRNA target sequence | |
| sgTop2b-2 | AGCTGTCCAAAAATTAAAGC | sgRNA target sequence | |
| sgControl | GCGACCAATACGCGAACGTC | sgRNA target sequence | |
| EGxxFP-Top2b-F | gctgcccgacaaccactgagTACCTTGATATCTTAGAGAGCTG | Cloning into pCAG-EGxxFP | |
| EGxxFP-Top2b-R | gggtcagcttgccgatatcgCTCGCGCATTGTCTTAGC | Cloning into pCAG-EGxxFP | |
| hU6-F | GAGGGCCTATTTCCCATGATT | Sequencing for sgRNA | |
Table 1: Sequences of oligos being used in this study.
Discussion
Using exo utero electroporation, we have previously reported siRNA-based in vivo functional analyses of Atoh1 at an early stage of cerebellar granule cell differentiation8. Due to siRNA dilution/degradation and exposure of embryos outside the uterine wall, phenotypic analysis of the electroporated granule cells was limited to embryonic stages. However, the current method enabled analysis of the phenotype of postnatal animals.
Our previous study demonstrated that Cas9-mediated knockout of tumor suppressor Ptch1 via in utero electroporation successfully induced medulloblastoma2. In comparison, the current approach exhibits two major advantages: 1) Cas9 does not have to be delivered with the plasmid, allowing for multiple gene targeting by carrying multiple sgRNA expression cassettes in a single plasmid instead; and 2) the target cells and their progenies are permanently labeled with GFP, enabling visualization of the live cells and analysis of the behavior of transforming tumor cells in vivo.
The most critical steps of this protocol are the proper injection of plasmid DNAs into the correct location and the delivery of electric pulses into the appropriate places in the brain. Cerebellar neurons are born from the cerebellar neuroepithelium sequentially in a birthdate-dependent manner. Not all types of cells in the cerebellar primordium can be targeted via electroporation, because only a specific time window at E12.5-14.5 is technically feasible. Deep cerebellar nuclei neurons and Purkinje cells have been known to arise at E10.5-11.5, when the extra-embryonic membranes are not transparent, and the embryo is not clearly visible for DNA injection. Later, unipolar brush cells are derived from E17.5 upper RL (uRL), when the fourth ventricle is too narrow to inject a sufficient amount of DNA in the vicinity of the uRL. Thus, our method is only applicable at E12.5-14.5, which mainly targets mid- and later-born progenitors, such as GNPs and inhibitory interneurons. Another drawback of the method is that a full knockout phenotype may not be feasible when compared to Cre/LoxP-mediated germline GEMMs, since only thousands of cerebellar cells can be targeted by electroporation in each embryo.
In an earlier study, approximately 80% of GFP-positive cerebellar granule cells lost expression of the targeted gene12. While the identity of the granule cells was confirmed by molecular markers and their distribution, this identification approach might not be always applicable, as genes of interest could be involved in the marker expression and neuronal migration. Conditional activation of Cre using a cell-specific promoter would solve the problem in this case. Therefore, the current ubiquitous Cbh promoter in the pU6-sgRNA-Cbh-Cre plasmid can be replaced with a cell-specific promoter.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We appreciate Laura Sieber, Anna Neuerburg, Yassin Harim, and Petra Schroeter for technical assistance. We also thank Drs. K. Reifenberg, K. Dell and P. Prückl for helpful assistance for animal experiments at DKFZ; the Imaging Core Facilities of the DKFZ and the Carl Zeiss Imaging Center in the DKFZ for confocal microscopy imaging. This work was supported by the Deutsche Forschungsgemeinschaft, KA 4472/1-1 (to D.K.).
Materials
| Alexa 488 Goat anti-Chicken | ThermoFisher | A11039 | 1:400 dilution |
| Alexa 568 Donkey anti-Mouse | Life Technologies | A-10037 | 1:400 dilution |
| Alexa 594 Donkey anti-Rabbit | ThermoFisher | A21207 | 1:400 dilution |
| Alexa 647 Donkey anti-Rabbit | Life Technologies | A31573 | 1:400 dilution |
| Alkaline Phosphatase (FastAP) | ThermoFisher | EF0654 | |
| Autoclave band | Kisker Biotech | 150262 | |
| BamHI (HF) | NEB | R3136S | |
| BbsI (FastDigest) | ThermoFisher | FD1014 | |
| Cellulose Filter Paper (Whatman) | Sigma-Aldrich | WHA10347525 | |
| Cloth | Tork | 530378 | |
| Confocal laser scanning microscope | Zeiss | LSM800 | |
| D-Luciferin | biovision | 7903-1 | |
| DAPI | Sigma-Aldrich | D9542 | 1:1000 dilution |
| Disposable plastic molds (Tissue-Tek Cyromold) | VWR | 4566 | |
| DMEM Glutamax | ThermoFisher | 31966047 | |
| Donkey serum | Sigma-Aldrich | D9663 | |
| EcoRI (HF) | NEB | R3101S | |
| Electro Square Porator | BTX | ECM830 | |
| Endofree Maxi Kit | Qiagen | 12362 | |
| Ethanol | Merck | 107017 | |
| Eye ointment (Bepanthen) | Bayer | 81552983 | |
| Fast Green | Merck | 104022 | |
| FBS | ThermoFisher | 10270-016 | |
| Filter (0.22 µm) | Merck | F8148 | |
| Fluorescent cell imager (ZOE) | Biorad | 1450031 | |
| Forceps straight | Fine Science Tools | 91150-20 | |
| Gauze (X100 ES-pads 8f 10 x 10 cm) | Fisher Scientific | 15387311 | |
| GFP antibody | Abcam | ab13970 | 1:1000 dilution |
| Gibson Assembly Master Mix | NEB | E2611S | |
| Glass Capillary with Filament | Narishige | GD1-2 | |
| Heating Pad | ThermoLux | 463265 / -67 | |
| Image Processing software (ImageJ and Fiji) | NIH | – | |
| Insulin syringe (B. Braun OMNICAN U-100) | Carl Roth | AKP0.1 | |
| Isoflurane | Zoetis | TU061219 | |
| IVIS Lumina LT Series III Caliper | Perkin Elmer | CLS136331 | |
| Kalt Suture Needles | Fine Science Tools | 12050-02 | |
| KAPA HIFI HOTSTART READY mix | Kapa Biosystems | KK2601 | |
| Ki67 antibody | Abcam | ab15580 | 1:500 dilution |
| Light Pointer | Photonic | PL3000 | |
| Liquid blocker pen | Kisker Biotech | MKP-1 | |
| Metamizol | WDT | – | |
| Microgrinder | Narishige | EG-45 | |
| Microinjector | Narishige | IM300 | |
| Micropipette Puller | Sutter Instrument Co. | P-97 | |
| Microscope software ZEN | Zeiss | – | |
| Non-sterile Silk Suture Thread (0.12 mm) | Fine Science Tools | 18020-50 | |
| O.C.T. Compound (Tissue-Tek) | VWR | 4583 | |
| p27 antibody | BD bioscience | 610241 | 1:200 dilution |
| Paraformaldehyde | Roth | 335.3 | |
| PBS (1x) | Life Technologies | 14190169 | |
| pCAG-EGxxFP | Addgene | 50716 | |
| Polyethylenimine | Sigma-Aldrich | 408727 | |
| pX330 plasmid | Addgene | 42230 | |
| QIAprep Spin Miniprep Kit | Qiagen | 27104 | |
| QIAquick Gel Extraction Kit | Qiagen | 28704 | |
| Quick Ligation Kit | NEB | M2200S | |
| Ring Forceps | Fine Science Tools | 11103-09 | |
| Slides (SuperFrost) | ThermoFisher | 10417002 | |
| Software for biostatistics (Prism 7) | GraphPad Software, Inc | – | |
| Spitacid | EcoLab | 3003840 | |
| Stereomicroscope | Nikon | C-PS | |
| Sucrose | Sigma-Aldrich | S5016 | |
| Surgical scissors | Fine Science Tools | 91460-11 | |
| Surgical scissors with blunt tip | Fine Science Tools | 14072-10 | |
| Suture (Supramid schwarz DS 16, 1.5 (4/0)) | SMI | 220340 | |
| T4 DNA Ligation Buffer | NEB | B0202S | |
| T4 PNK | NEB | M0201S | |
| Tissue scissors Blunt (11.5 cm) | Fine Science Tools | 14072-10 | |
| TOP2B antibody | Santa Cruz | sc13059 | 1:200 dilution |
| Trypsin (2.5 %) | ThermoFisher | 15090046 | |
| Tweezers w/5mm Ø disk electrodes Platinum | Xceltis GmbH | CUY650P5 | |
| Vaporizer | Drägerwerk AG | GS186 |
References
- Mikuni, T., Nishiyama, J., Sun, Y., Kamasawa, N., Yasuda, R. High-Throughput, High-Resolution Mapping of Protein Localization in Mammalian Brain by In Vivo Genome Editing. Cell. 165 (7), 1803-1817 (2016).
- Zuckermann, M., Hovestadt, V., Knobbe-Thomsen, C. B., Zapatka, M., Northcott, P. A., Schramm, K., et al. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat Commun. 6, 7391 (2015).
- Chen, F., Rosiene, J., Che, A., Becker, A., LoTurco, J. Tracking and transforming neocortical progenitors by CRISPR/Cas9 gene targeting and piggyBac transposase lineage labeling. Development. 142 (20), 3601-3611 (2015).
- Saito, T., Nakatsuji, N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Developmental biology. 240 (1), 237-246 (2001).
- Matsuda, T., Cepko, C. L. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci U S A. 101 (1), 16-22 (2004).
- Matsui, A., Yoshida, A. C., Kubota, M., Ogawa, M., Shimogori, T. Mouse in utero electroporation: controlled spatiotemporal gene transfection. J Vis Exp. (54), (2011).
- Kawauchi, D., Taniguchi, H., Watanabe, H., Saito, T., Murakami, F. Direct visualization of nucleogenesis by precerebellar neurons: involvement of ventricle-directed, radial fibre-associated migration. Development. 133 (6), 1113-1123 (2006).
- Kawauchi, D., Saito, T. Transcriptional cascade from Math1 to Mbh1 and Mbh2 is required for cerebellar granule cell differentiation. Developmental biology. 322 (2), 345-354 (2008).
- Saba, R., Nakatsuji, N., Saito, T. Mammalian BarH1 confers commissural neuron identity on dorsal cells in the spinal cord. Journal of Neuroscience. 23 (6), 1987-1991 (2003).
- Sato, T., Muroyama, Y., Saito, T. Inducible gene expression in postmitotic neurons by an in vivo electroporation-based tetracycline system. J Neurosci Methods. 214 (2), 170-176 (2013).
- Kawauchi, D., Ogg, R. J., Liu, L., Shih, D. J. H., Finkelstein, D., Murphy, B. L., et al. Novel MYC-driven medulloblastoma models from multiple embryonic cerebellar cells. Oncogene. , (2017).
- Feng, W., Kawauchi, D., Korkel-Qu, H., Deng, H., Serger, E., Sieber, L., et al. Chd7 is indispensable for mammalian brain development through activation of a neuronal differentiation programme. Nat Commun. 8, 14758 (2017).
- Platt, R. J., Chen, S., Zhou, Y., Yim, M. J., Swiech, L., Kempton, H. R., et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 159 (2), 440-455 (2014).
- Ran, F. A., Hsu, P. D., Wright, J., Agarwala, V., Scott, D. A., Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 8 (11), 2281-2308 (2013).
- Joung, J., Konermann, S., Gootenberg, J. S., Abudayyeh, O. O., Platt, R. J., Brigham, M. D., et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protocols. 12 (4), 828-863 (2017).
- Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 339 (6121), 819-823 (2013).
- Mashiko, D., Fujihara, Y., Satouh, Y., Miyata, H., Isotani, A., Ikawa, M. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Scientific Reports. 3, 3355 (2013).
- Gibson, D. G., Smith, H. O., Hutchison, C. A., Venter, J. C., Merryman, C. Chemical synthesis of the mouse mitochondrial genome. Nature. 7 (11), 901-903 (2010).
- Baumgart, J., Baumgart, N. Cortex-, Hippocampus-, Thalamus-, Hypothalamus-, Lateral Septal Nucleus- and Striatum-specific In Utero Electroporation in the C57BL/6 Mouse. J Vis Exp. (107), e53303 (2016).
- Martinez, S., Andreu, A., Mecklenburg, N., Echevarria, D. Cellular and molecular basis of cerebellar development. Frontiers in Neuroanatomy. 7, 18 (2013).

