Primary Cell Culture of Purified GABAergic or Glutamatergic Neurons Established through Fluorescence-activated Cell Sorting
Summary
This protocol describes a cell sorting based method for the purification and culture of fluorescent GABAergic or glutamatergic neurons from the neocortex and hippocampus of postnatal mice or rats.
Abstract
The overall goal of this protocol is to generate purified neuronal cultures derived from either GABAergic or glutamatergic neurons. Purified neurons can be cultured in defined media for 16 days in vitro and are amenable to any analyses typically performed on dissociated cultures, including electrophysiological, morphological and survival analyses. The major advantage of these cultures is that specific cell types can be selectively studied in the absence of complex external influences, such as those arising from glial cells or other neuron types. When planning experiments with purified cells, however, it is important to note that neurons strongly depend on glia-conditioned media for their growth and survival. In addition, glutamatergic neurons further depend on glia-secreted factors for the establishment of synaptic transmission. We therefore also describe a method for co-culturing neurons and glial cells in a non-contact arrangement. Using these methods, we have identified major differences between the development of GABAergic and glutamatergic neuronal networks. Thus, studying cultures of purified neurons has great potential for furthering our understanding of how the nervous system develops and functions. Moreover, purified cultures may be useful for investigating the direct action of pharmacological agents, growth factors or for exploring the consequences of genetic manipulations on specific cell types. As more and more transgenic animals become available, labeling additional specific cell types of interest, we expect that the protocols described here will grow in their applicability and potential.
Introduction
Cell sorting is a powerful tool for the isolation of living cells of interest from a heterogeneous mixture of cells. Cells can be sorted based on size and shape criteria, as well as the expression of fluorescent markers1,2. Often, fluorophore-conjugated antibodies are used to label distinct cell types by targeting cell-specific surface antigens3,4. Alternatively, transgenic animals or viral delivery systems have been engineered to express fluorophores under cell-specific promoters5,6. Historically, the development of transgenic tools and animals was costly and time consuming. More recently, decreasing costs and fewer technical difficulties have led to a dramatic increase in the number of available reporter lines. As the availability of transgenic tools and reporter animals continues to grow, so too should the usefulness and applicability of fluorescence-based cell sorting methods.
We recently demonstrated that cell sorting from transgenic animals can be routinely applied to purify primary neurons in preparation for cell culture7. By sorting cells from either rats or mice, we were able to isolate and culture fluorescent neurons, which express fluorescent reporter proteins specifically in either GABAergic or glutamatergic neurons6,8,9. By studying these purified neuronal cultures, we were able to identify an important difference in the way GABAergic and glutamatergic neurons depend on glia-secreted factors for the establishment of synaptic transmission7. Additionally, by co-culturing glial cells with purified neurons, we were able to extend on previous observations demonstrating the critical role that glial cells play in the growth and survival of neurons10,11. Thus, through a combination of cell sorting and cell culture, we were able to study not only the development of specific neuron types in isolation, but were able to investigate the influence of glial cells on their function.
We present here a protocol for the purification and culture of GABAergic and glutamatergic neurons from the cortex and hippocampus of transgenic mice or rats. We also present a method for the non-contact co-culture of purified neurons and glial cells, adapted from Geissler et al.12. In order to generate purified GABAergic cultures, we have sorted fluorescent neurons from VGAT-Venus-A Wistar rats8 or VGAT-Venus mice13, which selectively express an enhanced yellow fluorescent protein variant (Venus) in >95% of cortical GABAergic neurons. To generate purified glutamatergic cultures, we have sorted fluorescent neurons from NexCre;Ai9 mice6,9, which express tdTomato in cortical glutamatergic neurons. The entire sorting and culture procedure can be performed within 3-4 h and can be used to generate hundreds of replicate cultures suitable for electrophysiological, morphological and cell survival analysis. The method is simple, reproducible and can produce purified neuronal cultures that are greater than 97% pure for the cell type of interest.
Protocol
All procedures and animal maintenance were performed in accordance with institutional guidelines, the German Animal Welfare Act and the European Council Directive 86/609/EEC regarding the protection of animals, in the presence of permissions from local authorities (LaGeSo Berlin, T-0215/11).
NOTE: This protocol describes the culture of purified neurons from a single transgenic mouse or rat pup (postnatal day 0–2). All techniques should be performed under sterile conditions. All solutions should be sterilized using a 0.2 µm filter (see Table of Materials). Glass coverslips and dissection tools should be heat sterilized for 3 h at 185 °C.
1. Coating Glass Coverslips with Poly-L-lysine
- Prepare glass coverslips by defrosting a 5 mL aliquot of 200 µg/mL poly-L-lysine (PLL) solution. Dilute this stock solution to 20 µg/mL by adding 45 mL of pure injection grade water. Filter-sterilize the solution into a new 50 mL conical tube. Label this tube as “PLL Sterile”.
NOTE: Use water stored at room temperature to speed up the defrosting process. - Add 100 sterile, round, 12 mm glass coverslips to the sterile PLL solution. Agitate the tube every 5–10 min, for 2–3 min, to ensure even coating. Coat the coverslips in this way for 1 h.
NOTE: German-manufactured round glass coverslips (diameter = 12 mm) are preferred, as they provide a reliable culture surface and fit easily into the wells of a 24-well cell culture plate. - After 40 min of PLL coating, take 2 pieces of tissue paper and lay them flat in the flow cabinet. Sterilize the paper using 70% ethanol, then flatten to remove creases and leave to dry.
- After coating the glass coverslips for 1 h with PLL, remove excess PLL solution and add sterile injection grade water. Gently agitate the coverslips for 2–3 s to allow removal of excess PLL. Repeat this rinsing step 2 additional times.
- Remove excess water, and then transfer the coverslips to the sterile tissue paper. Carefully separate each coverslip using curved forceps and leave to dry.
NOTE: Coverslips that do not easily separate can be discarded. Alternatively, a small amount of water can be added to help separation. - Once dry, transfer the coverslips to a 24-well culture plate.
NOTE: Coverslips should be made ready approximately 30 min–1 h before starting the dissection process. Plates can be stored in an incubator at 37 °C and 5% CO2 until required.
2. Dissociation of Hippocampal and Cortical Tissue
- Preparation of cell culture solutions
- For the dissociation of neural tissue, first defrost a 40 mL aliquot of cell culture buffer (composition [in mM]: 116 NaCl, 5.4 KCl, 26 NaHCO3, 1.3 NaH2PO4, 1 MgSO4·7H2O, 1 CaCl2·2H2O, 0.5 EDTA·2Na·2H2O and 25 D-glucose, pH = 7.4). Measure out 12 mL of cell culture buffer to a 15 mL conical tube; label as “BSA”. Measure out 5 mL of cell culture buffer to a different 15 mL tube; label as “papain”. Incubate both tubes in a water bath for 15 min at 37 °C.
- Filter-sterilize the remaining cell culture buffer, label as “Buffer – Sterile” and store at 4 °C until required.
- Weigh out 120 mg of Bovine Serum Albumin (BSA) and add to the tube labeled “BSA”. Weigh out 7 mg of papain and add to the tube labeled “papain”. Return both tubes to the water bath for 15 min.
NOTE: It is helpful to add a small amount of buffer to the weigh boat to facilitate transfer of the papain powder. - Once all powders have dissolved, filter-sterilize each solution to a fresh 15 mL conical tube. For tube papain, label the filtered solution as “papain – Sterile”. For tube BSA, divide the sterile BSA solution into 3 tubes, each containing 4 mL of BSA solution. Label each tube as “BSA – Sterile” and either “1”, “2” or “3”. Return all tubes back to the water bath and continue to incubate at 37 °C until required.
- Preparation for tissue dissection
- Take a single piece of 35 mm filter paper (see Table of Materials) and sterilize with 70% ethanol; leave to dry in the lid of a 100 mm Petri dish, in a flow cabinet.
- Lay out the scissors, forceps, scalpel and spatula (see Table of Materials) required for the dissection of the hippocampus and cortex.
- Place two 35 mm Petri dishes and a 100 mm Petri dish containing sterile filter paper in an accessible location in center of the flow cabinet.
- Collect transgenic NexCre;Ai9 and VGAT Venus mouse pups that are to be dissected. Use a fluorescent lamp with appropriate excitation and emission filters (see Table of Materials) to discriminate fluorescent pups from wild type littermates.
- Immediately before dissecting animals, fill each 35 mm Petri dish with chilled, sterile cell culture buffer.
NOTE: One Petri dish is for cleaning the tools between dissections, and the other is for collecting the hippocampus and cortex.
- Tissue dissection
- As soon as the cell culture buffer is poured, decapitate a postnatal day 0–2 transgenic mouse or rat pup using large, sharp scissors and use a sterile 100 mm Petri dish to transfer the head into the flow cabinet. Use forceps, scissors and a spatula to carefully transfer the brain of a transgenic pup to sterile filter paper.
- Use a type 22 scalpel to dissect away the cerebellum and to separate the 2 hemispheres. Use a small spatula to roll the hemispheres laterally. On each hemisphere, press down gently so that the cortex comes into contact with the filter paper. Gently move midbrain regions to reveal the hippocampus and cortex.
NOTE: Be careful not to apply too much pressure when pressing down on the hemispheres or cells can become crushed. - Use a scalpel or spatula to separate the hippocampus and cortex from each hemisphere. Transfer dissected tissue to a 35 mm Petri dish containing chilled cell culture buffer.
NOTE: If dissecting multiple animals, store sterile cell culture buffer in a 50 mL conical tube on ice. After each dissection, transfer dissected tissue to the chilled cell culture buffer. - After successful dissection of cortico-hippocampal tissue, transfer the sterile papain solution from the water bath to the flow cabinet. Use a 3 mL Pasteur pipette to transfer the dissected hippocampus and cortex to the lid of a sterile 35 mm Petri dish. Chop in a criss-cross motion, using the flat part of a type 22 scalpel, until the tissue is in small pieces.
- Use a small amount of papain solution to transfer the chopped tissue from the lid of the Petri dish to the sterile papain tube. Return the papain tube to the water bath and incubate the tissue at 37 °C for 25 min.
- During papain incubation, make up complete fluorescence media solution. Defrost a 1 mL aliquot of B27, a 0.5 mL aliquot of Glutamax and a 0.5 mL aliquot of Pen-Strep. Aliquot 48.5 mL of Hibernate A low fluorescence media to a 50 mL conical tube. Add B27, Glutamax and Pen-Strep to the solution. Shake the solution well, then filter-sterilize and label as “Hibernate A complete media” (with date and initials). Incubate at 37 °C in a water bath.
- After papain incubation, transfer the papain-tube and sterile BSA-tubes to the flow cabinet. Use a 1 mL Pasteur pipette to remove only the cortico-hippocampal tissue from the papain tube into BSA-tube 1.
NOTE: Take care to minimize the transfer of excess papain solution.
- Dissociation of cells
- In order to break up any large clumps of tissue, triturate the tissue several times using a 1 mL Pasteur pipette. Following this, triturate the tissue 7 times using a fine tip Pasteur pipette. Leave the tissue to stand for 30 s, allowing larger pieces of tissue to sediment.
- After 30 s, transfer the lower 1 mL of solution and tissue from BSA-tube 1 to BSA-tube 2. Triturate the tissue in BSA-tube 2 several times using a fine tip Pasteur pipette.
- After trituration, transfer again the lower 1 mL of tissue and solution from BSA-tube 1, but now to BSA-tube 3. Triturate the tissue in BSA-tube 3 several times using a fine tip Pasteur pipette.
- After trituration, transfer all solution and tissue from tubes 2 and 3 into BSA-tube 1. Triturate a further 2-3 times and centrifuge at 3,000 x g for 3 min.
- During centrifugation, make up complete Neural Basal A media (NBA media). Aliquot 48.5 mL of NBA media to a 50 mL conical tube and incubate at 37 °C in a water bath. Label this tube “NBA only”. In addition, defrost a 1 mL aliquot of B27, a 0.5 mL aliquot of Glutamax and a 0.5 mL aliquot of Pen-Strep at room temperature.
- Following centrifugation, carefully remove the supernatant from the pelleted tissue and re-suspend the cells in 2 mL of complete media. Use a P1000 pipette to triturate the tissue up and down 20 times.
NOTE: Take care not to pipette the cells too vigorously. If too much pressure is applied, the cells may become damaged. - Use a 1 mL Pasteur pipette to filter the cell suspension through a 30 µm cell sieve into a polystyrene sample tube.
NOTE: If the cell suspension does not pass immediately through the cell sieve, it may be necessary to press on top of the sieve with a sterile glove to start the flow. This important step prevents clogging of the cell sorter. - For collecting neurons after sorting, prepare collection tubes by pipetting 300 µL of complete media to the required number of polypropylene tubes.
- The cell suspension is now ready to be taken to the cell sorter for sorting. Take the cell suspension, extra collection tubes and spares complete media in case dilution of the sample is required.
3. Cell Sorting of Purified GABAergic or Glutamatergic Neurons
NOTE: To minimize the chance of bacterial contamination during sorting rinse the sample tubing of the sorter with 70% ethanol for at least 5 min prior to sorting. Detailed sorting parameters vary among instruments, fundamental considerations are as follows.
- During the first cell sort, establish levels of background fluorescence from unlabeled cells by sorting and comparing cells from a wild type animal.
- For each fluorescent cell type to be sorted, choose the appropriate excitation and emission filters. Excite Venus and TdTomato proteins using 488 nm or 531 nm excitation wavelengths, respectively. Detect emitted light through 530/40 and 575/30 emission filter sets, respectively.
- Add polypropylene collection tubes, containing 300 μL of complete media, to the cell sorter for cell collection.
- For high purity, sort brightly labeled fluorescent cells (see Figure 1B for example dot plots of sorted cells).
- Following sorting, for optimal results, perform a purity test of the sorted cells to estimate purity levels. Note that the typical recovery rate of cells following sorting is between 70–80%.
- Note down the number of cells sorted in each collection tube. This value is given by the cell sorting instrument.
NOTE: Cell sorting can be performed using a 70 µm nozzle (70 psi sheath fluid pressure) or a 100 µm nozzle (20 psi sheath fluid pressure).
4. Culturing of Sorted Neurons
- Following cell sorting, transfer collected cells to 2 mL round bottom centrifuge tubes, using a 1 mL Pasteur pipette. Centrifuge the cells at 3,000 x g for 3 min to form a cell pellet.
NOTE: To help identify the location of the cell pellet, orientate the round bottom centrifuge tubes with the hinge of the lid facing outward, which allows the cell pellet to form on the back of the centrifuge tube (i.e., on the same side of the tube as the hinge). - During centrifugation, add 1 mL of B27, 0.5 mL of Glutamax and 0.5 mL of Pen-Strep to the 48.5 mL of pre-warmed NBA media. Shake the solution well, then filter sterilize and label as “NBA complete media” (with date and initials). Return to water bath until required.
- Following centrifugation, use a 1 mL Pasteur pipette to transfer the supernatant to a different centrifuge tube (retain the supernatant incase the cell pellet was disturbed). Re-suspend the cell pellet in the required amount of pre-warmed complete NBA media to achieve a cell density of 1,000 cells/µL. Re-suspend the cells by pipetting up and down with a P200 or P1000 pipette.
- To confirm the presence of dissociated cells, check the cell solution under a microscope, using a 4x or 10x objective lens. Alternatively, pipette a small amount of cell solution onto a coverslip and check under the microscope. If large numbers of cells are present, the supernatant from step 4.3 can be discarded.
- Before plating the cells, vortex on a medium speed for 2–3 s to ensure an even cell suspension. Following vortexing, quickly pipette 10 µL of the cell suspension to the center of each coverslip. After pipetting the cells, quickly return each plate to the incubator (5% CO2/37 °C) and incubate for 1 h.
NOTE: If adding cells to multiple 24-well plates, ensure even cell densities by vortexing cells between plates. - After 1 h, feed the cells with 500 µL of pre-warmed complete NBA media and return to the incubator.
NOTE: When feeding the cells, it is important to pipette the media gently down the side of the well to avoid cell detachment. For short experiments (<16 days), feeding at the above density is not necessary. When plating a greater density of cells, more frequent feeding intervals may be required (see Discussion).
5. Astrocyte Enriched Glial Support Cultures
NOTE: The production of glial cultures14 and the use of cell culture inserts has been described previously12. In brief, astrocyte enriched glia cultures are derived from cortico-hippocampal tissue (with meninges removed; P2–P5), which have been cultured for one week on a 20 µg/mL PLL-coated 6-well plates, in serum supplemented DMEM media.
- To co-culture purified neurons and glial cells, passage confluent glial cells to the required number of cell culture inserts (0.4 µm pore size — see Table of Materials). To passage the cells, remove a 6-well plate containing confluent glial cells from the incubator and wash 2 wells, 2 times, with 2 mL of pre-warmed (37 °C) phosphate buffered saline (PBS).
- Aspirate the PBS solution and use a P1000 pipette to add 1 mL of pre-warmed (37°C) Trypsin (1:250)/EDTA (0.25%/0.02%) solution to each well.
- Return the 6-well plate to the incubator and check every 2–3 min for cell detachment.
- Once significant cell detachment has occurred, pipette the cells and cell solution gently up and down using a fine tip Pasteur pipette, to help further detachment and separation of individual cells.
- Transfer the cell solution to a 15 mL conical tube and centrifuge at 3,000 x g for 3 min.
- Re-suspend the cells using a P1000 pipette in 5 mL of pre-warmed (37 °C) NBA complete media by gently pipetting up and down until the cell pellet is in solution.
- Perform a cell count using a haemocytometer or an automated cell counter.
NOTE: After 7 days in culture, approximately 750,000–1,000,000 cells can be harvested from each well of a 6-well plate. - Plate 40,000 glial cells to each cell culture insert in a 500 µL droplet (80 cells/µL).
- After allowing the cells to adhere for 1 h, use sterile forceps to transfer glial-covered cell culture inserts to 24-well plates containing purified neurons. Remove excess media from the surface of the cell culture inserts (approximately 300 µL).
NOTE: As air bubbles can often get trapped below the cell culture inserts, it may be necessary to gently tilt the inserts to dislodge these bubbles. If more glial cells are added to the cell culture insert, further washing and centrifuge steps may be required to remove excess trypsin, which can be detrimental to cell survival.
Representative Results
The dissociation and cell sorting of fluorescent neurons from transgenic mouse or rat cortico-hippocampal tissue can be performed in approximately 3–4 h. The result is a population of highly pure fluorescent neurons, which can be grown in culture for 16 days.
To generate purified cultures, transgenic animals were first identified using a fluorescent lamp with appropriate filter sets (examples of fluorescent neonatal VGAT Venus and NexCre;Ai9 mouse pups are shown in Figure 1A). Following the identification of transgenic animals, dissected tissue was dissociated, and the most strongly fluorescent neurons were sorted to produce a purified cell population. Example fluorescence intensity dot plots for NexCre;Ai9 and VGAT Venus mouse neurons, obtained during FACS, are shown in Figure 1B. When optimized, it is possible to harvest between 500,000 and 800,000 cells from the cortex and hippocampus of individual P0 -2 NexCre;Ai9 or VGAT Venus mice. Cells can be sorted at a speed upwards of 600 events/s. Estimates of cell purity were performed using DAPI as a nuclear marker (Figure 1C). By sorting strongly positive cells, a purity of more than 97% can be routinely achieved7.
Following successful sorting, plated neurons should appear round in shape, have a smooth membrane and should be seen to sprout neurites after approximately 1 h in vitro (Figure 2A, B). By 7 days in vitro, although some cell death may be apparent, viable cells should be present in all culture conditions (Figure 2C, D). At 12–16 days in vitro, using whole-cell patch-clamp recordings and biocytin-filling15, it is possible to investigate the morphological and electrophysiological development of purified neurons (Figure 3). Analysis of purified cultures reveals that both glutamatergic and GABAergic neurons were able to extend axons and dendrites from their cell bodies (Figure 3A, B) and retain the ability to generate action potentials in response to suprathreshold depolarizing current injections (Figure 3C, D). Notably, however, following purification, only GABAergic neurons received significant amounts of spontaneous synaptic transmission, and glutamatergic neurons receive very little synaptic transmission in the absence of glial cells (Figure 3E, F)7.
As we have previously reported, cells in purified cultures tend to survive more poorly than those in unsorted cultures and have significantly smaller dendrites and axons7. To overcome these deficits, we have adapted and applied methods for supporting the development of purified cultures with glial cells12,14. The cellular composition of our glial support cultures (DIV7) is shown in Figure 4A. These cultures contained predominantly glial fibrillary acidic protein (GFAP) positive astrocytes, but also contained some cluster of differentiation molecule (CD11b) positive microglia and myelin basic protein (MBP) positive oligodendrocytes. After DIV 7, these cells can be passaged to cell culture inserts to provide non-contact support of purified neurons (Figure 4B, C). Analysis of glia-neuron co-cultures reveals that approximately 40,000 glial cells were sufficient to improve the long-term survival and growth of both purified glutamatergic and GABAergic neurons (Figure 4D, E)7. Furthermore, electrophysiological analysis reveals that glutamatergic neurons, co-cultured with glial cells, were highly active and had strongly increased network activity (percentage of glial supported glutamatergic neurons firing bursts of action potentials: 62%; n = 28; Figure 4F, G). Glial support is therefore important not only for promoting neuron growth and survival, but also for promoting synaptic transmission and the establishment of network activity in glutamatergic cultures.
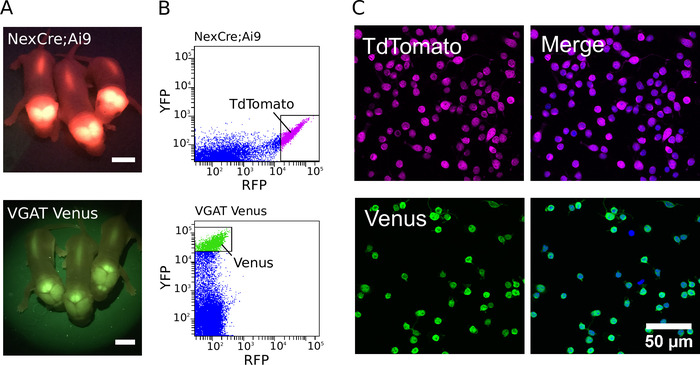
Figure 1: Purification of glutamatergic and GABAergic neurons. (A) Images showing fluorescent signal from the transgene expression of TdTomato in NexCre;Ai9 mice (top) and Venus in VGAT Venus mice (bottom). Scale bars = 5 mm. (B) Intensity scatter plots of the TdTomato and Venus fluorescence of cortico-hippocampal dissociated cells from NexCre;Ai9 mice (top) and VGAT Venus mice (bottom). Strongly fluorescent TdTomato or Venus neurons were selected for sorting (indicated by the gating boxes). (C, left) Confocal images of sorted TdTomato (top) and Venus (bottom) positive neurons. (C, right) Merged image showing cells co-stained with DAPI (in blue pseudocolour). TdTomato fluorescence is endogenous and remains strong despite fixation (in magenta pseudocolor). Venus expression is enhanced using a combination of a primary antibody directed against GFP and Alexa Fluor-488-conjugated secondary antibody (in green pseudocolor). Please click here to view a larger version of this figure.
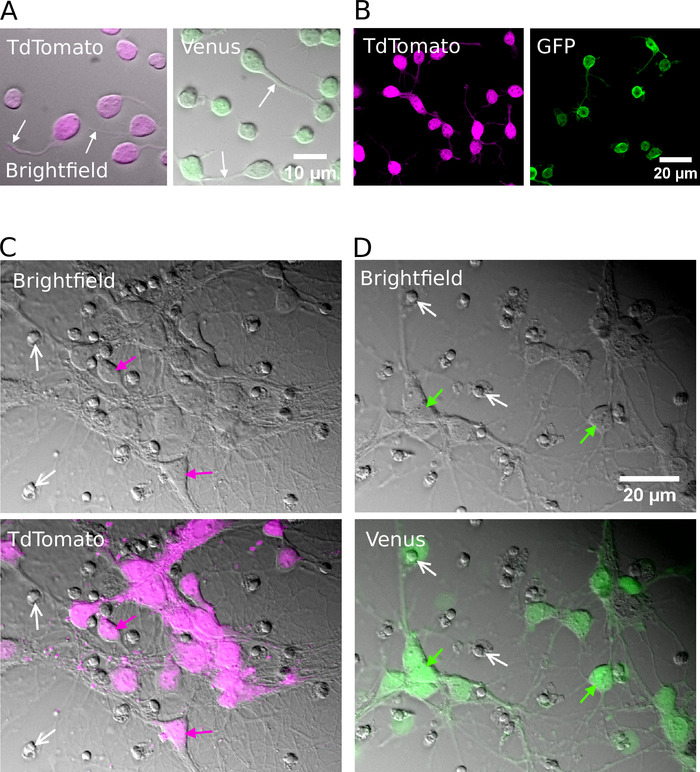
Figure 2: Cell culture of purified glutamatergic or GABAergic neurons. (A) Combined infrared (bright field) images and superimposed fluorescent signal from TdTomato (left) and Venus (right) positive neurons cultured for 1 h in vitro. White arrows indicate the location of growing neurites. (B) Confocal images of TdTomato (left) and Venus (right) positive neurons cultured for 1 h in vitro. (C, D) Bright field (top) and combined bright field and fluorescent images (bottom) of TdTomato (C) and Venus positive neurons (D) cultured for 7 days in vitro (DIV). White arrows show the location of condensed nuclei from dead cells. Colored arrows identify fluorescently labeled neurons. Please click here to view a larger version of this figure.
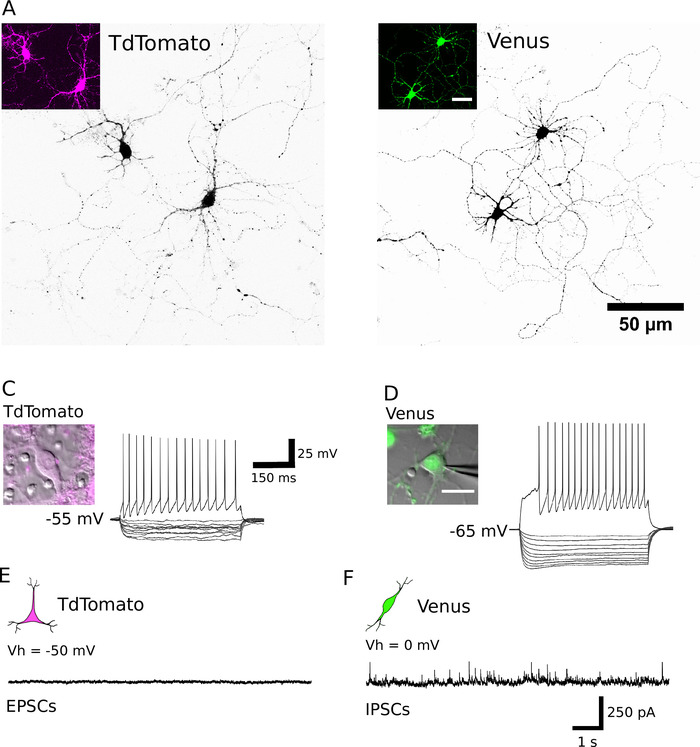
Figure 3: Morphological, electrophysiological and synaptic properties of purified neurons. (A, B) Confocal images of TdTomato (A) and Venus positive neurons (B) at DIV 13 and DIV 14, respectively. Cells are presented in black using an inverted look-up table to aid neurite visualization. Insets show the fluorescent signal expressed by the identified neurons. (C, D) The voltage response from a TdTomato (C) and Venus positive neuron (D) to hyperpolarizing (200 to -20 pA, in 20 pA steps) and depolarizing (200 pA) current pulses, obtained by whole-cell patch-clamp recordings. Insets show the corresponding recorded neurons. The resting membrane potential of each cell is indicated to the left of the recording trace. (E, F) Representative voltage-clamp recordings (10 s) obtained from TdTomato (E) and Venus positive neurons (F). Spontaneous excitatory postsynaptic events (EPSCs) were recorded from TdTomato positive neurons, maintained at a holding potential of 50 mV. Spontaneously occurring inhibitory postsynaptic events (IPSCs) were recorded from Venus positive neurons maintained at a holding potential of 0 mV. Please click here to view a larger version of this figure.
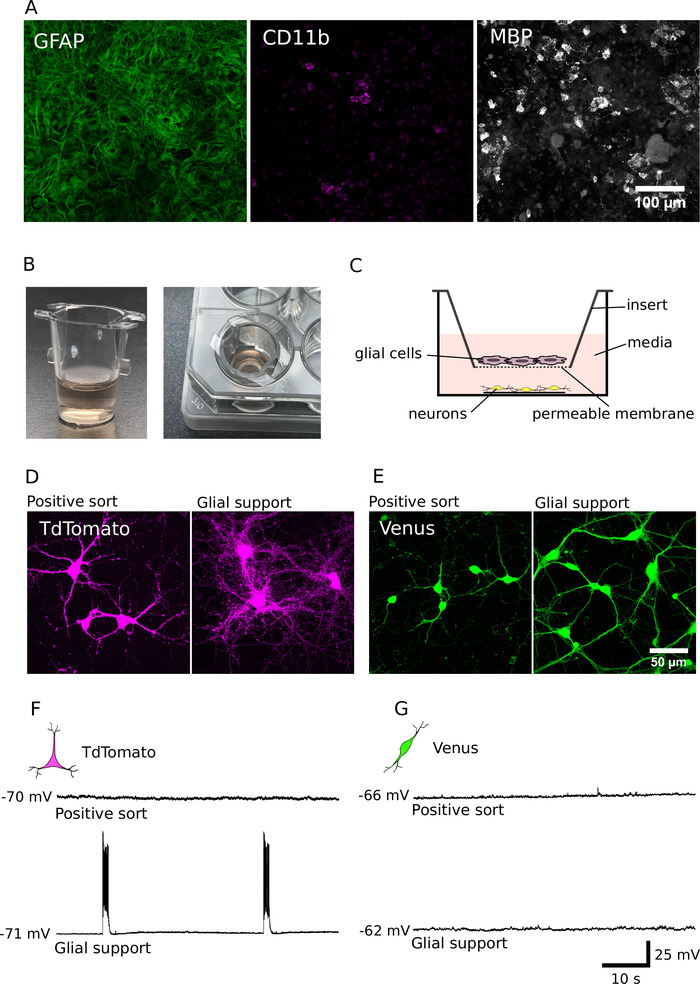
Figure 4: Supporting neuron development with glial co-culture. (A) Confocal images of glial cultures immunolabeled for glial fibrillary acidic protein (GFAP, left), cluster of differentiation molecule (CD11b, middle) and myelin basic protein (MBP, right). Glial cells were cultured for DIV 7. (B) Images of cell culture inserts used to co-culture glial cells with purified neurons in a non-contact arrangement. (C) A schematic of the spatial arrangement used to co-culture neurons and glia. (D, E) Confocal images of TdTomato (D) and Venus positive neurons (E), grown for 14 days, in the absence (left) or presence (right) of glial support. (F, G) Current-clamp recordings (60 s) from TdTomato (F) and Venus positive neurons (G) cultured for 14 days in the absence (top) or presence (bottom) of glial support. Neurons were recorded at their resting membrane potential (presented to the left of each recording trace). Please click here to view a larger version of this figure.
Discussion
We describe here a method that combines the sorting and culturing of primary neurons to generate purified neuronal cell cultures. This method takes around 1 h longer than a typical primary dissociated neuronal culture protocol yet allows for the generation of hundreds of replicate cultures containing specific neuronal types. Purified neurons, which can be grown in isolation for at least 16 days, extend axons and dendrites and can fire repetitive trains of action potentials (Figure 2 and Figure 3). Importantly, these cells are amenable to the same experimental analyses as regular primary dissociated neuronal cultures, including electrophysiological, morphological and survival analyses. A major benefit of working with these purified cultures is that the development of specific cell types can be studied in isolation. To support the development of purified cultures, we also present a protocol for co-culturing purified neurons with glial cells. As shown previously, co-culturing purified neurons with glial cells improves their survival, growth and can promote network formation7 (Figure 4). Thus, we present here a combination of methods that should further the study of glia-neuron interactions and may prove useful for studying the development and interaction between of other cell types of interest.
Studies on cultures of purified glutamatergic neurons have revealed fundamental insights into the way glial cells secrete factors that promote the development of neuronal networks and synapse formation16,17,18. In general, methods for purifying specific neuron types have been more successfully applied to studying the development of glutamatergic neurons rather than GABAergic neurons. This has led to a disparity in our understanding of how these two cell types develop. Given that GABAergic and glutamatergic neurons differ significantly in terms of their anatomy, physiology and developmental origins, it is vital that we study GABAergic neurons in their own right, to better understand their function and physiology. Using the protocol presented here, we have previously identified important differences in the way GABAergic and glutamatergic neurons depend on glia-secreted factors for the establishment of synaptic transmission7. By publishing this protocol we hope that others can make further insights into the important interactions between neurons and glial cells.
In this protocol, we describe a flow cytometry based cell sorting method, which we have used to purify GABAergic or glutamatergic neurons from different transgenic rodent lines. Venus positive GABAergic neurons were sorted from VGAT Venus mice13 or rats8 and TdTomato positive glutamatergic neurons were sorted from NexCre;Ai9 mice (bred originally from NexCre9 and Ai9 reporter lines6, see Turko et al., 20187). In recent years, due to technological advancements, the generation of transgenic animals has become significantly easier. As such, the availability of animals expressing fluorescent molecules, in many different cell types, has grown rapidly. This has, in turn, increased the use and applicability of fluorescent activated cell sorting. While alternative methods for isolating cells of interest currently exist16,19,20, they are somewhat hindered by their dependence on the availability of suitable antibodies to naturally occurring surface antigens. This limits their versatility when compared with fluorescence-based cell sorting methods, which can already be used to sort cells from the many cell specific transgenic reporter animals that are already available. Nevertheless, when optimized, antibody-based sorting methods can be rapid and high yielding, and may even better preserve cell anatomy, by allowing purification of cells with their axons and dendrites intact21. Antibody sorting methods should therefore still be considered when deciding on a sorting strategy. Ultimately, the cell type of interest, the age at which cells are to be sorted, the availability of transgenic animals or surface antigens and the number of cells needed will be the determining factors when choosing the most suitable sorting strategy.
Although fluorescence-based cell sorting is a simple and reproducible method for purifying cells, care should be taken during certain steps of the protocol to preserve cell quality. For instance, following each centrifugation step, it is important to make sure that the cell pellet is re-suspended as quickly as possible and that cells have been successfully recovered. Occasionally, the cell pellet can be disturbed when removing the supernatant. It is therefore advised, between centrifugation steps, to check the presence of cells under the microscope to rule out any extensive cell loss. Following cell plating, cells should be allowed to adhere for at least 1 h before feeding. If cells are fed too soon, some cells may become dislodged from the coverslip, thereby reducing the culture density. After 1 h in culture, it is prudent to assess cell health. If cells do not appear healthy (an example of healthy cells is shown in Figure 2A), or there is significant cell death, then this may be an indication of a problem during the dissociation or sorting procedure. A further important consideration, when culturing any cell type, is to take care when managing the cell culture media. NBA media contains phenol red, which acts as a pH indicator22. If the media becomes too yellow in color, then this indicates that the pH is too acidic; if the media becomes too pink, then this indicates that the solution is too alkaline. Stock solutions open for long periods, particularly media aliquoted into conical tubes, tend to become more alkaline over time. It is therefore advised to make up fresh complete NBA media each week and complete media every two weeks (the medium buffers under atmospheric conditions and should therefore be more stable). Taking these points into consideration it should be possible to establish purified cell cultures in any laboratory with access to cell sorting and cell culture equipment.
In the majority of our experiments we produced purified cultures from a single animal. However, when purifying cells for biochemical analyses, it may be necessary to pool together multiple animals for sorting. We have successfully sorted up to 8 embryonic mice (embryonic day 13 animals) using the above protocol (data not shown). However, if more animals are to be sorted, it may be necessary to increase the volume of both papain and BSA solutions (described in steps 2.1.1–2.1.4) to accommodate the increase in tissue amount. Additionally, if more cells are plated in culture, then a more frequent feeding schedule may be required. As a starting point, cells can be fed every 7 days by removing 100 µL of conditioned media and adding 200 µL of fresh complete NBA media. For the sake of neuron viability, if sorting more animals, thought should be given to minimizing the sort time as much as possible. This often requires the careful optimization of downstream analyses for the efficient use of purified cells. We have routinely sorted mouse neurons at rates of 600 events/s, up to 500,000–800,000 cells per animal (postnatal day 0–2). However, this was without exhaustive optimization of sorting speeds and conditions. Therefore, further improvements in sorting speed and yield should be possible.
Purified neurons require glia-conditioned media for their survival. This has been demonstrated previously by culturing neurons and glial cells separately, before treating neurons with glial conditioned media10. In our experiments, we chose to support purified neurons by culturing glial cells on semi-permeable cell culture inserts, which are placed inside the cell culture plate. This method has been successfully applied to study glia-derived extracellular matrix proteins and their interaction with neurons12. The non-contact, but continuous support provided by this method has a number of advantages when compared to the separate culture of cells. Most notably, the co-culture of glial cells with neurons allows for the continuous regulation and conditioning of the cell culture media, which more closely resembles the in vivo situation. In addition, continuous co-culture of cells allows for potential feedback signaling between neurons and glia, which is not possible in separated cultures. In our protocol, if required, the continuous co-culture method can be easily omitted and a classical treatment of neurons with glia-conditioned media can be performed.
In summary, the protocol presented here aims to provide the reader with a solid foundation from which they can establish their own purified cell culture experiments. We predict that the availability of transgenic animals and viral constructs will continue to increase for the foreseeable future. Therefore, cell sorting techniques based on fluorescence are likely to become even more widely used and valuable.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank the excellent technical support provided by Jenny Kirsch and Ana Teichmüller at the Flow Cytometry Core Facility, Deutsches Rheuma-Forschungszentrum, Berlin. We would like to thank Jie Song for his help with survival analysis. We would also like to thank Rita Loureiro for her help capturing the images of fluorescent mice and Christian Ebner for critical reading of the protocol. VGAT-Venus transgenic rats were generated by Drs. Y. Yanagawa, M. Hirabayashi and Y. Kawaguchi in National Institute for Physiological Sciences, Okazaki, Japan, using pCS2-Venus provided by Dr. A. Miyawaki. This work was supported by the German Research Council (Deutsche Forschungsgemeinschaft, DFG EXC 257 to IV).
Materials
| Neural Basal A media (NBA) | ThermoFisher Scientific | 10888022 | Cell Culture Buffer |
| B27 | ThermoFisher Scientific | 17504001 | Culture supplement |
| Glutamax | ThermoFisher Scientific | 35050-038 | Culture supplement |
| Penicillin Streptomycin (10,000 U/mL) | ThermoFisher Scientific | 15140-122 | Antibiotic |
| Poly-L-Lysine | SIGMA | P1399 | Coverslip coating |
| Papain | SIGMA | P4762-1G | Enzyme |
| Bovine Serum Albumin | SIGMA | A3294-100G | Serum |
| Hibernate A low fluorescence media | Brain Bits Ltd | HALF | Cell Transport media |
| Dulbeccos Modified Eagles Medium (DMEM) | Biochrom | F0435 | Glial Culture Buffer |
| Fetal Calf Serum (FCS) | Biochrom | S0115 | Serum |
| Trypsin / EDTA | Biochrom | L2163 | Enzyme |
| Fine Tip Pasteur Pipette | Neo Labs | – | Used for trituration of cells |
| 24-well plates | BD | 353047 | Culture plate |
| 50 mL Falcon tubes | BD | 352070 | – |
| 15 mL Falcon tubes | BD | 352096 | – |
| Glass coverslips: 12 mm round | Roth | P231.1 | – |
| 35 mm Petri dish | Corning | 353001 | – |
| 100 mm Petri dish | Corning | 353003 | – |
| 30 µm CellTrics Cell Sieve | sysmex | 04-004-2326 | To remove cell clumps before cell sorting |
| Round bottom polystyrene tubes | BD | 352054 | Transport tube for sorted cells |
| Round bottom polypropylyne tubes | BD | 352063 | Collection tube for sorted cells |
| Cell culture inserts – 0.4 µm transparent PET | Falcon | 353 095 | For the co-culture of neurons and glia |
| Extra fine Bonn Scissors | Fine Scientific Tools | 14084-08 | To remove overlying skin and bone of mice |
| Extra narrow Scissors | Fine Scientific Tools | 14088-10 | To remove overlying skin and bone of rats |
| Forceps | Fine Scientific Tools | 11242-40 | To hold the head in place |
| Spatula (130 mm long / 5 mm tip width) | Fine Scientific Tools | 3006.1 | To remove the brain to filter paper |
| Scalpel Blades | Swan-Morton | #0308 | To mechanically dissociate neural tissue |
| Haemocytometer (Neubauer Imroved) | Optik Labor | – | To cell count dissociated cells |
| Fluorescent Miners Goggles (excitation light) | Biological Laboratory Equipment Maintenance and Servce Ltd (BLS) |
FHS/LS-1G | To excite TdTomato |
| Fluorescent Miners Goggles (emission filter) | Biological Laboratory Equipment Maintenance and Servce Ltd (BLS) |
FHS/EF-4R2 | Td Tomato compatible emission filter |
| Fluorescent Miners Goggles (excitation light) | Biological Laboratory Equipment Maintenance and Servce Ltd (BLS) |
FS/ULS-02B2 | To excite Venus |
| Fluorescent Miners Goggles (emission filter) | Biological Laboratory Equipment Maintenance and Servce Ltd (BLS) |
FS/TEF-3GY1 | Venus compatible emission filter |
| Mouse-Anti-GFP primary antibodies | UC Davis | 75-132 | To enhance Venus signal following fixation |
| Mouse-Anti-GFAP primary antibodies | SIGMA | G-3893 | The identification of reactive astrocytes |
| Rabbit-Anti-CD11b primary antibodies | Southern Biotech | 1561-15 | The identification of microglial cells |
| Rabbit-Anti-MBP primary antibodies | Millipore | AB980 | The identification of oligodendrocyes |
References
- Radcliff, G., Jaroszeski, M. J. Basics of flow cytometry. Methods in Molecular Biology. 91, 1-24 (1998).
- Feher, K., Kirsch, J., Radbruch, A., Chang, H. D., Kaiser, T. Cell population identification using fluorescence-minus-one controls with a one-class classifying algorithm. Bioinformatics. 30 (23), 3372-3378 (2014).
- Sergent-Tanguy, S., Chagneau, C., Neveu, I., Naveilhan, P. Fluorescent activated cell sorting (FACS): a rapid and reliable method to estimate the number of neurons in a mixed population. Journal of Neuroscience Methods. 129 (1), 73-79 (2003).
- Adan, A., Alizada, G., Kiraz, Y., Baran, Y., Nalbant, A. Flow cytometry: basic principles and applications. Critical Reviews in Biotechnology. 37 (2), 163-176 (2017).
- Guez-Barber, D., et al. FACS purification of immunolabeled cell types from adult rat brain. Journal of Neuroscience Methods. 203 (1), 10-18 (2012).
- Madisen, L., et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature Neuroscience. 13 (1), 133-140 (2010).
- Turko, P., Groberman, K., Browa, F., Cobb, S., Vida, I. Differential Dependence of GABAergic and Glutamatergic Neurons on Glia for the Establishment of Synaptic Transmission. Cerebral Cortex. , (2018).
- Uematsu, M., et al. Quantitative chemical composition of cortical gabaergic neurons revealed in transgenic venus-expressing rats. Cerebral Cortex. 18, 315-330 (2008).
- Goebbels, S., Bormuth, I., Bode, U., Hermanson, O., Schwab, M. H., Nave, K. -. A. Genetic targeting of principal neurons in neocortex and hippocampus of NEX-Cre mice. Genesis. 44, 611-621 (2006).
- Banker, G. A. Trophic interactions between astroglial cells and hippocampal neurons in culture. Science. 209, 809-810 (1980).
- Lindsay, R. M. Adult rat brain astrocytes support survival of both NGF-dependent and NGF-insensitive neurones. Nature. 282 (5734), 80-82 (1979).
- Geissler, M., et al. Primary hippocampal neurons, which lack four crucial extracellular matrix molecules, display abnormalities of synaptic structure and function and severe deficits in perineuronal net formation. Journal of Neuroscience. 33 (18), 7742-7755 (2013).
- Wang, Y., et al. Fluorescent labeling of both gabaergicand glycinergic neurons in vesicular GABA transporter (VGAT)-venus transgenic mouse. 神经科学. 164 (3), 1031-1043 (2009).
- Höltje, M., et al. Role of Rho gtpasein astrocyte morphology and migratory response during in vitro wound healing. Journal of Neurochemistry. 95 (5), 1237-1248 (2005).
- Booker, S. A., Song, J., Vida, I. Whole-cell Patch-clamp Recordings from Morphologically- and Neurochemically-identified Hippocampal Interneurons. Journal of Visualized Experiments. , e51706 (2014).
- Pfrieger, F. W., Barres, B. A. Synaptic efficacy enhanced by glial cells in vitro. Science. 277 (5332), 1684-1687 (1997).
- Ullian, E. M., Sapperstein, S. K., Christopherson, K. S., Barres, B. A. Control of synapse number by glia. Science. 291, 657-661 (2001).
- Christopherson, K. S., et al. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 120, 421-433 (2005).
- Berghuis, P., et al. Brain-derived neurotrophic factor controls functional differentiation and microcircuit formation of selectively isolated fast-spiking gabaergic interneurons. European Journal of Neuroscience. 20 (5), 1290-1306 (2004).
- Liddelow, S. A., et al. Activated microglia induce neurotoxic reactive astrocytes via Il-1α, tnfα, and c1q. Nature. 541, 481-487 (2017).
- Tomlinson, M. J., Tomlinson, S., Yang, X. B., Kirkham, J. Cell separation: Terminology and practical considerations. Journal of Tissue Engineering. 4, (2013).
- Brewer, G. J., Torricelli, J. R., Evege, E. K., Price, P. J. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. Journal of Neuroscience Research. 35 (5), 567-576 (1993).

