Construction of Synthetic Phage Displayed Fab Library with Tailored Diversity
Summary
This protocol describes a detailed procedure for the construction of a phage-displayed synthetic antibody library with tailored diversity. Synthetic antibodies have broad applications from basic research to disease diagnostics and therapeutics.
Abstract
Demand for monoclonal antibodies (mAbs) in basic research and medicine is increasing yearly. Hybridoma technology has been the dominant method for mAb development since its first report in 1975. As an alternative technology, phage display methods for mAb development are increasingly attractive since Humira, the first phage-derived antibody and one of the best-selling mAbs, was approved for clinical treatment of rheumatoid arthritis in 2002. As a non-animal based mAb development technology, phage display bypasses antigen immunogenicity, humanization, and animal maintenance that are required from traditional hybridoma technology based antibody development. In this protocol, we describe a method for construction of synthetic phage-displayed Fab libraries with diversities of 109-1010 obtainable with a single electroporation. This protocol consists of: 1) high-efficiency electro-competent cell preparation; 2) extraction of uracil-containing single-stranded DNA (dU-ssDNA); 3) Kunkel's method based oligonucleotide-directed mutagenesis; 4) electroporation and calculation of library size; 5) protein A/L-based enzyme-linked immunosorbent assay (ELISA) for folding and functional diversity evaluation; and 6) DNA sequence analysis of diversity.
Introduction
mAbs have broad applications ranging from basic research to disease diagnostics and therapeutics. As of 2016, more than 60 mAbs have been approved by the United States Food and Drug Administration (USFDA) for clinical treatment of autoimmune diseases, cancer, and infectious diseases1,2.
In 1975, Kohler and Milstein reported a technique for the continuous generation of antibodies of a single clonal specificity from a cellular source referred to as 'hybridomas' and this technique has subsequently become a cornerstone in medicine and industry3,4. Generation of mAbs by this method requires various steps including antigen production, mouse immunization, extraction of B lymphocytes, fusion of B cells with myeloma cells to form immortal hybridoma cells, clone selection, and for therapeutic applications, humanization is required to avoid human anti-mouse antibody (HAMA)4,5. However, for this technology, antigens including toxins, pathogens, and highly conserved proteins are relatively ineffective in triggering an in vivo immune response for mAb production5.
In 1978, Hutchison et al. reported the use of an oligonucleotide to direct mutagenesis of a residue in a single-stranded bacteriophage virus6. In 1985, Smith reported that foreign gene fragments can be fused in frame with the gene encoding phage coat protein III and can thus be displayed on the phage surface without compromising its infectivity7. These pioneering works laid a foundation for the subsequent construction of phage-displayed antibody libraries in immune, naïve, and synthetic forms with the formats of single-chain variable fragment (scFv) and antigen-binding fragment (Fab) for therapeutic mAb development8,9. From the technical point of view, phage display-based antibody development offers a complementary approach to hybridoma-based mAb development that can help to circumvent the limitations some antigens can pose and the humanization process that hybridoma-derived antibodies often require5. As of 2016, 6 phage display-derived mAbs have been approved in the market including Humira, one of the most successful mAbs used for treatment of rheumatoid arthritis, and many phage display-derived antibody candidates are currently at various stages of clinical investigation10.
For immune and naïve phage antibody libraries, the diversity of complementarity-determining regions (CDRs) in light and heavy chain is derived from the natural immune repertoire (i.e., from B cells). In contrast, the diversity of CDRs in synthetic phage antibody libraries is entirely artificial. Synthetic approaches to library construction provide precise control over the design of sequence diversity and offer opportunities for mechanistic studies of antibody structure and function11,12. Moreover, the framework for synthetic libraries can be optimized before library construction to facilitate downstream, large-scale industrial development11,12.
In 1985, Kunkel reported a single-stranded DNA (ssDNA) template-based mutagenesis approach to introduce site-directed mutations into M13 bacteriophage efficiently13. This approach was subsequently used widely for construction of phage-displayed libraries. Chemically synthesized DNA oligonucleotides designed to introduce diversity into Fab CDRs are incorporated into a phagemid with an antibody backbone template. In this process, the phagemid is expressed as a uracil-containing ssDNA (dU-ssDNA) and the oligonucleotides are annealed onto the CDRs and extended to synthesize double-stranded DNA (dsDNA) in the presence of T7 DNA polymerase and T4 DNA ligase. Finally, generated ds-DNA can be introduced into Escherichia coli by electroporation.
For high diversity, phage-displayed library construction, high-voltage electroporation of a two-component mixture of electro-competent cells and covalently closed circular dsDNA (CCC-dsDNA) should be prepared carefully. Sidhu et al. modified the preparation of electro-competent cells and DNA from traditional methods and greatly improved library diversity14.
In this protocol, we describe a method for construction of synthetic phage-displayed Fab libraries with diversities of 109-1010 obtainable with a single electroporation. Figure 1 shows an overview of library construction including: 1) high-efficiency electro-competent cell preparation; 2) extraction of dU-ssDNA; 3) Kunkel's method based oligonucleotide-directed mutagenesis; 4) electroporation and calculation of library size; 5) protein A/L-based ELISA for folding and functional diversity evaluation; and 6) DNA sequence analysis of diversity. All the reagents, strains and equipment are listed in the Material's Table. Table 1 shows the reagent setup.
Protocol
NOTE: Filter sterile tips must be used throughout when dealing with phage to avoid contamination to pipette gun and surrounding area. Aseptic area or hood must be used when handling with bacteria and phage experiments. Phage experiment area must be cleaned up using 2% sodium dodecyl sulfate (SDS) followed by 70% ethanol to avoid phage contamination. For making serial dilutions in this protocol, new tips should be used for each dilution.
1. E. Coli SS320 Electro-competent Cell Preparation
- Pre-warm LB/tet agar plate (prepared and stored at 4 °C for less than 1-week old) at 37 °C incubator for 1 h. Use a sterile inoculation loop or sterile tip to streak out a glycerol stock of E. coli SS320 cells onto the pre-warmed LB/tet agar plate in a zig-zag direction gently along the surface of the plate. Incubate the plate at 37 °C incubator overnight for around 12-15 h.
- The following day, use a sterile inoculation loop or sterile tip to pick a well-separated single colony along the zig-zag line and inoculate into 10 mL of 2YT/tet medium in a 50-mL round bottom tube by dipping the loop into the medium and stirring briefly.
- Incubate at 37 °C with shaking at 200 rpm for around 3-4 h until OD600 is reached at around 0.4-0.8 (log phase growth). Monitor OD600 at 2 h. During this period, pre-warm 5 LB/tet agar plates (prepared and stored at 4 °C for less than 1-week old) in a 37 °C incubator for at least 1 h.
- Add 20 μL M13KO7 helper phage from lab stock with titer around 1 x 1013 colony forming unit (cfu)/mL into 180 μL of sterile 1X PBS in autoclaved 1.5-mL tubes to prepare 10 ten-fold serial dilutions.
- Aliquot 4 mL autoclaved and liquid 2YT top agar into 5 X 14 mL round bottom tubes, label each tube with one dilution from the fifth to the ninth of the ten-fold dilution, respectively, and incubate in a 65 °C incubator to maintain the 2YT top agar in the liquid state.
- Mix 500 μL of log phase E. coli SS320 cells into the M13KO7 dilution tube (choose the fifth ten-fold dilution to the ninth ten-fold dilution) and incubate in a 37 °C incubator for 5-10 min.
- During the incubation of step 1.6, transfer 2YT top agar from step 1.5 to room temperature (RT) to cool down for around 5 min to about 42 °C. Use inner wrist to test temperature of 2YT top agar; it should remain as liquid. Transfer each dilution mixture from step 1.5 to each corresponding 2YT top agar tube. Tighten the lid of each tube and mix by turning upside down several times gently and briefly to avoid bubble generation.
- Carefully pour each mixture along the edge of a pre-warm LB/tet agar plate (from step 1.3) and slant the plate slightly to fully and evenly fill the plate with the mixture while avoiding the introduction of bubbles. Keep the plates at RT for around 5-10 min to solidify the top agar within each plate and incubate the 2YT top agar plates at 37 °C overnight for around 15-18 h.
- Choose the dilution plate after overnight growth from step 1.8 with around 100-200 average-sized, single, well-separated plaques for plaque inoculation. Use one hand to hold the agar plate against a light source, and the other hand to hold a pipette gun loaded with a long sterile pipette tip to vertically stab into the top agar, and collect a single and well-separated plaque.
- Slant the pipette tip slightly to separate top agar with plaque. Pipette up and down several times to dislodge the agar with plaque into a 14-mL round bottom culture tube pre-loaded with 1 mL of 2YT/kan/tet medium (see Table 1).
- Repeat this procedure to pick 3-5 plaques in total. The purpose of picking several plaques is to ensure successful inoculation, as a plaque can be faint and relatively small when compared with a bacteria colony. Grow the plaques for 8 h at 37 °C with shaking at 200 rpm.
- Transfer a tube of growing culture into 50 mL of 2YT/kan/tet medium in a 250-mL baffled flask. Grow at 37 °C with shaking at 200 rpm overnight for around 12 h.
- Inoculate three 2-L baffled flasks containing 900 mL of superbroth/tet/kan medium each with 5 mL of the overnight culture. Incubate at 37 °C with shaking at 200 rpm for 6-7 h to OD600 around 0.6-0.8.
- Chill the three flasks of bacterial culture in an ice bath for 5 min with gentle constant swirling by hand. The following steps should be done in a cold room, on ice, with prechilled solutions and equipment.
- Use absorbance tissue towel to dry the outer glass of each flask; use one hand to slant the 1-L autoclaved centrifuge bottle on bench and the other hand to pour the medium gently from each flask into each centrifuge bottle.
- Spin at 5,000 × g and 4 °C for 10 min to pellet the bacteria.
- Following centrifugation, gently decant the supernatant into a 5-L autoclaved waste beaker.
- Fill each bottle with 100 mL of sterile-filtered 1.0 mM HEPES, pH 7.4, and add an autoclaved magnetic stir bar to each bottle to aid in pellet resuspension at 200 rpm. Swirl and dislodge the entire pellet from the bottle wall every several minutes during the magnetic stirring. Once the pellet is dissolved, fill each bottle with an additional 400 mL of sterile-filtered 1.0 mM HEPES, pH 7.4.
- Centrifuge at 5,000 × g and 4 °C for 10 min. Decant the supernatant, retaining the stir bar in the bottle.
- Repeat steps 1.16 to 1.17 once.
- Resuspend each pellet in 500 mL of sterile-filtered, 10% ultrapure glycerol with the aid of the stir bars. Swirl and dislodge the entire pellet from the bottle wall every several minutes during the magnetic stirring.
- Centrifuge and decant as in step 1.17. Repeat step 1.19 once. Use long-arm autoclaved forceps to remove the stir bar.
- Centrifuge at 5,000 × g and 4 °C for 15 min. Decant the supernatant and remove any remaining traces of supernatant from each centrifuge bottle with a pipette.
- Add 3.0 mL of 10% ultrapure glycerol to one bottle and gently resuspend the pellet by pipetting. Transfer the suspension to the next bottle and repeat until all of the pellets are resuspended and combined in one bottle. Approximately 6 mL of highly concentrated cells are obtained with a titer of around 3 x 1011 cfu/mL.
- Pre-chill 1.5-mL autoclaved microcentrifuge tubes and one 96-well tube storage box without dividers at -80 °C for at least 1 h before this step. Transfer the pre-chilled microcentrifuge tubes to the cold room on ice before pipetting aliquots of the cell pellet suspension. Use a pipette to aliquot 350 μL of cell suspension into each 1.5 mL microcentrifuge tube.
- Transfer the aliquots into a foam box container filled with liquid nitrogen for flash freezing (3-5 min).
- Transport the foam box with liquid nitrogen to a -80 °C freezer (see step 1.23).
- Remove the 1.5-mL microcentrifuge tube storage box from the -80 °C freezer (see step 1.23) and put on ice.
- Quickly use a metal mesh to sieve the aliquots from liquid nitrogen and place them in the tube storage box. Store at -80 °C.
CAUTION: Liquid nitrogen can cause burns and care must be taken for safety protection.Use a Fab backbone phagemid to check the efficiency of the prepared electro-competent cells (the Fab backbone phagemid stock should be in ultrapure (MilliQ) water at 400 ng/µL). Thaw 1 μg of the Fab backbone phagemid in 10 µL ultrapure water and a 350 μL aliquot of electro-competent SS320 on ice, and prechill a 0.2-cm gap electroporation cuvette on ice.
- Add the 350 μL electro-competent SS320 cells to the 10 µL DNA after thawing and mix by pipetting several times while keeping the mixtures on ice. Avoid introducing bubbles.
- Pre-warm 15 mL of SOC medium in a 50-mL tube in a 37 °C water bath for at least 30 min before electroporation. Transfer the 360 μL mixture to the cuvette and perform electroporation following the manufacturer's instructions.
- Immediately rescue the cells after electroporation by adding 1 mL of pre-warmed SOC medium (from step 1.27) and pipetting. Transfer the medium from the cuvette to a 125-mL flask pre-loaded with 5 mL of pre-warmed SOC.
- Rinse the cuvette twice, each time with 1 mL of pre-warmed SOC medium, and transfer the medium to the 125-mL flask (see step 1.27). Add pre-warmed SOC medium to a final volume of 10 mL to the 125-mL flask.
- Incubate the 10-mL cell culture for 30 min at 37 °C with shaking at 200 rpm.
- Make serial dilutions to determine the transfection efficiency and M13KO7 pre-infection rate.
- Use a multi-channel pipette to add 180 μL of 2YT media to each well of a single column of a 96-microwell plate.
- Make 8 tenfold serial dilutions: transfer 20 μL of culture from step 1.29 to the first well of the plate, mix by pipetting, and transfer 20 μL of the mixture to the next well. Repeat this step to the end of the serial dilution.
- Plate 10 μL of each serial dilution on the LB/carb, LB/kan, and LB/tet plates in duplicate.
- Incubate overnight at 37 °C.
- Efficiency calculation formula: Assume that M is the average colony number counted from the diluted fold 10N (N is from 1-8). The E. coli SS320 electro-competent cell transfection efficiency of the Fab backbone phagemid from LB/carb plate is equal to M X 10N+3 cfu/µg. The M13KO7 pre-infection rate is estimated from the ratio of the colonies in LB/kan and LB/tet. The percentage of E. coli SS320 competent cells transfected with the Fab backbone phagemid is estimated from the ratio of colonies in LB/carb and LB/kan.
2. Preparing Uracil-containing ssDNA (dU-ssDNA) from the Phagemid Template
NOTE: A previously reported Fab backbone phagemid was used as the template for dU-ssDNA preparation15. The architecture of the Fab backbone phagemid is shown in Figure 2. A plasmid spin kit (QIAprep Spin M13) is used for extraction of dU-ssDNA with slight modifications.
- Pre-warm a LB/cmp plate (prepared and stored at 4 °C for less than 1-week old) in a 37 °C incubator for 1 h. Streak out a glycerol stock of E. coli CJ236 cells (or another dut−/ung− strain) with sterile loops or tips onto the pre-warmed LB/cmp plate. Incubate the plate at 37 °C overnight for around 12-15 h.
- Pick a single colony with a sterile tip and inoculate into 2 mL of 2YT/cmp medium in a 14-mL polypropylene round-bottom tube.
- Incubate the medium at 37 °C with shaking at 200 rpm for 3-4 h until OD600 is reached at around 0.4-0.8 (log phase growth).
- Add 5-10 μL phage of Fab backbone template into the culture, and incubate at 37 °C with shaking at 200 rpm for 30 min to allow phage infection.
- After incubation, use a sterile tip to streak out 10 μL of culture onto a pre-warmed LB/carb plate at 37 °C. Incubate at 37 °C overnight for around 12-15 h.
- Pick a single colony of the E. coli CJ236 containing Fab backbone phagemid to inoculate a starter culture of 3 mL 2YT/carb/cmp in a 14-mL polypropylene round-bottom tube, and grow at 37 °C with shaking at 200 rpm overnight for around 12 h.
- Inoculate the 0.3 mL starter culture into 30 mL 2YT/carb/cmp in a 250-mL baffled bottle.
- Incubate the cell culture at 37 °C with shaking at 200 rpm for 3-4 h until OD600 is reached at around 0.4-0.8 (log phase growth).
- Add the M13KO7 (lab stock, titer of approximately 1 x 1013 cfu/mL) to the culture from step 2.8 with a multiplicity of infection (MOI) of approximately 10 and the final titer of M13KO7 is approximately 1 x 1010 cfu/mL.
- Incubate at 200 rpm and 37 °C for 1 h.
- Pellet the culture by centrifugation in a 50-mL round-bottom tube at 5,000 × g and 25 °C for 20 min.
- Decant the supernatant and resuspend the pellet with 30 mL of fresh 2YT/carb/kan/uridine. Transfer the resuspension into a new 250-mL baffled bottle.
- Incubate at 200 rpm and 25 °C for 22-24 h.
- Transfer the culture from step 2.13 into a 50-mL round-bottom tube and centrifuge the culture at 12,000 × g for 20 min to separate the phage supernatant from bacterial cell pellet. Transfer the phage supernatant into a new 50-mL round-bottom tube and add 1/5 the final volume of PEG/NaCl solution to precipitate the phage. Mix well and incubate on ice for 30 min to precipitate the phage particles.
- Centrifuge at 12,000 × g and 4 °C for 30 min. Decant the supernatant and centrifuge at 4,000 × g and 4 °C for 2 min. Aspirate the remaining supernatant.
- Resuspend the phage pellet in 2 mL of sterile-filtered 1X PBS and transfer to 1.5-mL microcentrifuge tubes. Centrifuge at 12,000 × g for 5 min in a benchtop microcentrifuge to remove any remaining bacterial debris and transfer the phage supernatant to new 1.5-mL microcentrifuge tubes and store at 4 °C.
- Check the uracil incorporation efficiency in E. coli CJ236 through side by side comparison with E. coli SS320.
- Add 180 μL of 2YT media to each well of a single row of a 96-well plate.
- Make ten 10-fold serial dilutions: transfer 20 μL of the phage supernatant to the first well of the plate, mix by pipetting, and transfer 20 μL of the mixture to the next well. Repeat this step to the end of the serial dilution.
- Add 10 μL of phage from last eight serial dilutions to infect 90 μL of E. coli CJ236 and E. coli SS320 in log phase (OD600 = 0.4-0.8). Incubate at 37 °C for 30 min with gentle shaking.
- Plate 10 μL from the serial dilution of each infection in E. coli CJ236 and E. coli SS320 on 2YT/Carb plates in duplicate.
- Incubate overnight at 37 °C.
- Titer calculation formula: Assume that M is the average colony number counted from the diluted fold 10N (N is from 1-10). The titer from E. coli CJ236 or E. coli SS320 is equal to M X 10N+2 cfu/mL. Estimate the efficiency of uracil incorporation from the titer ratio of E. coli CJ236 and E. coli SS320.
- Add 1/100 volume of the phage precipitation buffer MP into the phage supernatant in 1.5-mL microcentrifuge tubes, and turn upside down gently to mix for several times. Incubate at RT for at least 2 min. Phage particles are precipitated from the culture medium, and thus, a cloudy solution should be visible at this point.
- Apply the sample from step 2.18 to a plasmid spin column (e.g., QIAprep) in a 1.5-mL microcentrifuge tube. The binding capacity of one spin column for ssDNA can reach at least 10 µg. Centrifuge at 6,000 × g and 25 °C for 30 s in a benchtop microcentrifuge. Discard the flow-through which is in the 1.5-mL microcentrifuge tube. Phage particles remain bound to the column matrix at this stage.
- Add 0.7 mL of the UT-MLB phage lysis and binding buffer (see Discussion) to the column and incubate at RT for at least 1 min. Centrifuge at 6,000 x g and 25 °C for 30 s and discard the flow-through.
- Add another 0.7 mL of the UT-MLB buffer and incubate at RT for at least 1 min.
- Centrifuge at 6,000 × g for 30 s. Discard the flow-through. At this stage, the phage coat protein is separated from the dU-ssDNA, which remains bound to the column matrix.
- Add 0.7 mL of wash buffer PE containing ethanol following the manufacturer's instructions. Centrifuge at 6,000 × g for 30 s and discard the flow-through.
- Repeat step 2.23, then centrifuge once more at 6,000 × g for 30 s to remove residual buffer PE.
- Transfer the column to a new 1.5-mL microcentrifuge tube and add 100 μL of elution buffer EB (10 mM Tris·CL, pH 8.5) to the center of the column membrane.
- Incubate at RT for 10 min and centrifuge at 6,000 × g for 1 min to elute the dU-ssDNA. Approximately, 1.5-2.5 μg dU-ssDNA/mL culture can be obtained.
- Analyze the eluted DNA by electrophoresing 1 μL on a 1% agarose TAE gel. The DNA should appear as a predominantly single band with no smearing.
- Determine the DNA concentration by measuring absorbance on a nanodrop spectrophotometer at 260 nm (A260 = 1.0 for 33 ng/μL of ssDNA). Typical dU-ssDNA concentrations are within 200-500 ng/μL.
3. Kunkel's Method Based Oligonucleotide-directed Mutagenesis
Notes: It is advisable to conduct small-scale reactions prior to full scale reactions to ensure the quality of the mutagenic oligonucleotide and reaction components. A cartoon of Kunkel's method based oligonucleotide-directed mutagenesis is shown in Figure 3. Various amino acid diversities are introduced into CDRH1, CDRH2, CDRH3, and CDRL3 regions with IMGT numbering nomenclature16 (Table 2). The theoretical amino acid diversity of each CDR, total theoretical amino acid diversity, and oligonucleotide sequences are listed in Table 2.
- Oligonucleotide phosphorylation with T4 polynucleotide kinase
- Combine 0.6 μg of mutagenic oligonucleotides, designed to mutate a single CDR, with 2 μL 10X TM buffer, 2 μL 10 mM ATP, and 1 μL 100 mM DTT. Add ultrapure H2O to a total volume of 18 μL in a 1.5-mL tube. For the library construction, 4 separate and parallel phosphorylation reactions are set up corresponding to CDRH1, CDRH2, CDRH3, and CDRL3, respectively.
- Add 20 U (2 uL) of T4 polynucleotide kinase to each tube and incubate for 1 h at 37 °C (Reaction 1 in Table 3). Use immediately for annealing.
- Annealing of the oligonucleotides to the template
- To 20 μg of dU-ssDNA template, add 25 μL of 10X TM buffer, 20 μL of each phosphorylated oligonucleotide solution, and ultrapure H2O to a final volume of 250 μL in a 0.5-mL tube. Mix well and transfer into 0.20 mL PCR tubes (50 μL each) as Reaction 2 in Table 3. These DNA quantities provide a molar ratio of 3:1 between oligonucleotide and template, assuming that the length ratio of oligonucleotide and template is 1:100.
- Incubate the reaction in a PCR machine at 90 °C for 3 min, 50 °C for 3 min, and 20 °C for 5 min.
- Enzymatic synthesis of CCC-dsDNA
- To the 250 μL annealed oligonucleotide/template mixture, add 10 μL of 10 mM ATP, 10 μL of 25 mM dNTP mix, 15 μL of 100 mM DTT, 30 Weiss units T4 DNA ligase, and 30 U T7 DNA polymerase as Reaction 3 in Table 3.
- Incubate the reaction in the 1.5-mL tube at 20 °C overnight.
- Wash and concentrate the synthesized CCC-dsDNA in a 0.5-mL centrifugal filter device with a 30 kDa pore size membrane at RT.
- Transfer the overnight reaction mixture to the filter device and add ultrapure H2O to 400 µL final volume. Spin at 14,000 × g for 10 min; the volume is less than 50 μL.
- Discard the flow through, add 400 µL of ultrapure H2O into the filter, and spin at 14,000 × g for 10 min.
- Repeat step 3.3.3.2 once more.
- Place the filter upside down in a clean microcentrifuge tube to recover the CCC-dsDNA. Spin at 1,000 × g for 2 min; the recovered volume is generally around 20-40 μL. The recovered CCC-dsDNA can be used immediately for electroporation of E. coli or frozen at -20 °C for later use. Normally 20-40 μg CCC-DNA can be obtained.
- Electrophorese 1.0 µL of the eluted reaction product alongside the dU-ssDNA template to visualize the outcome of the reaction.
4. Electroporation and Calculation of the Library Size
- Chill the purified CCC-dsDNA (20 μg in a maximum volume of 50 μL) in a 1.5-mL microcentrifuge tube and a 0.2-cm gap electroporation cuvette on ice.
- Pre-warm 20 mL of SOC media in a 50-mL polypropylene conical centrifuge tube in 37 °C water bath for at least 30 min.
- Thaw a 350 μL aliquot of electro-competent E. coli SS320 on ice. Add the cells to the DNA and mix thoroughly by pipetting several times. Avoid introducing bubbles.
- Transfer the mixture to the cuvette and perform electroporation following the manufacturer's instructions. For example, when the BTX ECM-630 electroporation system is used, the relevant settings are 2.5 kV field strength, 125 Ω resistance, and 50 µF capacitance.
- Immediately rescue the electroporated cells by adding 1 mL of pre-warmed SOC medium and transferring into 17 mL of SOC medium in a 125-mL flask. Rinse the cuvette twice with 1 mL of SOC medium and transfer to the same flask (final volume is 20 mL).
- Incubate for 30 min at 37 °C with shaking at 200 rpm.
- Determine the electroporation efficiency.
- Add 180 μL of 2YT media to each well of a single column of a 96-microwell plate.
- Make 8 ten-fold serial dilutions: transfer 20 μL of the 20-mL culture to the first well of the plate, mix with pipetting, and transfer 20 μL of the mixture to the next well. Repeat this step to the end of the serial dilution.
- Plate 10 μL of each of the serial dilutions on one LB/carb plate in duplicate. Plate 100 µL remaining from each of the serial dilutions onto separate LB/carb plates for colony count cross-check. These plates will also provide single clones for ELISA and sequence analysis (see section 5).
- Incubate overnight at 37 °C.
- Count the colonies from the 10 μL duplicates on the LB/carb plate. Assume that M is the average colony number counted on 10N fold LB/carb plate (N is from 1-8). The total library size is equal to 2M X 10N+3 colonies.
- Aliquot the culture from step 4.6 equally into two 2-L baffled flasks, each containing 500 mL of 2YT/carb/kan medium for phage library generation.
- Incubate at 37 °C with shaking at 200 rpm overnight for around 16 h.
- Transfer the culture to two 1-L autoclaved centrifuge bottles and centrifuge for 30 min at 12,000 × g at 4 °C.
- Transfer the supernatant to two new 1-L autoclaved centrifuge bottles and add 1/5 the final volume of PEG/NaCl solution to precipitate the phage. Incubate on ice for 30 min.
- Centrifuge for 30 min at 12,000 × g and 4 °C. Carefully decant the supernatant and avoid disturbing of the phage pellet. Spin for 1 min at 4,000 x g and remove the remaining supernatant with a pipette.
- Resuspend the phage pellet with 20 mL of sterile-filtered 1X PBS buffer and transfer to a new 50-mL tube.
- Pellet the insoluble matter by centrifuging for 5 min at 12,000 × g and 4 °C. Transfer the supernatant to a new 50-mL tube.
- Measure the phage concentration by spectrophotometer (OD268 = 1.0 for a solution of 5 X 1012 phage/mL).
- Adjust the phage concentration to 5 X 1012 phage/mL in 1X PBS with 10% ultrapure glycerol.
- Aliquot 1 mL of phage solution per 1.5-mL microcentrifuge tube. Use the libraries immediately for panning or store at -80 °C.
5. Quality Assessment by Protein A/L Direct Binding ELISA Assay and Sequencing
- Randomly pick 96 single colonies on LB/carb plate from step 4.7.3 into a 96-deep well culture plate containing 800 μL of 2YT/carb in each well. Incubate for 3-4 h at 37 °C with shaking at 1,000 rpm to OD600 = 0.4-0.8.
- Add 100 μL of M13KO7 (1 X 1011 cfu/mL) to each well of a 96-deep well culture plate with a multichannel pipette. Incubate at 37 °C with shaking at 1,000 rpm for 1 h.
- Add 100 μL of 2YT containing 10X concentration of kanamycin (500 μg/mL) to each well with a multichannel pipette. Incubate overnight at 37 °C with shaking at 1,000 rpm.
- Dissolve protein L to 1 µg/mL in 1X PBS. Coat 3/4 of the wells in a 384-well high protein-binding polystyrene plate with 30 µL/well of the protein L solution. Incubate overnight at 4 °C.
- Discard the overnight protein L coating solution from step 5.4. Add 50 µL of M-PBST (the blocking solution) to all of the wells in the 384-well high protein-binding polystyrene plate with a multichannel pipette. Incubate the plates on a microplate shaker for 1 h at RT.
- Spin down the overnight culture from step 5.3 in a deep 96-well culture plate at 3,000 x g for 10 min at 4 °C. The phage is in the supernatant.
- Discard the blocking solution from step 5.5. Add 15 μL of M-PBST and 15 μL of each phage supernatant (step 5.6) to 3 of the protein L coated wells as triplicate, and 1 non-coated well as negative control using a multichannel pipette. Incubate at RT for 1 h with shaking at 200 rpm.
- Discard the phage solution. Wash the plate 6 times with 80 μL of PBST by an automated plate washer.
- Add 15 μL of M-PBST and 15 μL of Protein A-HRP (1:1,500 diluted in 1X PBS) to each well with a multichannel pipette, and incubate at RT for 1 h with shaking at approximately 200 rpm.
- Discard the Protein A-HRP/M-PBST solution. Wash the plate 6 times with 80 μL of PBST.
- Add TMB substrate (30 μL/well) with a multichannel pipette and incubate for 2-3 min until the color develops. Stop the reaction with 1.0 M H3PO4 (30 μL/well).
- Read the plates spectrophotometrically at 450 nm. Positive clones are defined as those that exhibit an average ratio of OD450 absorbance of protein L wells to the M-PBST well greater than 3.0.
- In a 96-deep well, infect 50 μL of SS320 in 2YT/tet at log phase with 5 μL of the same phage used for the ELISA (step 5.6) for 30 min at RT.
- Add 950 μL of 2YT/carb into the 96-deep well from step 5.13 and incubate overnight at 37 °C with shaking at 1,000 rpm.
- Extract the phagemid DNA by mini-prep DNA extraction kit. Use the upstream primers of VL (5'-TCGCTTTGTTTTTATTTTTTAATGTA-3') and VH (5'-GACTACTAATAACATAAAGTCTACGCCG-3') for sequence analysis to estimate the library sequence diversity.
Representative Results
Following the flow chart of the Fab library construction (see Figure 1), we prepared M13KO7 helper phage pre-infected E. coli SS320 electro-competent cells. The efficiency of these electro-competent cells is estimated as 2 X 109 cfu/µg when the Fab phagemid backbone for library construction was used (Figure 4).
The uracil incorporation efficiency by comparison of titer in both E. coli CJ236 and E. coli SS320 cells was checked. The E. coli CJ236 and E. coli SS320 cells were infected by phage harboring dU-ssDNA. E. coli SS320 has enzymes (dUTPase and uracil glycosylase) that can degrade uracil-containing DNA, while E. coli CJ236 lacks these enzymes and cannot degrade uracil-containing DNA. To achieve an acceptable uracil incorporation efficiency, titers from E. coli CJ236 need to be at least 104 times higher than those from E. coli SS320. Otherwise the wild-type population will increase in the constructed antibody library due to inefficient uracil incorporation. Figure 5 showed that the titer from E. coli CJ236 is approximately 3 X 105 times higher than that from E. coli SS320, indicating an efficient uracil incorporation into phage ssDNA.
Next, we prepared and extracted dU-ssDNA. The dU-ssDNA purity is checked by agarose gel electrophoresis (Figure 6). Then the oligonucleotide-directed mutagenesis was conducted and the efficiency of the dU-ssDNA conversion to CCC-DNA was evaluated (Figure 6). Three products with lower motility than dU-ssDNA can be visualized on the gel including the fastest-running band (CCC-dsDNA), the middle weak band (nicked band), and the slowest-running band (strand-displaced DNA).
After electroporation into E. coli SS320, the library size was estimated from the overnight incubation plate (see step 4.7.5). The average library size was 5 X 109 from duplicate serial dilutions on LB/carb plates (Figure 7). However, the estimated size at this step may contain phage that do not display Fabs due to the presence of a frameshift or stop codon, or display misfolded Fabs. Sequencing and ELISA were used to estimate the functional diversity of constructed library. 96 randomly picked single clones were sent for sequencing analysis. Table 4 shows that 90 out of the 96 randomly picked single clones were successfully sequenced, which contains 70 clones without a premature stop codon (53 clones with at least one CDR mutant and 17 clones with the wild-type sequence) and 20 clones with a premature stop codon at different regions. Within the 70 clones, mutant rates of CDRH1, CDRH2, CDRH3, and CDRL3 are 50%, 57%, 53%, and 56%, respectively, while the mutant rate with at least one CDR is 76%. In the 20 clones with a premature stop codon (90%), the premature stop codon was mainly derived from the frameshift of oligonucleotide mutagenesis primers, including 45% (CDRH1), 10% (CDRH2), 15% (CDRH3), and 20% (CDRL3).
To detect the display of properly folded Fabs, a protein A/L based ELISA was employed as it is known that protein A and protein L can recognize proper folding of the VH framework and VL framework, respectively17,18. In agreement with the sequencing analysis, the ELISA assay in triplicate (Figure 8) showed that the 20 clones with a premature stop codon were all negative while the 17 clones with a wild-type sequence were all positive when the positive ratio was empirically set at 3.0. For the 53 clones with at least one CDR mutant, 43 clones were positive in ELISA while 10 clones were negative; this indicates that most of the clones were well folded while the CDRs from the 10 clones can have detrimental effects on Fab folding. In total, 43 clones out of the 90 clones (48%) were well folded and contained at least one CDR mutant. Thus, the functional amino acid diversity of the constructed library based on protein A/L ELISA and sequence analysis was estimated to be 2.4 X 109 (i.e., 48% of 5 X 109).
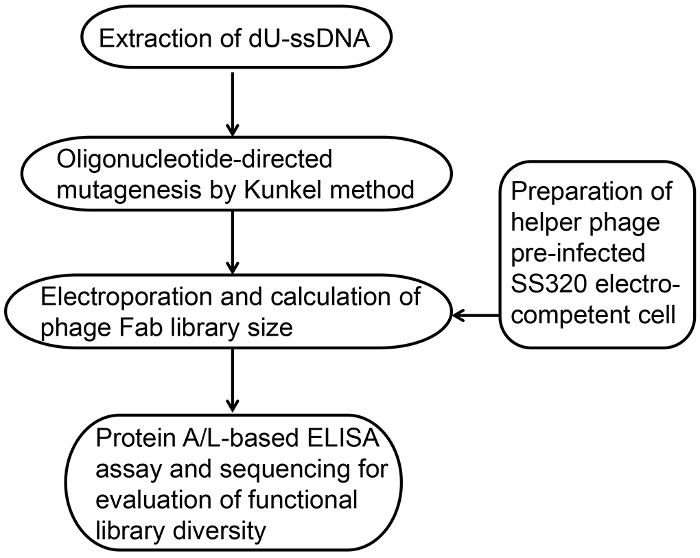
Figure 1: Overview of the phage-displayed Fab library construction. Phage-displayed Fab library construction follows a basic series of steps. It involves preparation of high-efficiency electro-competent bacterial cells, extraction of dU-ssDNA, Kunkel's method based oligonucleotide-directed mutagenesis, electroporation and calculation of phage Fab library size, functional evaluation by protein A/L ELISA, and DNA sequence analysis. Please click here to view a larger version of this figure.
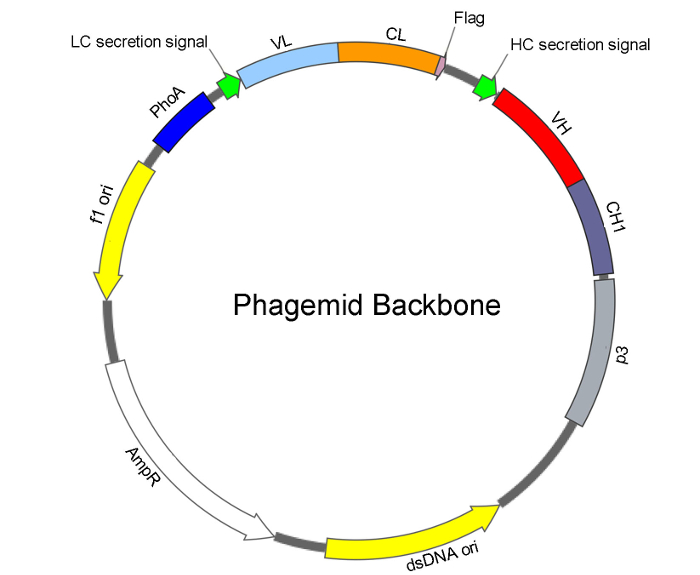
Figure 2: Phagemid architecture for the Fab library construction. The basic features of the phagemid backbone consist of origins of single-stranded (f1 ori) and double-stranded (dsDNA ori) DNA replication, and an ampicillin/carbenicillin resistance gene (AmpR). For Fab display, under the control of the alkaline phosphatase promoter (PhoA), the phagemid contains a bicistronic cassette to drive expression and secretion of: light chain (LC) consisting of a secretion signal, VL (variable region of light chain), CL (constant region of light chain), and C-terminal flag tag; and heavy chain (HC) consisting of a secretion signal, VH (variable region of heavy chain), and CH1 (constant region 1 of heavy chain) fused with a p3 phage minor coat protein. Assembly of the light chain and heavy chain into Fab within the E. coli periplasm directs the display of Fab on the phage surface. Please click here to view a larger version of this figure.
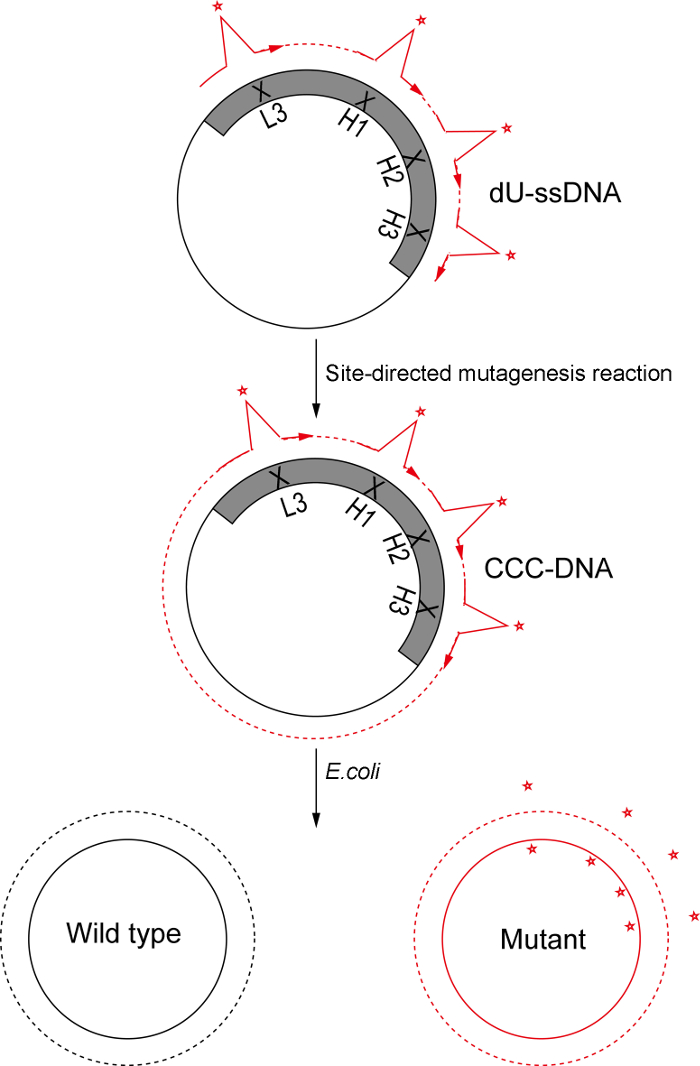
Figure 3: Schematic of Kunkel's method based oligonucleotide-directed mutagenesis. In this protocol, we used Kunkel's method to prepare dU-ssDNA template. Oligonucleotides for CDRH1, CDRH2, CDRH3, and CDRL3 with designed diversity are phosphorylated, annealed to the template, and used to convert ss-DNA to CCC-dsDNA. Following electroporation into E. coli SS320 electro-competent cells, the heteroduplex DNA is repaired to either the wild type or the mutant form; Due to the presence of uracil in the wild type strand, the repair process favors the mutant form, and thus, the mutant form dominates the library. Please click here to view a larger version of this figure.
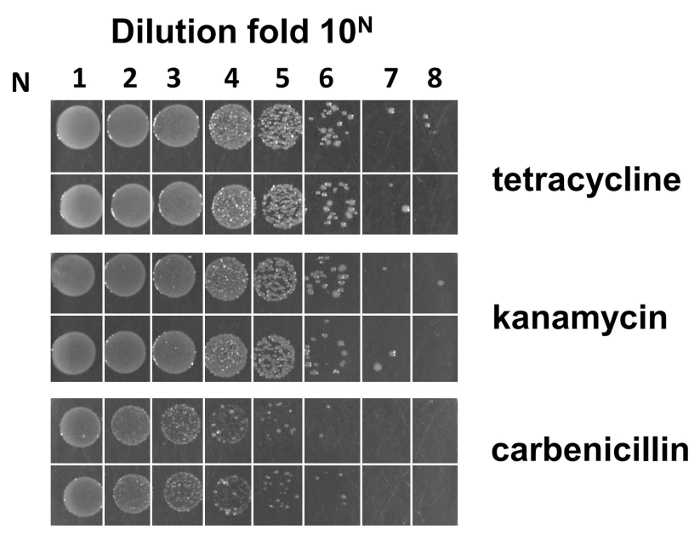
Figure 4: Estimation of the M13KO7 pre-infected E. coli SS320 electro-competent cell efficiency. A phagemid backbone vector was used to check the electroporation efficiency of the competent cells. Formula to calculate the efficiency is as follows: assume that M is the average colony number counted from the most diluted fold 10N (N is from 1-8) in duplicate. E. coli SS320 efficiency from LB/carb plate is equal to M X 10N+3 cfu/µg. The efficiency of the electro-competent cells is around 2 X 109 cfu/µg. Please click here to view a larger version of this figure.
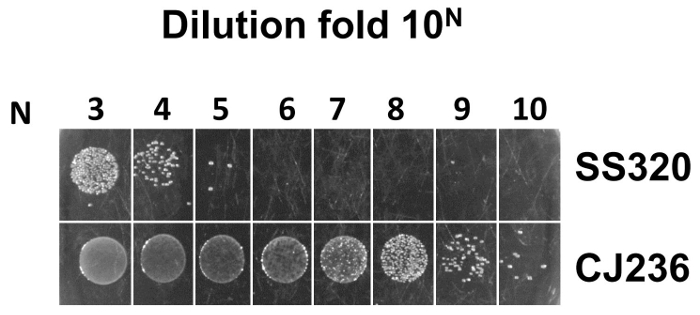
Figure 5: Assessment of uracil incorporation into ssDNA by phage infection of E. coli CJ236 and E. coli SS320 cells. Based on Kunkel's method, the uracil incorporation efficiency is checked by comparison of phage infection titer in both E. coli CJ236 and E. coli SS320 cells. The titer calculation formula is as follows: assume that M is the average colony number counted from the most diluted fold 10N (N is from 1-10), and that the titer from E. coli CJ236 or E. coli SS320 is equal to M X 10N+2 cfu/mL. The efficiency of uracil incorporation can be estimated from the titer ratio of E. coli CJ236 and E. coli SS320. The titer in E. coli CJ236 was 9 X 1012 cfu/mL while the titer in E. coli SS320 was 3 X 107 cfu/mL. The titer ratio of E. coli CJ236 and E. coli SS320 was 3 X 105. Please click here to view a larger version of this figure.
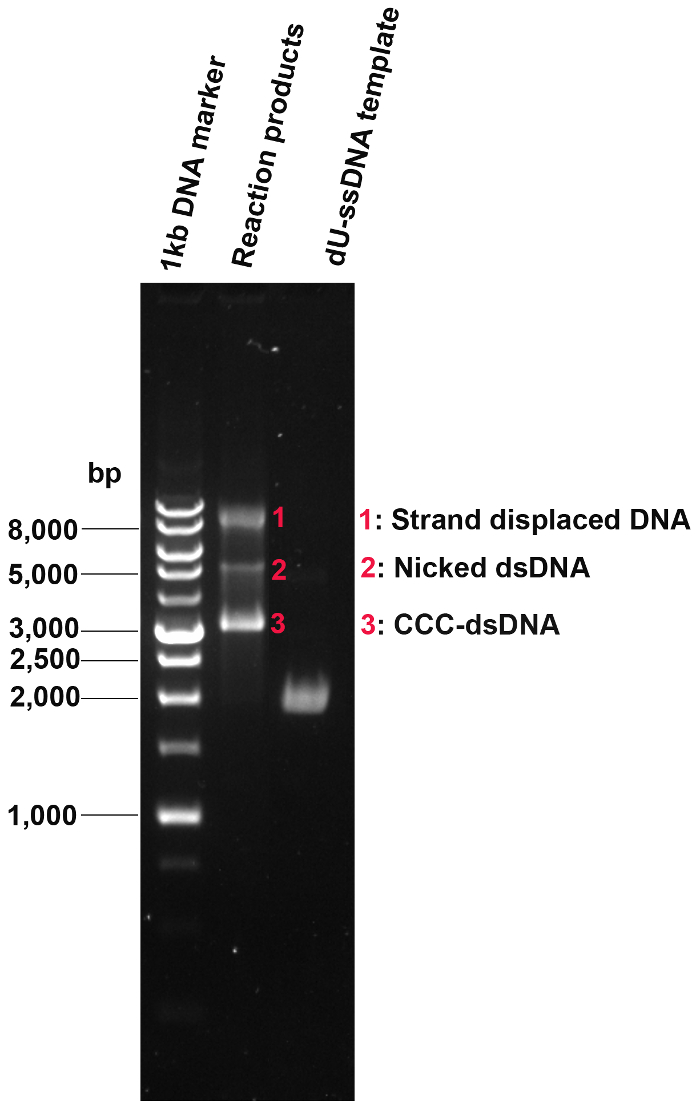
Figure 6: Conversion of dU-ssDNA to CCC-DNA by oligonucleotide-directed mutagenesis. Following oligonucleotide-directed mutagenesis, the efficiency of dU-ssDNA conversion to CCC-DNA was evaluated. dU-ssDNA was completely converted to dsDNA. The dominant band is CCC-dsDNA while there is a minor portion of nicked dsDNA and strand-displaced DNA. Please click here to view a larger version of this figure.
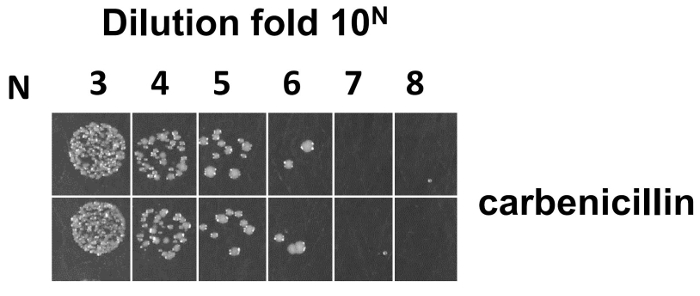
Figure 7: Phage titration for calculation of library size. After electroporation into E. coli SS320, the library size was estimated from serial dilutions on LB/carb plates. The size calculation formula is as follows: assume M is the average colony number counted from the most diluted fold 10N from a 2YT/Carb plate (N is from 1-8), size is equal to 2M X 10N+3. Please click here to view a larger version of this figure.
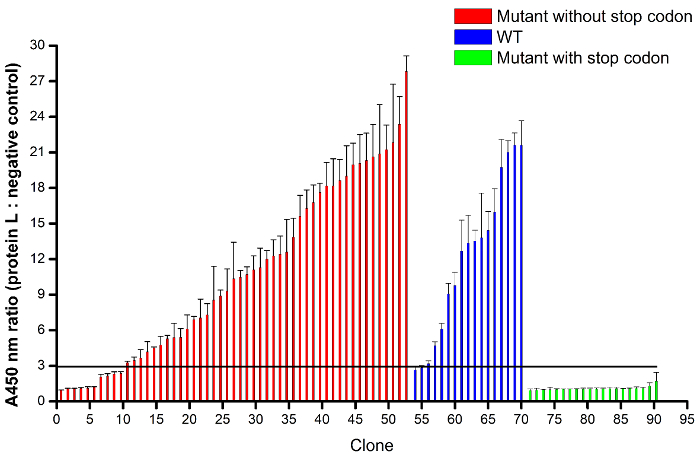
Figure 8: Protein A/L direct binding phage ELISA. Protein L can recognize the framework of well folded kappa light chain VL and protein A can recognize the framework of well folded heavy chain VH. Binding of Fab with protein L and A indicates proper folding of both heavy chain and light chain. In brief, protein L in triplicate and the negative control M-PBST were coated to the plate, Fab phage supernatants from different clones were incubated with protein L and M-PBST, then after wash, protein A-HRP was used to capture bound Fab phage. Phage ELISA readings showed 90 randomly picked clones with successful sequencing readout. A threshold line representing the clone as positive was empirically defined where the ratio of OD450 absorbance value from protein L (average of triplicate with error bar) versus negative control was more than 3.0. Three groups based on sequencing analysis were shown, corresponding to a mutant without stop codon in red (53 clones), wild type (WT) in blue (17 clones), and mutant with stop codon in green (20 clones). Please click here to view a larger version of this figure.
| Reagent setup | Component | Amount | comments/description |
| 2YT medium | Yeast extract | 10 g | Add ultrapure water to make up the volume to 1.0 L, adjust pH to 7.0, autoclave. |
| Tryptone | 16 g | ||
| NaCl | 5 g | ||
| 2YT top agar | Yeast extract | 10 g | Add ultrapure water to make up the volume to 1.0 L and adjust pH to 7.0, heat to dissolve, autoclave. |
| Tryptone | 16 g | ||
| NaCl | 5 g | ||
| Granulated agar | 7.5 g | ||
| 2YT/carb/cmp medium | Carbenicillin | 100 μg/mL | |
| Chloramphenicol | 10 μg/mL | ||
| 2YT/carb/kan/uridine medium | Carbenicillin | 100 μg/mL | |
| Kanamycin | 50 μg/mL | ||
| Uridine | 0.25 μg/mL | ||
| 2YT/carb/tet medium | Carbenicillin | 100 μg/mL | |
| Tetracycline | 10 μg/mL | ||
| 2YT/carb medium | Carbenicilin | 100 μg/mL | |
| 2YT/kan medium | Kanamycin | 50 μg/mL | |
| 2YT/kan/tet medium | Kanamycin | 50 μg/mL | |
| Tetracycline | 10 μg/mL | ||
| 2YT/tet medium | Tetracycline | 10 μg/mL | |
| 2YT/cmp medium | Chloramphenicol | 10 μg/mL | |
| LB medium agar | Yeast extract | 5 g | Add ultrapure water to make up the volume to 1.0 L, adjust pH to 7.0, autoclave. For LB agar, add 20 g of granulated agar, autoclave. |
| Tryptone | 10 g | ||
| NaCl | 10 g | ||
| LB/carb plates | LB agar | 1L | |
| Carbenicillin | 100 μg/mL | ||
| LB/tet plates | LB agar | 1 L | |
| Tetracycline | 10 μg/mL | ||
| LB/cmp plates | Chloramphenicol | 10 μg/mL | |
| LB/kan plates | Kanamycin | 50 μg/mL | |
| SOC medium | Yeast extract | 5 g | Add ultrapure water to make up the volume to 1.0 L and adjust pH to 7.0, autoclave. |
| Tryptone | 20 g | ||
| NaCl | 0.5 g | ||
| KCl | 0.2 g | ||
| 2.0 M MgCl2 | 5.0 mL | ||
| 1.0 M glucose | 20 mL | ||
| Superbroth medium | Tryptone | 12 g | Add ultrapure water to 900 mL, autoclave, add 100 mL of autoclaved 0.17 M KH2PO4, 0.72 M K2HPO4. |
| Yeast extract | 24 g | ||
| Glycerol | 5 mL | ||
| Superbroth kan/tet medium | Kanamycin | 50 μg/mL | |
| Tetracycline | 10 μg/mL | ||
| 1X PBS | NaCl | 137 mM | Adjust pH to 7.2, autoclave. |
| KCl | 3 mM | ||
| Na2HPO4 | 8 mM | ||
| KH2PO4 | 1.5 mM | ||
| TAE/agarose gel | TAE buffer | ||
| Agarose | 1% (w/v) | ||
| GelRed | 1:10000 (v/v) | ||
| TMB substrate | TMB | 50% (v/v) | |
| H2O2 peroxidase substrate | 50% (v/v) | ||
| M-PBST buffer | 1X PBS | 100 ml | |
| Tween-20 | 0.05% (v/v) | ||
| NON-Fat Powdered Milk | 5% (v/v) | ||
| 5X PEG/NaCl | PEG-8000 | 20% (w/v) | Add ultrapure water to make up the volume to 1L, and autoclave. |
| NaCl | 2.5 M | ||
| PBST buffer | 1X PBS | 1 L | 0.22 μm filter-sterilize. |
| Tween-20 | 0.05% (v/v) | ||
| 10X TM buffer | MgCl2 | 0.1 M | Adjust pH to 7.5. |
| Tris | 0.5 M | ||
| 1.0 mM HEPES, pH 7.4 | 1.0 M HEPES | 4.0 mL | 0.22 μm filter-sterilize. |
| Ultrapure water | 4.0 L | ||
| 10% (v/v) ultrapure glycerol | Ultrapure glycerol | 100 ml | 0.22 μm filter-sterilize. |
| Ultrapure water | 900 mL | ||
| Ultrapure water | H20 | Dnase-free, Rnase-free, Pyrogen-free. |
Table 1: Reagent setup.
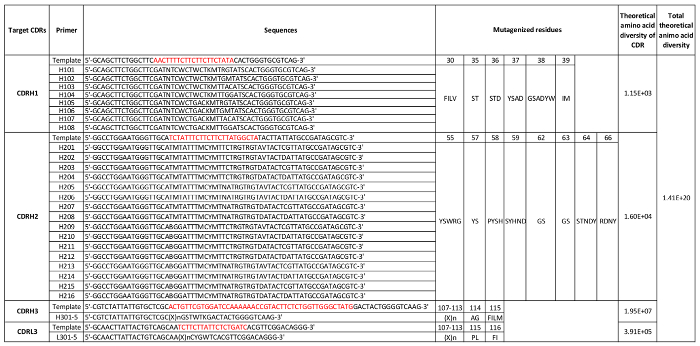
Table 2: CDR diversities and mutagenesis primers. The DNA sequences of the CDR regions to be mutated are shown in red; sequences are formatted using the IUPAC nucleotide code. "X" indicates tri-nucleotide from a mixture designed to contain different amino acid sets; "n" indicates different number of X. Five primers with a different number of X were used to diversify CDRL3 or CDRH3, respectively, to generate variable length of CDRL3 and CDRH3. The residue numbers are defined by the IMGT nomenclature. Please click here to download this table.
| Kunkel's method based mutagenesis | ||
| Reaction 1. Oligonucleotide phosphorylation with T4 polynucleotide kinase | ||
| Component | Amount | Final |
| mutagenic oligonucleotides | 0.6 μg | |
| 10X TM buffer | 2 μL | 1X |
| 10 mM ATP | 2 μL | 1 mM |
| 100 mM DTT | 1 μL | 5 mM |
| T4 polynucleotide kinase (10 U/μL) | 2 μL | 20 U |
| Ultrapure H20 | Up to 20 μL | |
| Reaction setting | ||
| Step 1. | 37 °C for 1 h | |
| Reaction 2. Annealing of the oligonucleotides to the template | ||
| Component | Amount | Final |
| dU-ssDNA template | 20 μg | 20 μg |
| 10X TM buffer | 25 μL | 1X |
| phosphorylated CDRH1 oligonucleotides | 20 μL | 0.6 μg |
| phosphorylated CDRH2 oligonucleotides | 20 μL | 0.6 μg |
| phosphorylated CDRH3 oligonucleotides | 20 μL | 0.6 μg |
| phosphorylated CDRL3 oligonucleotides | 20 μL | 0.6 μg |
| Ultrapure H20 | Up to 250 μL | |
| Reaction setting | ||
| Step 1. | 90 °C for 3 min | |
| Step 2. | 50 °C for 5 min | |
| Step 3. | 20 °C for 5 min | |
| Reaction 3. Enzymatic synthesis of CCC-dsDNA | ||
| Component | Amount | Final |
| annealed oligonucleotides/template mixtures | 250 μL | |
| 10 mM ATP | 10 μL | 346 µM of each nucleotide |
| dNTP mix (25 mM of each nucleotide) | 10 μL | 865 µM of each nucleotide |
| 100 mM DTT | 15 μL | 5 mM |
| T4 DNA ligase | 1 μL | 30 Weiss units |
| T7 DNA polymerase | 3 μL | 30 U |
| Reaction setting | ||
| Step 1. | 20 °C for overnight | |
Table 3: Procedures and components of Kunkel's method based reaction.
| Group | Clone number | Region | Percentage | ||
| No premature stop codon | 70 | CDRH1 mutation | 50% (35/70) | ||
| CDRH2 mutation | 57% (40/70) | ||||
| CDRH3 mutation | 53% (37/70) | ||||
| CDRL3 mutation | 56% (39/70) | ||||
| At least one CDR mutation | 76% (53/70) | ||||
| Premature stop codon | 20 | CDRH1 defect | 45% (9/20) | ||
| CDRH2 defect | 10% (2/20) | ||||
| CDRH3 defect | 15% (3/20) | ||||
| CDRL3 defect | 20% (4/20) | ||||
| Other defect | 10% (2/20) | ||||
Table 4: Sequence analysis of CDRH1, CDRH2, CDRH3, and CDRL3 from the synthetic Fab library.
Discussion
To construct high diversity, phage-displayed Fab libraries, quality control check points are needed to monitor various stages of the construction process, including the competency of electro-competent cells, quality of the dU-ssDNA template, efficiency of CCC-dsDNA synthesis, titer after electroporation, Fab folding, and amino acid diversity of CDRs by sequence analysis of Fab-phage clones.
High yield and purity of dU-ssDNA is essential for high mutagenesis rate. In our experience, phage induction at 25 °C overnight can yield more dU-ssDNA than that from phage induction at 37 °C overnight. This is in agreement with a previous report19. Regarding ssDNA extraction, the initial plasmid Spin kit (QIAprep) contained MLB for phage lysis and binding. Later, MLB was discontinued with unknown reason and replaced by PB. We found that the yield of dU-ssDNA is much lower from PB treatment as compared with that from MLB treatment. In this protocol, we used a reagent named UT-MLB20 to replace MLB and found the yield of dU-ssDNA is similar to that from the initial Spin Kit.
As CDRH3 and CDRL3 are the most diverse regions for antigen recognition21, to introduce a tailored diversity with a specific set of amino acid combinations and ratios, and to remove redundancy bias and stop-codons introduced by degenerate codons such as NNK (N, equimolar of A/C/G/T; K, equimolar of G/T), trimer codon phosphoramidite-based oligonucleotides22 with exactly one trimer codon corresponding to one specific amino acid were designed for CDRH3 and CDRL3. Moreover, variable lengths of CDRH3 and CDRL3 oligonucleotides were used to further increase diversities.
After enzymatic synthesis of CCC-dsDNA, generally three bands are observed by agarose gel electrophoresis and the bands should be clear without smear. Among them, the fastest-running band is the CCC-dsDNA that can yield a high mutation rate (around 80%) after electroporation23. The slowest-running band is the strand-displaced DNA that arises from inherent strand-displacement activity of T7 DNA polymerase and has a low mutation rate (around 20%)23. The middle weak band is nicked DNA after extension due to insufficient T4 DNA ligase activity or insufficient oligonucleotide phosphorylation.
A small sequencing sample pool was used to estimate the library diversity though not accurate24. To estimate the real diversity accurately, next generation sequencing (NGS) may be a good option in mining the diversity depth of the constructed library25. In practice, due to the current challenges of NGS technology including read length, accuracy, cost, and high throughput, the sequencing of the Fab phage library used in this protocol with the length of around 950 bp spanning CDRH1, CDRH2, CDRH3, and CDRL3 is not achievable; however, it is possible to estimate the scFv (around 700 bp) library diversity within the range of millions24,25. Another key standard to evaluate the diversity of constructed library is to use the library to pan against many different types of antigens and calculate the positive clones captured since library diversity is directly correlated with the successful rate of antigen panning26. High throughput selection platform is well suited for this purpose and readers can refer to a detailed protocol reported by Miersch et al.27
Theoretically, phage-displayed synthetic antibody libraries with tailored diversity can be used to target any antigen and thus have broad applications. Currently, companies including Cambridge Antibody Technology (CAT), MedImmune, Genentech, Dyax, Bioinvent, Pfizer, and MorphoSys rely heavily on phage display platforms for therapeutic antibody development28. Moreover, many phage display core technology patents have expired29. Undoubtedly, this will unleash the maximum potential of phage-displayed antibody technology.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors appreciate Dr. Frederic Fellouse from the Sidhu lab for critical comments on Kunkel's method based synthetic Fab phage library construction. The authors appreciate Mrs. Alevtina Pavlenco and other members from the Sidhu lab for valuable help of preparing high-efficiency electro-competent E. coli cells and high quality dU-ssDNA. This work was supported by National Natural Science Foundation of China (Grant No.: 81572698, 31771006) to DW and by ShanghaiTech University (Grant No.: F-0301-13-005) to Laboratory of Antibody Engineering.
Materials
| Reagents | |||
| 1.0 M H3PO4 | Fisher | AC29570 | |
| 1.0 M Tris, pH 8.0 | Invitrogen | 15568-025 | |
| 10 mM ATP | Invitrogen | 18330-019 | |
| 100 mM dithiothreitol | Fisher | BP172 | |
| 100 mM dNTP mix | GE Healthcare | 28-4065-60 | solution containing 25 mM each of dATP, dCTP, dGTP and dTTP. |
| 3,3’,5,5’-tetramethylbenzidine (TMB) | Kirkegaard & Perry Laboratories Inc | 50-76-02 | |
| 50X TAE | Invitrogen | 24710030 | |
| Agarose | Fisher | BP160 | |
| Carbenicillin, carb | Sigma | C1389 | 100 mg/mL in water, 0.22 μm filter-sterilize, work concentration: 100 μg/mL. |
| Chloramphenicol, cmp | Sigma | C0378 | 100 mg/mL in ethanol, 0.22 μm filter-sterilize, work concentration: 10 μg/mL. |
| EDTA 0.5 M, pH 8.0 | Invitrogen | AM9620G | |
| Granulated agar | VWR | J637-500G | |
| H2O2 peroxidase substrate | Kirkegaard & Perry Laboratories Inc | 50-65-02 | |
| K2HPO4 | Sigma | 795488 | |
| Kanamycin, kan | Fisher | AC61129 | 50 mg/mL in water, 0.22 μm filter-sterilize, work concentration: 50 μg/mL. |
| KH2PO4 | Sigma | P2222 | |
| Na2HPO4 | Sigma | 94046 | |
| NaCl | Alfa Aesar | U19C015 | |
| Nanodrop | Fisher | ND2000C | |
| NaOH | Fisher | SS256 | ! CAUTION NaOH causes burns. |
| NON-Fat Powdered Milk | Sangon Biotech | A600669 | |
| PEG-8000 | Fisher | BP233 | |
| Protein A-HRP conjugate | Invitrogen | 101123 | |
| QIAprep Spin M13 Kit | Qiagen | 22704 | |
| QIAquick Gel Extraction Kit | Qiagen | 28706 | |
| QIAquick PCR Purification Kit | Qiagen | 28104 | |
| Recombinant Protein L | Fisher | 77679 | |
| T4 DNA polymerase | New England Biolabs | M0203S | |
| T4 polynucleotide kinase | New England Biolabs | M0201S | |
| T7 DNA polymerase | New England Biolabs | M0274S | |
| Tetracycline, tet | Sigma | T7660 | 50 mg/mL in water, 0. 22 μm filter-sterilize, work concentration: 10 μg/mL. |
| Tryptone | Fisher | 0123-07-5 | |
| Tween-20 | Sigma | P2287 | |
| Ultrapure glycerol | Invitrogen | 15514-011 | |
| Uridine | Sigma | U3750 | 25 mg/mL in ethanol, work concentration: 0.25 μg/mL. |
| Yeast extract | VWR | DF0127-08 | |
| Name | Company | Catalog Number | Comments |
| Strains | |||
| E.coli CJ236 | New England Biolabs | E4141 | Genotype: dut– ung– thi-1 relA1 spoT1 mcrA/pCJ105(F' camr). Used for preparation of dU-ssDNA. |
| E.coli SS320 | Lucigen | 60512 | Genotype: [F'proAB+lacIq lacZΔM15 Tn10 (tetr)] hsdR mcrB araD139 Δ(araABC-leu)7679 ΔlacX74 galUgalK rpsL thi. Optimized for high-efficiency electroporation and filamentous bacteriophage production. |
| M13KO7 | New England Biolabs | N0315S | |
| Name | Company | Catalog Number | Comments |
| Equipment | |||
| 0.2-cm gap electroporation cuvette | BTX | ||
| 96-well 2mL Deep-well plates | Fisher | 278743 | |
| 96-well Maxisorp immunoplates | Nunc | 151759 | |
| Baffled flasks | Corning | ||
| Benchtop centrifuge | Eppendorf | 5811000096 | |
| Centrifuge bottles | Nalgene | ||
| ECM-630 electroporator | BTX | ||
| Magnetic stir bars | Nalgene | ||
| Thermo Fisher centrifuge | Fisher | ||
| High speed shaker | TAITEK | MBR-034P | |
| Microplate shaker | QILINBEIER | QB-9002 | |
| Liquid handler for 96 and 384 wells | RAININ | ||
| Mutil-channel pipette | RAININ | E4XLS | |
| Amicon concentrator | Merck | UFC803096 |
References
- Singh, S., et al. Monoclonal Antibodies: A Review. Curr Clin Pharmacol. , (2017).
- Reichert, J. M. Antibodies to watch in 2017. MAbs. 9 (2), 167-181 (2017).
- Kohler, G., Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 256 (5517), 495-497 (1975).
- Milstein, C. The hybridoma revolution: an offshoot of basic research. Bioessays. 21 (11), 966-973 (1999).
- Gray, A. C., Sidhu, S. S., Chandrasekera, P. C., Hendriksen, C. F., Borrebaeck, C. A. Animal-Friendly Affinity Reagents: Replacing the Needless in the Haystack. Trends Biotechnol. 34 (12), 960-969 (2016).
- Hutchison, C. A., et al. Mutagenesis at a specific position in a DNA sequence. J Biol Chem. 253 (18), 6551-6560 (1978).
- Smith, G. P. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 228 (4705), 1315-1317 (1985).
- Sidhu, S. S. . Phage Display in Biotechnology and Drug Discovery. , (2005).
- Weiss, G. A., Watanabe, C. K., Zhong, A., Goddard, A., Sidhu, S. S. Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc Natl Acad Sci U S A. 97 (16), 8950-8954 (2000).
- Frenzel, A., Schirrmann, T., Hust, M. Phage display-derived human antibodies in clinical development and therapy. MAbs. 8 (7), 1177-1194 (2016).
- Sidhu, S. S., Fellouse, F. A. Synthetic therapeutic antibodies. Nat Chem Biol. 2 (12), 682-688 (2006).
- Adams, J. J., Sidhu, S. S. Synthetic antibody technologies. Curr Opin Struct Biol. 24, 1-9 (2014).
- Kunkel, T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 82 (2), 488-492 (1985).
- Sidhu, S. S., Lowman, H. B., Cunningham, B. C., Wells, J. A. Phage display for selection of novel binding peptides. Methods Enzymol. 328, 333-363 (2000).
- Persson, H., et al. CDR-H3 diversity is not required for antigen recognition by synthetic antibodies. J Mol Biol. 425 (4), 803-811 (2013).
- Lefranc, M. P., et al. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev Comp Immunol. 27 (1), 55-77 (2003).
- Graille, M., et al. Complex between Peptostreptococcus magnus protein L and a human antibody reveals structural convergence in the interaction modes of Fab binding proteins. Structure. 9 (8), 679-687 (2001).
- Graille, M., et al. Crystal structure of a Staphylococcus aureus protein A domain complexed with the Fab fragment of a human IgM antibody: structural basis for recognition of B-cell receptors and superantigen activity. Proc Natl Acad Sci U S A. 97 (10), 5399-5404 (2000).
- Huang, R., Fang, P., Kay, B. K. Improvements to the Kunkel mutagenesis protocol for constructing primary and secondary phage-display libraries. Methods. 58 (1), 10-17 (2012).
- Chen, G., Sidhu, S. S. Design and generation of synthetic antibody libraries for phage display. Methods Mol Biol. 1131, 113-131 (2014).
- Padlan, E. A. Anatomy of the antibody molecule. Mol Immunol. 31 (3), 169-217 (1994).
- Virnekas, B., et al. Trinucleotide phosphoramidites: ideal reagents for the synthesis of mixed oligonucleotides for random mutagenesis. Nucleic Acids Res. 22 (25), 5600-5607 (1994).
- Fellouse, F. A., Sidhu, S. S. . Making and Using Antibodies: A Practical Handbook. , 157-180 (2006).
- Fantini, M., et al. Assessment of antibody library diversity through next generation sequencing and technical error compensation. PLoS One. 12 (5), e0177574 (2017).
- Glanville, J., et al. Deep sequencing in library selection projects: what insight does it bring?. Curr Opin Struct Biol. 33, 146-160 (2015).
- Perelson, A. S., Oster, G. F. Theoretical studies of clonal selection: minimal antibody repertoire size and reliability of self-non-self discrimination. J Theor Biol. 81 (4), 645-670 (1979).
- Miersch, S., et al. Scalable high throughput selection from phage-displayed synthetic antibody libraries. J Vis Exp. (95), e51492 (2015).
- Ponsel, D., Neugebauer, J., Ladetzki-Baehs, K., Tissot, K. High affinity, developability and functional size: the holy grail of combinatorial antibody library generation. Molecules. 16 (5), 3675-3700 (2011).
- Petering, J., McManamny, P., Honeyman, J. Antibody therapeutics – the evolving patent landscape. N Biotechnol. 28 (5), 538-544 (2011).

