Use of High-Throughput Automated Microbioreactor System for Production of Model IgG1 in CHO Cells
Summary
A detailed protocol for the concurrent operation of 48 parallel cell cultures under varied conditions in a microbioreactor system is presented. Cell culture process, harvest and subsequent antibody titer analysis are described.
Abstract
Automated microscale bioreactors (15 mL) can be a useful tool for cell culture engineers. They facilitate the simultaneous execution of a wide variety of experimental conditions while minimizing potential process variability. Applications of this approach include: clone screening, temperature and pH shifts, media and supplement optimization. Furthermore, the small reactor volumes are conducive to large Design of Experiments that investigate a wide range of conditions. This allows upstream processes to be significantly optimized before scale-up where experimentation is more limited in scope due to time and economic constraints. Automated microscale bioreactor systems offer various advantages over traditional small scale cell culture units, such as shake flasks or spinner flasks. However, during pilot scale process development significant care must be taken to ensure that these advantages are realized. When run with care, the system can enable high level automation, can be programmed to run DOE's with a higher number of variables and can reduce sampling time when integrated with a nutrient analyzer or cell counter. Integration of the expert-derived heuristics presented here, with current automated microscale bioreactor experiments can minimize common pitfalls that hinder meaningful results. In the extreme, failure to adhere to the principles laid out here can lead to equipment damage that requires expensive repairs. Furthermore, the microbioreactor systems have small culture volumes making characterization of cell culture conditions difficult. The number and amount of samples taken in-process in batch mode culture is limited as operating volumes cannot fall below 10 mL. This method will discuss the benefits and drawbacks of microscale bioreactor systems.
Introduction
Monoclonal antibodies (mAbs) were first produced in mouse hybridoma cells in 19751. Since then, an increase in the development of recombinant protein production has taken place to humanize mAbs to increase in vivo safety and efficacy2,3,4. Most of the recombinant protein production processes employ Chinese Hamster Ovary (CHO) cells for the ease with which they can be adapted to serum free media, their ability to produce proteins with similar post-translational modifications to that of an innate human protein and their dependability as host cells5,6.
Demand is growing to deliver product faster, and for larger patient populations with consistent quality. In addition to the economic benefits, the repertoire of diseases treated by mAbs is increasing, which now includes autoimmune diseases, post-transplantation complications, arthritis, and cancers7. Average yields for modern commercial mAb production lines are typically in the range of 5-6 g/L and continue to rise5. In part, this has been accomplished through CHO cell engineering and improved production line screening using high-throughput bioreactors8. However, most increases in protein production have been attributed to process improvements, including advancements in media optimization, cell culture conditions, and enhanced feeding strategies7,9,10. Nutrient supplementation is essential not only for proper cell growth but also for efficient production of high quality protein. Furthermore, cells require the stoichiometric addition of specific nutrients, requiring additional understanding for feeding strategy optimization6,11. Traditional optimization methods include individual media component titration and media blending with mixture designs. However, these methods are time consuming, labor intensive, and involve risks associated with human error12,13.
Media optimization studies previously relied on shake flasks and 1-2 L bioreactors which may be prohibitively expensive in terms of raw materials and human capital. Microplates have also been used but these methods provide limited scalability. Furthermore, this may still require multiple time-consuming runs that introduce batch-to-batch variability which obscures the CQA variability caused by media composition and feeding strategy14,15,16. Thus, the need for high-throughput and highly consistent parallel bioreactor systems emerged17,18,19,20.
With the significant expense associated with the operation of traditional bench-scale bioreactors (0.5-5 L), microbioreactors offer a cost-reducing alternative for assessing the production of biologically derived drugs.21 Bench-scale stirred-tank bioreactors are dependable and provide dense data via sensory arrays. Feedback control systems allow for easy oversight of operation. However, the assembly, calibration, cleaning, labor costs, substrate costs, and sterilization requirements make bench-scale stirred-tank bioreactors expensive and labor intensive to operate. Shake flasks and microtiter plates remove some of the cost and labor problems associated with larger scale bioreactors, but these alternatives provide weak control over processing conditions and produce low density data, often only end-point measurements.22
Alternatively, microbioreactors utilize a small working volume to provide a scale-down approach to cell line and upstream process development. The scale of microbioreactor experiments can significantly decrease running cost through lower utilization of power, substrate, labor, space, and utilities.23 Microbioreactors are like shake flasks in that they are easy to handle due to their size, but they retain the advantages of traditional bench-scale bioreactors through their online feedback control of pH, temperature, dissolved oxygen, and acid/base consumption as well as their real-time data output of quality parameters including off gas composition. Microbioreactor scale allows for high-throughput screening capability, which can be useful for clone selection and process development.24
The Advanced Microscale Bioreactor has been shown to be an effective tool for CHO cell culture process characterization and development18. Herein, an automated ambr15 system, consisting of 48 microbioreactors in parallel, that have been shown to be comparable to classical stirred tank reactors in scale up studies,25 was used in a manner analogous to prior work that optimized the media composition for a CHO-DG44 cell line producing a model chimeric IgG16. The effects of varying media conditions on growth and titer were compared, and analyzed. In this paper a general guideline to run the microbioreactor system and analysis of crude media samples has been presented.
Protocol
1. Seed Train Expansion
Note: This protocol uses 1 mL recombinant DG44 CHO cell stocks that have been stored at a density of ~ 3 x 107 cells/mL. Dilutions and timelines for individual CHO cell lines will vary. Measure growth curves of the cell line to be used beforehand and adjust accordingly. The cells are initially thawed into shake flasks and later transferred to a spinner flask. Determine the number of shake flasks and spinner flasks needed for the experiment based on the number of microbioreactors that will be run and the target seeding density.
- Rapidly thaw stock vial(s) of CHO cells by immersing in a 37 °C water bath until only a small sliver of ice remains. Decontaminate the outside of the vial using 70% ethanol solution and a lint-free tissue. Transfer to a biosafety cabinet.
- Re-suspend cells by gently pipetting up and down and transfer 1 mL to a sterile 125 mL vented shake flask containing 29 mL pre-warmed media supplemented with 8 mM L-glutamine and 1x Penicillin/Streptomycin.
Note: Unless otherwise indicated, the term "media" in this protocol will hereafter be defined as OptiCHO media. - Place shake flask(s) in an incubator maintained at 37 °C and 8% CO2. Use an orbital shaker to agitate cells at a speed of 130 rpm.
- Monitor the viable cell density (VCD) every day using an automated cell counting device or manually with a hemocytometer and trypan blue. Subculture (dilute) the cells 72 hours after inoculation in fresh media (Pre-warm the media to 37 °C every time it must be added to cells) such that the final volume is 100 mL in a 125 mL spinner flask. Incubate spinner cultures at the same conditions used for shake flask cultures at a speed of 70 rpm.
Note: After subculture, cells should be at a density of 0.7-1 x 106 cells/mL. Ensure that final density is not below 0.5 x 106 cells/mL. Subculture after 96 h if the target cell density for inoculation in spinners cannot be reached. - On day three (one day prior to inoculum preparations), add fresh, pre-warmed media to spinner-flask(s) to maintain ≥ 90% viability. Do not exceed 125 mL total volume.6
2. Running the automated microbioreactor system
Prerequisites: User must have received the appropriate training from the manufacturer and must be familiar with the safety and operating conditions for the system.
- Initializing and Connecting to Cell Counter
- Initialize the system operating software. Before opening the software, make sure the cell counter (see Table of Materials) is running along with the respective software. The cell counter is integrated with the system.
- Connect to Cell counter remote connection before opening the software. Click on the remote desktop icon and click on "Connect." Once the remote desktop is connected, open the cell counter software. Install a new reagent pack, empty the trypan blue waste and prime the system.
- Minimize the remote connection and open the micro bioreactor software using an existing experiment as a template to create a new experiment. Name and save the experiment. Ensure that the automated cell counter is connected under the status tab in the software. The status can also be checked on the cell counter software.
- Defining Plates and Loading Vessels
Note: Please refer to Figure 1 to orient loading of consumables and reagents on the culture stations and decks. The clamp plates should be autoclaved on a gravity cycle (used for solid materials) prior to use.- Before starting a run, define the plates used during the run in the mimic section of the software. Name each plate used and designate each plate to a deck on the microbioreactor culture station.
- Use a 24-well plate for media charging and place on the designated deck of the culture station. Place 1 mL and 4 mL pipet tip boxes in the sections as indicated in Figure 1.
- Place a 24-well inoculation plate on the respective deck of the culture station. Place the single-well 1X phosphate buffered saline (PBS) plate on deck 1. Place the single-well 1 M NaOH plate on deck 2 and the 24-well antifoam plate on deck 7 (Positions as indicated in Figure 1). The deck numbers are diagrammatically shown under the "Mimic" tab in the software.
- Unpack the culture vessels (equipped with sparger) inside the safety cabinet hood. Place twelve sterile culture vessels per culture station. Place autoclaved clamp plates on the top of the vessels. Ensure that the holes in inserts provided for the stirrers all face the same direction for easier placement.
Note: Using a permanent marker, categorize the culture vessels before placing them in the culture station to save time and avoid confusion at the time of harvest. - Clamp plate O-rings are the first part to wear down after repeated autoclaving, therefore carefully check them prior to autoclaving before each experiment. Keeping a sterile clamp plate as back-up in case of O-ring failure is recommended.
- Place the stir plates on top of the clamp plates, ensuring each pin is securely inserted. Secure the clamp plates with the screws and knobs provided. Tighten the knobs till they are hand tight. Tighten left and right knobs alternatively for even placement of the clamp plate.
Note: If the clamp is not flush against the surface or if the screws at the end of the clamp plate are unevenly tightened, the vessels will experience Dissolved Oxygen (DO) control problems. If these adjustments do not correct the DO problem, check the O-rings as incomplete sealing of plates can lead to inefficient gassing of culture.
- Running the Automated Micro Bioreactor Software
Note: Use "Process Steps" tab to edit or create new steps. Steps need to be individually programmed to start and stop any process. Click "Insert Step" button under the process steps tab to create new steps or double click on an existing step to edit that step. The program steps are divided into ten major sections: Startup, Media Charging, Antifoam Addition, DO/pH Control, Background Base Additions, Paused pH, Inoculation, 5X Cell Count, 10X Cell Count, Nutrient Analyzer Sampling and Culture Station Shutdown. The run duration is usually between 7-9 days when run in batch mode.- The system initializes and checks for the presence of the appropriate vessels. Scan the barcode provided with the culture vessels. The same barcode can be applied for culture stations with empty vessels or vessels not being used.
- The system will commence with the designed program after checking DO control, gassing and other connections.
Note: The user can proceed with the program even if an error occurs, but do so at the user's risk and if proceeding forward does not harm the system and will not interrupt the experiment. For example, gassing may be turned off for certain vessels not used in the experiment which will show up as an error but can be bypassed.
- Startup and Media Charging
- The first day setting up the system or the media charging day is designated as day 0 of culture time.
- Under the startup section, first load pipet tips, both 1 mL and 4 mL tips, as programmed. If already placed, click on continue. Place the media plate, 1X PBS, 1 M NaOH, antifoam plate, media charging plate and inoculation plate in the designated decks or, if already placed, press continue to move to the next step.
- As part of the startup protocol, begin temperature control and set temperature to 37 °C. Switch on the stirring at 1000 rpm and the DO/pH monitor.
- Next, execute the media charging program. The liquid handler of the automated microbioreactor system will dispense the media from the media plate to the culture vessels as mapped in the program. The media added is OptiCHO with varying process conditions.
Note: Two wells from a 24-well plate are required to fill one culture vessel to capacity (13 mL), as each well can only accommodate 8 mL of media. Use 7-8 mL in each well to prevent drawing air when media is being mixed by the liquid handler prior to charging. - Once the media charging is done, 35 µL of Ex-Cell antifoam will be added from the antifoam plate to the culture vessel. Allow 30 minutes for culture medium to be well mixed and the optical DO/pH sensors to be hydrated.
Note: The same volume of antifoam is added intermittently throughout culture time when foaming is detected.6 Foaming is detected visually, and reactor vessels are checked every day for foaming. Antifoam is added immediately upon detection. - The DO/pH control is then switched on to start monitoring and recording the DO and pH of the culture medium. Allow the DO to reach the set point of 50%, which takes at least 2 h.
- After DO equilibration, turn on background base additions to attain a pH set point of 7.1 for all culture vessels. Allow DO and pH to equilibrate overnight and to be completely hydrated.
- Paused pH – Day 1 pH Offset
- The next day (marked as day 1), execute the paused pH step whereby select culture vessels are analyzed and a pH correction offset is determined.
Note: The number of pH vessels to be sampled for offsetting the pH is user determined. A sample from every reactor vessel may not be necessary if you have a good representation of the bioreactor population. - Place the sample tube holder plates on designated decks and load micro centrifuge tubes with caps open and tucked beside them. The liquid handler will dispense 600 µL of the cell culture fluid into the tube as mapped in the program.
- Immediately remove the sample and measure pH on a nutrient bioanalyzer that has been properly calibrated and for which the QCs have been run (see below, "Daily Nutrient and Metabolite Analysis").
Note: Samples are run individually, immediately after drawing, to avoid pH changes due to exposure to air, as degassing of CO2 from the sample can cause pH changes. - Hit "continue" after each sample, otherwise the next step will not be executed.
- Enter the external, FLEX-derived pH values in the software, "compiling" the offsets for the sampled vessels automatically. Manually average the offsets obtained for the sample vessels and enter the number under the user pH offset column under the tab "Vessel Data". The average offset would then be applied to the all the vessels. Allow the pH to equilibrate for at least 2-3 hours, after which the culture vessels can be inoculated.
- The next day (marked as day 1), execute the paused pH step whereby select culture vessels are analyzed and a pH correction offset is determined.
- Inoculation
- After measuring VCD, transfer entire contents of spinner-flask(s) into a sterile, 250 mL conical centrifuge tube and pellet cells by centrifuging for 10 min at 140 x g at room temperature. Decant the old media and re-suspend in enough fresh media such that the final density should be 1×106 cells/mL after adding the inoculum to the culture vessel.
- Add the suspended cells into the designated well of a sterile lidded 24-well plate. Add 3 mL of inoculum to each well, out of which 2 mL will be removed as inoculum for each culture vessel.
- Place the inoculum plate on the designated deck inside the hood. Make sure to spray the outside of the lidded inoculum plate with 70% alcohol before placing it inside the hood.
- Cell Counting Using Automated Cell Counter
- After inoculation, let the culture vessels equilibrate for at least an hour. After an hour, execute the 5X cell count step in the program. 5X indicates the dilution factor used to read the cell count.
Note: 5X cell count is used initially when the cells are in the lag phase. After the cells reach their exponential phase the 10X cell count will be used. Based on the sample and diluent volumes the instrument automatically accounts for the dilution factor and adjusts the final value accordingly. - The liquid handler first adds 480 µL of 1X PBS to the cell counter cup followed by the addition of 120 µL of the cell culture fluid from the culture vessel. The cell count is then read using the automated cell counter. This step is repeated for all the vessels.
- The cell count is the last step for day 1 of the culture time. Along with the DO and pH data the cell count data (viable cell density and viability) is also recorded daily by the software.
Note: Clean the cell counter cup at least twice during the duration of the run with 70% IPA to prevent clogging of lines. Lines and flow-cell are automatically cleaned after every cell count.
- After inoculation, let the culture vessels equilibrate for at least an hour. After an hour, execute the 5X cell count step in the program. 5X indicates the dilution factor used to read the cell count.
- Paused pH – Day 2 pH Offset
- On day 2 of culture time, repeat the "Paused pH" step using culture vessels other than the ones that were sampled from during the initial paused pH step.
Note: Sampling more of the vessel population for paused pH will provide a better offset for pH correction. Hence, do not sample the same vessels used for paused pH from the previous day .
- On day 2 of culture time, repeat the "Paused pH" step using culture vessels other than the ones that were sampled from during the initial paused pH step.
- Daily Nutrient and Metabolite Analysis
- Samples will be taken for Nutrient analysis from Day 2 of culture until the end of the experiment and analyzed using nutrient bioanalyzer.
- Place the sample tube holder plates on designated decks and load micro centrifuge tubes with caps open and tucked beside them. The liquid handler will dispense the cell culture fluid into the tube as mapped in the program.
Note: For the initial few days (days 2 – 4) the amount of sample taken from the culture vessel is 300 µL. This is diluted with 300 µL of 1X PBS dispensed by the liquid handler. This is done to conserve the culture volume and prevent the level from dropping below 10 mL. Additionally, nutrient values for the first few days fall within the instrument detection range after dilution. - Place the samples in the analyzer tray to perform the nutrient analysis.
Note: Freeze any samples that will not be analyzed on the same day.
- System Shutdown
- To terminate the run, first turn off the temperature control followed by the agitation. Second, stop the DO/pH control and background base additions. Third, stop all other controls. Lastly, stop the system monitor.
- Unscrew the clamp and stir plates and remove culture vessels. Clean the inside of the culture station with a lint-free wipe. Place the drying plates on the culture station and screw them in.
Note: Drying cycle is required for the cooled version of the system and is not necessary for the standard version of the system. - Execute the 2-hour drying cycle on the program. Clean the clamp plates using ultrapure water followed by 70% Isopropyl Alcohol (IPA) to remove possible condensed liquid in the lines. Flush with air to remove any residual liquid.
- Click on "Stop" in the bioreactor software once the drying cycle has finished.
3. Cell Culture Harvest
- Transfer the cell culture fluid from the reactor vessels and pellet cells at 1,962 x g for 5 mins at room temperature.
- Decant the supernatant. Using a 0.22 µm PVDF filter sterile filter the decanted cell culture fluid.
- Aliquot 1 mL of the sterile cell culture fluid into 1.5 mL Eppendorf tubes for titer analysis. Store the 1 mL tubes at -20 °C. Purify the rest of the harvested cell culture fluid using fast protein liquid chromatography system.6
4. Measuring IgG Titers
Note: This is a cursory overview of running and analyzing samples using the proteinA biosensor system. All assay parameters ( e.g. temperature, read time, rpm, etc. ) must be determined empirically for each sample type.
- Turn on the system and allow the lamp to warm up for at least 1 h. Remove samples from freezer to thaw and equilibrate to room temperature.
- In the system software, set the plate temperature to 26 °C. Pre-soak the number of Protein A tips to be used in sample matrix (e.g. cell media) for at least 30 minutes.
Note: The optimal temperature needs to be determined empirically. A temperature of 26 °C is used here to minimize sample evaporation during longer analysis times and allow the instrument to hold a consistent temperature (by being several degrees above the ambient temperature). Samples must be pre-equilibrated to the chosen assay temperature prior to measurement by incubating the sample plate on the sample stage for ≥ 10 minutes. - Build a protein standard curve using the same antibody concentrated to 10 mg/mL and serially dilute in the sample matrix (i.e. media) in the range that needs to be detected.
Note: It is important to obtain a high concentration of antibody to minimize matrix effects due to dilution, but it is also important to not over concentrate the antibody and induce aggregation. Running an in-plate standard curve for each sample plate is preferable. Minimally, a positive control must be used to account for tip-to-tip and plate-to-plate variability. A new standard curve must be generated for each new lot of protein A tips used. - Design the sample plate (for an example, please see Figure 2). In a protein A assay that uses regeneration, up to 80 samples can be analyzed, which includes unknown culture samples, standards, and controls. For each plate analyzed, a single protein A tip will be used to measure up to 10 samples across the plate (columns 1-10), with a regeneration cycle in between each sample.
Note: It is recommended that the entire first row of the plate (row A) be used as negative control and reference. In the assay discussed here, rows B and C are used for two positive controls; one at the lower and one at the upper limit of the linear response. This ensures each tip measures negative and positive controls before analyzing samples. This set-up also simplifies reference subtraction in the analysis software. The remaining wells are then used for unknown samples. If there is a sample gap in one of the rows, there must be a matrix in that well to prevent drying of the tip (i.e. Wells A2 through G2 have sample while H2 does not. Ensure H2 contains sample matrix to ensure tip does not dry out before proceeding to sample H3). Lastly, do not exhaust tips by using one set to analyze multiple plates. Using a new, non-regenerated set for every plate is recommended. - Prepare samples for analysis by gently mixing, either by inversion or pipetting, followed by a pulse spin to collect sample at the bottom of the tube. For concentrated samples, create appropriate dilution replicates by diluting in sample matrix.
Note: The linear range of binding for an antibody for protein A bio-layer interferometry (BLI) measurements will vary by both the antibody, assay conditions, and matrix. This should be determined empirically beforehand to ensure appropriate sample dilutions are used. - Prepare 96-well sample plates as close to assay time as possible. Ensure there are no air bubbles in any of the wells. Remove air bubbles with a clean pipette tip or by centrifugation.
- Load plate into the system and run assay using the default "High Sensitivity Assay with Regeneration" in the Data Acquisition software. Analyze using HT Data Analysis software.
Note: If plates are to be prepared ahead of time due to constraints, seal securely with film to prevent evaporation. Depending on expected titers, acquisition rates and times may need to be adjusted. Refer to the manual for guidance. - Using the Data Analysis software, reference subtract and calculate the concentration of the unknown samples using standard curve and the initial slope (IS) function with a linear point-to-point fit.
Representative Results
Monitoring critical process parameters and other cell culture parameters throughout the cell cultures' operation is an important aspect in bioprocessing. The cell counter and nutrient analyzer were used to quantify five attributes that characterize cell growth, nutrient consumption and byproduct formation. The cell counts were obtained daily for all culture conditions. The average viable cell densities and viabilities are as seen in Figure 3 along with their ±1 SD interval. The nutrient and byproduct profiles are also shown with the ±1 SD interval through the stationary phase of the cultures. The slope of this profile represents the average glucose and glutamine consumption, and lactate production, rates. Overall, these results demonstrate the feasibility of monitoring these attributes in the microbioreactor system; as well as the capability of the microbioreactor system to maintain these parameters within a tight range.
The total productivity of the cell cultures was quantified using a proteinA biosensor system after the harvested cell culture media was passed through a 0.22 micron PVDF filter. The specific productivity per cell ranged from 0.87 pg/cell-d to 1.15 pg/cell-d as seen in Figure 4. Based on these results a wide array of conditions can be investigated to select media composition and feeding strategies that maximize the amount of protein produced for a minimal investment in experimental procedures.
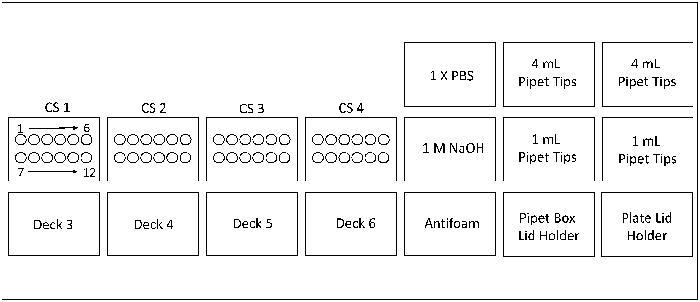
Figure 1: Layout of the microbioreactor system with 4 culture stations (CS) having 12 reactor vessels each. Please click here to view a larger version of this figure.
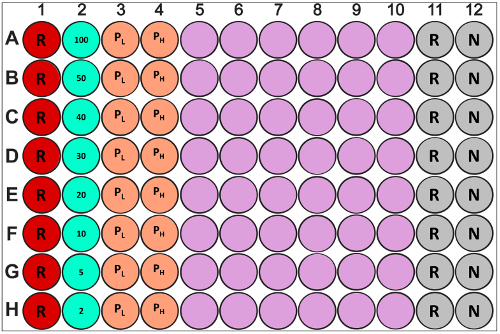
Figure 2: Example sample plate set-up for a basic quantitation with regeneration experiment on the proteinA biosensor system. row 1 (red "R") is reserved for reference sample (i.e. matrix absent analyte); row 2 (aquamarine) is a set of standards (concentrations are in µg/mL); rows 3 and 4 (orange) are sets of low and high positive controls ("PL" and "PH," respectively); rows 5 through 10 (purple) contain unknown samples; rows 11 and 12 (grey) are program-default positions for regeneration ("R") and neutralization ("N") buffers. Please click here to view a larger version of this figure.
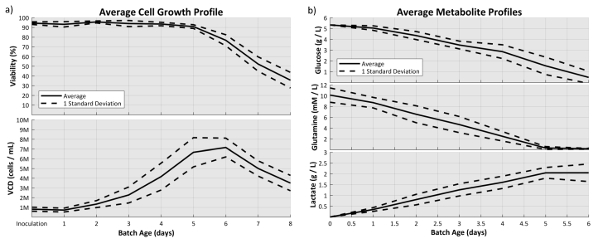
Figure 3: a) Average viable cell density and viability profiles over the age of a batch. ± 1 SD is also shown to indicate the tight control of cell growth in the microbioreactor systems. b) Average nutrient profile for glucose and glutamine as well as the average byproduct profile for lactate. ± 1 SD is also shown to indicate tight control over media composition in the microbioreactor systems. (N=3, all conditions were run in triplicate). Please click here to view a larger version of this figure.
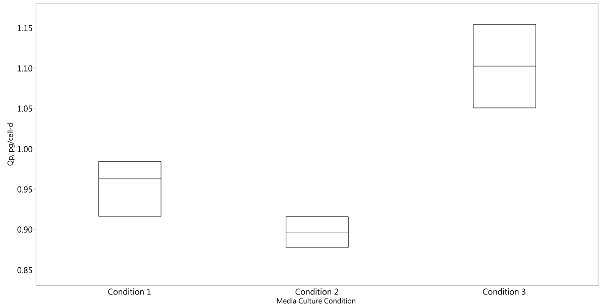
Figure 4: Representative box-plot of average specific productivity of the various media conditions. (N=3, all conditions were run in triplicate) Please click here to view a larger version of this figure.
Discussion
Running the automated micro bioreactor system properly and efficiently involves the timely execution of multiple automated steps. One of the most important parts of running the system is programming the software. If there is any error while writing the program, there will be serious errors in the experiment that may result in unexpected changes in the process, feeding strategy, sampling strategy, or final product quality which may invalidate the findings of the study. Another important aspect of running the system is to place and tighten the clamp plate correctly to ensure proper DO control. The most common indication that the clamp plate has been tightened unevenly is unexpected variations in DO measurements for vessels 1, 6, 7 and 12 (corner reactor vessels). Overall DO instability indicates a loosening of gaskets at the gas inlet lines in the clamp plates. This scenario may impede reaching the DO set point. Another common pitfall to avoid when starting an experiment is letting the cells sit too long during the inoculation step, causing them to settle. The less time the cells spend sitting, the less chance there is that progressively lower inoculum cell counts are added chronologically to the reactor vessels which can induce significant bias that unwittingly harms study results. It is better to inoculate in multiple stages, i.e. inoculate each culture station one after another with pause steps in between so the cells are not sitting in the inoculation plate for longer than 15 minutes.
Concerning day-to-day use, maintaining sterility is vital. Although the system is in a biological safety cabinet, sterility is not guaranteed due to frequent movement in and out of the hood. Consequently, everything that goes in the hood must be sprayed with 70% IPA. Secondly, it is essential to ensure that minimal foaming occurs during the culture; media can clog gassing and exhaust lines, leading to damage of the clamp plate and even core components below. Preventive anti-foam addition steps are critical in any micro bioreactor program design. In the event of a "foam out" it would be beneficial to follow the manufacturers cleaning protocol and can prevent permanent damage of clamp plates. Alternatively, use of non-sparged vessels may be beneficial for lower cell densities or when running in batch mode as higher surface to volume ratio enables efficient oxygen even with lack of a sparger. However, non-sparged vessels might not be useful for high cell density or perfusion cultures as the head space is insufficient to keep up with the cultures growing consumption of oxygen.
There are numerous advantages provided by the microbioreactor system, as it enables multiple controlled cultures to be run in parallel at a small scale with greater control than shake flasks.17 Therefore, the system facilitates the execution of screening studies, DoEs, high throughput clone studies and transfection studies. Automated liquid handling also reduces analyst-to-analyst variability while simultaneously minimizing tedious and time-intensive labor for trained personnel. While there are several advantages to the system, there are several key disadvantages that should be considered. First, a culture volume of 15 mL significantly limits in-process sampling and final harvest material, and multiple alternative small scale bioreactors (up to 500 mL) have recently become available. One recent advancement to the system is the integration of the automated microscale bioreactor with the BioProfile FLEX 2 analyzer from Nova Biomedical, which mitigates the in process-sampling issue by reducing sample volume for cell density and nutrient analysis. Benefits can include quick setup and virtually no cleaning leading to operational savings, however the cost of disposable units should be considered for long term projects as it may be costlier to purchase the units than the reusable conventional systems.
The method discussed in this paper is primarily suitable for batch mode cell culture, but can be modified depending on the needs of the user. Each culture station has independent control of temperature, while DO and pH can be varied at the level of individual reactor vessels. Sartorius also offers DoE planning software designed specifically to allow experiments to be tailored for the micro bioreactor system. Large scale DoE studies using the new DoE software provided by the manufacturer can help in media and supplement optimization. Although not used here, the microbioreactor system also enables fed-batch studies. The system has not yet been optimized for perfusion cell cultures. However, there have been limited studies and trials to mimic perfusion cell culture operation in the current micro bioreactor system.26 This method can be modified to mimic high density perfusion cultures by cell settling. By varying the height to which the pipet is inserted into the reactor and by optimizing settling time, the media can be removed and replenished to mirror profusion mode of culture. There are new products in this developing area that may work better than the system presented here if perfusion mode of culture is desired.
In summary, this study demonstrates the use of automated micro-bioreactors and associated analytical for CHO cell culture operations to produce and characterize a model IgG1 Monoclonal antibody. It emphasizes the role small scale micro-bioreactors play in bioprocess manufacturing and their impact on cell culture development and media screening. While there are many advantages to using an automated small scale system, to fully realize their benefits process understanding and analytical characterization is imperative. This study provides the user with a guideline for using an automated microscale reactor system, that can be developed and improved per individual research needs.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank Scott Lute for the analytical support they provided. Partial internal funding and support for this work was provided by the CDER Critical Path Program (CA #1-13). This project was supported in part by an appointment to the Internship/Research Participation Program at the Office of Biotechnology Products, U.S. Food and Drug Administration, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and FDA.
Materials
| CHO DG44 Cell Line | Invitrogen | A1100001 | |
| ambr 15 automated microbioreactor system | Sartorius | 001-2804 | automated micro bioreactor |
| ambr 15 Cell Culture 24 Disposable Bioreactors – Sparged | Sartorius | 001-2B80 | |
| 1 mL disposable pipette tips, sterilized | Sartorius | A-0040 | |
| 5 mL disposable pipette tips, sterilized | Sartorius | A-0039 | |
| 24 Well deep well plates | Sartorius | A-0038 | |
| 1 Well plates | Sartorius | A-0068 | |
| Vi-Cell XR cell counter | Beckman Coulter | 731050 | automated cell counter |
| EX-CELL Antifoam (gamma irradiated) | Sigma-Aldrich | 59920C-1B | |
| CD OptiCHO AGT Medium | Thermo Fisher Scientific | A1122205 | |
| 200 mM L-glutamine | Corning | 25-005-CV | |
| 100X Penicillin/Streptomycin | Corning | 30-001-CI | |
| 125 mL F-Bottom Shake Flasks (Sterile, Vented) | Fisher Scientific | PBV12-5 | |
| 125 mL glass Spinner Flasks | Corning Life Sciences Glass | 4500-125 | |
| 250 mL PP Conical Centrifuge Tubes (Sterile) | Nalgene (Thermo Scientific) | 376814 | |
| TC20 Automated Cell Counter | BioRad Laboratories, Inc. | 1450103 | |
| Trypan Blue | Sigma-Aldrich | T8154 | |
| 10x PBS | Corning | 46-013-CM | |
| BioProfile FLEX Analyzer | Nova Biomedical | 49418 | Nutrient Analyzer |
| Octet Red 96 | Pall FortéBio | 99-0042 | Protein A Biosensor |
| Protein A Dip and Read Biosensors | Pall FortéBio | 18-5010 | |
| Polypropylene 96-well Microplate, F-bottom, Chimney-style, Black | Greiner Bio-One | 655209 |
References
- Köhler, G., Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 256, 495-497 (1975).
- Buss, N. A., Henderson, S. J., McFarlane, M., Shenton, J. M., de Haan, L. Monoclonal antibody therapeutics: history and future. Current Opinion in Pharmacology. 12 (5), 615-622 (2012).
- Jakobovits, A. Production of fully human antibodies by transgenic mice. Current Opinion in Biotechnology. 6 (5), 561-566 (1995).
- Maksimenko, O. G., Deykin, A. V., Khodarovich, Y. M., Georgiev, P. G. Use of Transgenic Animals in Biotechnology: Prospects and Problems. Acta Naturae. 5 (1), 33-46 (2013).
- Jayapal, K. P., Wlaschin, K. F., Hu, W., Yap, M. G. Recombinant protein therapeutics from CHO cells-20 years and counting. Chemical Engineering Progress. 103 (10), 40 (2007).
- Velugula-Yellela, S. R., et al. Impact of media and antifoam selection on monoclonal antibody production and quality using a high throughput micro-bioreactor system. Biotechnology Progress. , (2017).
- Foltz, I. N., Karow, M., Wasserman, S. M. Evolution and Emergence of Therapeutic Monoclonal Antibodies. Circulation. 127 (22), 2222-2230 (2013).
- Kondragunta, B., Drew, J. L., Brorson, K. A., Moreira, A. R., Rao, G. Advances in clone selection using high-throughput bioreactors. Biotechnology Progress. 26 (4), 1095-1103 (2010).
- Altamirano, C., Paredes, C., Cairo, J., Godia, F. Improvement of CHO cell culture medium formulation: simultaneous substitution of glucose and glutamine. Biotechnology Progress. 16 (1), 69-75 (2000).
- Selvarasu, S., et al. Combined in silico modeling and metabolomics analysis to characterize fed-batch CHO cell culture. Biotechnology and Bioengineering. 109 (6), 1415-1429 (2012).
- Seth, G., Ozturk, S., Zhang, C., Hu, W. -. S. . Cell Culture Bioprocess Engineering. , 97-126 (2012).
- McKeehan, W. M. K., Ham, R., Ham, R., Waymouth, C., Chapple, P. . The Growth Requirements of Vertebrate Cells In vitro. , 223-243 (1981).
- Jordan, M., et al. Cell culture medium improvement by rigorous shuffling of components using media blending. Cytotechnology. 65 (1), 31-40 (2013).
- Castro, P. M. L., Hayter, P. M., Ison, A. P., Bull, A. T. Application of a statistical design to the optimization of culture medium for recombinant interferon-gamma production by Chinese hamster ovary cells. Applied Microbiology and Biotechnology. 38 (1), 84-90 (1992).
- Liu, C. -. H., Chu, I. M., Hwang, S. -. M. Factorial designs combined with the steepest ascent method to optimize serum-free media for CHO cells. Enzyme and Microbial Technology. 28 (4-5), 314-321 (2001).
- Parampalli, A., et al. Developement of serum-free media in CHO-DG44 cells using a central composite statistical design. Cytotechnology. 54 (1), 57-68 (2007).
- Hsu, W. -. T., Aulakh, R. P., Traul, D. L., Yuk, I. H. Advanced microscale bioreactor system: a representative scale-down model for bench-top bioreactors. Cytotechnology. 64 (6), 667-678 (2012).
- Janakiraman, V., Kwiatkowski, C., Kshirsagar, R., Ryll, T., Huang, Y. -. M. Application of high-throughput mini-bioreactor system for systematic scale-down modeling, process characterization, and control strategy development. Biotechnology Progress. 31 (6), 1623-1632 (2015).
- Kim, B. J., Diao, J., Shuler, M. L. Mini-scale bioprocessing systems for highly parallel animal cell cultures. Biotechnology Progress. 28 (3), 595-607 (2012).
- Legmann, R., et al. A predictive high-throughput scale-down model of monoclonal antibody production in CHO cells. Biotechnology and Bioengineering. 104 (6), 1107-1120 (2009).
- Schäpper, D., Alam, M. N. H. Z., Szita, N., Lantz, A. E., Gernaey, K. V. Application of microbioreactors in fermentation process development: a review. Analytical and Bioanalytical Chemistry. 395 (3), 679-695 (2009).
- Hegab, H. M., ElMekawy, A., Stakenborg, T. Review of microfluidic microbioreactor technology for high-throughput submerged microbiological cultivation. Biomicrofluidics. 7 (2), 021502 (2013).
- Rameez, S., Mostafa, S. S., Miller, C., Shukla, A. A. High-throughput miniaturized bioreactors for cell culture process development: Reproducibility, scalability, and control. Biotechnology Progress. 30 (3), 718-727 (2014).
- Xu, P., et al. Characterization of TAP Ambr 250 disposable bioreactors, as a reliable scale-down model for biologics process development. Biotechnology Progress. 33 (2), 478-489 (2017).
- Delouvroy, F., et al. Evaluation of the advanced micro-scale bioreactor (ambr™) as a highthroughput tool for cell culture process development. BMC Proceedings. 7 (6), 1-3 (2013).
- Kelly, W., et al. Optimizing performance of semi-continuous cell culture in an ambr15 microbioreactor using dynamic flux balance modeling. Biotechnology Progress. , (2017).

