HOX Loci Focused CRISPR/sgRNA Library Screening Identifying Critical CTCF Boundaries
Summary
A CRISPR/sgRNA library has been applied to interrogating protein-coding genes.However, the feasibility of a sgRNA library to uncover the function of a CTCF boundary in gene regulation remains unexplored. Here, we describe a HOX loci specific sgRNA library to elucidate the function of CTCF boundaries in HOX loci.
Abstract
CCCTC-binding factor (CTCF)-mediated stable topologically associating domains (TADs) play a critical role in constraining interactions of DNA elements that are located in neighboring TADs. CTCF plays an important role in regulating the spatial and temporal expression of HOX genes that control embryonic development, body patterning, hematopoiesis, and leukemogenesis. However, it remains largely unknown whether and how HOX loci associated CTCF boundaries regulate chromatin organization and HOX gene expression. In the current protocol, a specific sgRNA pooled library targeting all CTCF binding sites in the HOXA/B/C/D loci has been generated to examine the effects of disrupting CTCF-associated chromatin boundaries on TAD formation and HOX gene expression. Through CRISPR-Cas9 genetic screening, the CTCF binding site located between HOXA7/HOXA9 genes (CBS7/9) has been identified as a critical regulator of oncogenic chromatin domain, as well as being important for maintaining ectopic HOX gene expression patterns in MLL-rearranged acute myeloid leukemia (AML). Thus, this sgRNA library screening approach provides novel insights into CTCF mediated genome organization in specific gene loci and also provides a basis for the functional characterization of the annotated genetic regulatory elements, both coding and noncoding, during normal biological processes in the post-human genome project era.
Introduction
Recent genome interaction studies revealed that the human nuclear genome forms stable topologically associating domains (TADs) that are conserved across cell types and species. The organization of the genome into separate domains facilitates and restricts interactions between regulatory elements (e.g., enhancers and promoters). The CCCTC-binding factor (CTCF) binds to TAD boundaries and plays a critical role in constraining interactions of DNA elements that are located in neighboring TADs1. However, genome wide CTCF binding data revealed that although CTCF mostly interacts with the same DNA-sites in different cell types, it often functions as a chromatin barrier at a specific site in one cell type but not in the other, suggesting that CTCF functions together with other activities in the formation of chromatin boundaries2. What remains unknown is whether the boundary elements (CTCF-binding sites) are directly linked to the biological function of CTCF, and how these links occur. Therefore, we hypothesize that specific CTCF binding sites in the genome directly regulate the formation of TADs and control promoter/enhancer interactions within these domains or between neighboring domains. The completion of the human and mouse genome sequencing projects and subsequent epigenetic analyses have uncovered new molecular and genetic signatures of the genome. However, the role of specific signatures/modifications in gene regulation and cellular function, as well as their molecular mechanism(s), have yet to be fully understood.
Multiple lines of evidence support that the CTCF-mediated TADs represent functional chromatin domains3,4,5. Although CTCF mostly interacts with the same DNA-sites in different cell types, genome wide CTCF ChIP-seq data revealed that CTCF often functions as a chromatin barrier in one cell type but not in the other2. CTCF plays an essential role during development by mediating genome organization4,6,7. Disruption of CTCF boundaries impaired enhancer/promoter interactions and gene expression, leading to developmental blockage. This suggests that CTCF mediated TADs are not only structural components, but also regulatory units required for proper enhancer action and gene transcription5,8,9.
HOX genes play critical roles during embryonic development and they are temporally and spatially restricted in their expression patterns. The HOXA locus forms two stable TADs separating anterior and posterior genes by a CTCF-associated boundary element in both hESCs and IMR90 cells1. Recent reports demonstrated that HoxBlinc, a HoxB locus associated lncRNA, mediates the formation of CTCF directed TADs and enhancer/promoter interactions in the HOXB locus. This leads to anterior HOXB gene activation during ESC commitment and differentiation10. Furthermore, at specific gene loci including the HOXA locus, alteration of CTCF mediated TAD domains changed lineage specific gene expression profiles and was associated with the development of disease states11,12. The evidence supports a primary function for CTCF in coordinating gene transcription and determining cell identity by organizing the genome into functional domains.
Despite its role in the embryonic development, during hematopoiesis, HOX genes regulate hematopoietic stem and progenitor cell (HS/PC) function. This is done by controlling the balance between proliferation and differentiation10,13,14,15. The expression of HOX genes is tightly regulated throughout the specification and differentiation of hematopoietic cells, with highest expression in HS/PCs. HOX gene expression gradually decreases during maturation, with its lowest levels occurring in differentiated hematopoietic cells16. HOX gene dysregulation is a dominant mechanism of leukemic transformation by dysregulating self-renewal and differentiation properties of HS/PCs leading to leukemic transformation17,18. However, the mechanism of establishing and maintaining normal vs. oncogenic expression patterns of HOX genes as well as associated regulatory networks remains unclear.
CRISPR-Cas9 sgRNA library screening has been widely used to interrogate protein-coding genes19 as well as non-coding genes, such as lncRNA20 and miRNA21 in different species. However, the cost to use the CRISPR-Cas9 sgRNA library to identify new genomic targets remains high, because high-throughput genome sequencing is often applied to verify the sgRNA library screening. Our sgRNA screening system is focused on the specific genome loci and evaluates the targeting sgRNAs through one-step RT-PCR according to the marker gene expression, such as HOXA9. Additionally, Sanger sequencing confirmed that the sgRNA was integrated into the genome, and Indel mutations can be detected to identify the sgRNA targeting site. Through the loci-specific CRISPR-Cas9 genetic screening, the CBS7/9 chromatin boundary has been identified as a critical regulator for establishing oncogenic chromatin domain and maintaining ectopic HOX gene expression patterns in AML pathogenesis12. The method can be widely applied to identify not only specific function of CTCF boundary in embryonic development, hematopoiesis, leukemogenesis, but also CTCF boundary as potential therapeutic targets for future epigenetic therapy.
Protocol
1. CTCF sgRNALibrary Design Using an Online Tool
- Design the sgRNA targeting CTCF binding sites in the human HOX loci using the genetic perturbation platform (GPP) designer tool (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design).
- Synthesize a total of 1,070 sgRNAs consisting of sgRNAs targeting 303 random targeting genes, 60 positive controls, 500 non-Human-targeting controls, and 207 CTCF elements or lncRNA targeting genes (Figure 1, Table 1). Each targeting DNA element is targeted by 5-10 different sgRNAs.
2. sgRNA Library Cloning
- Clone the synthesized oligonucleotides into the CRISPR lentiviral backbone vector (lentiCRISPRv2).
- Digest the LentiCRISPRv2 vector with BsmBI restriction enzyme at 37 °C for 2 h.
- Look for the presence of the larger band (around 12,873 bp) on the gel after BsmBI digestion, and then purify it with the gel extraction kit.
NOTE: A 2kb small filler piece is also present on the gel after digestion, but this should be ignored. - Ligate the synthesized oligonucleotides and digested LentiCRISPR vector with 150 ng of digested LentiCRISPR DNA, 1 µL of 10 µM oligos, 2 µL of 10x T4 ligase buffer, 1 µL of T4 ligase, and then incubate them at 16 °C overnight.
- Transform the lentiviral CRISPR/sgRNA library into electro-competent cells for amplification.
- Prepare the electroporator at 1.8 kV, 200 Ω and 25 µF. Then pre-warm the recovery SOC media in a 37 °C water bath, and pre-warm LB ampicillin antibiotic plates at 37 °C.
- Thaw the competent cells on ice for 10 min.
- Prepare 1.5 mL micro-centrifuge tubes and 1 mm electroporation cuvettes on ice.
- Mix 1 µL of a 10 ng/µL library plasmid DNA into 25 µL of competent cells in a 1.5 mL micro-centrifuge tube, and gently mix by flicking the bottom of the tube a few times manually.
- Once the cuvette is cold enough, transfer the DNA/competent cell mixture to it. Tap twice on the countertop and wipe any water droplets from the exterior of cuvette with a tissue paper. Then place the cuvette in the electroporation module and press pulse.
- Immediately add 975 µL of 37 °C pre-warmed SOC media. Mix by pipetting up and down and transfer to a 15 mL tube.
- Rotate and incubate at 37 °C for 1 h.
- Dilute 100 µL cells into 900 µL of SOC media and place 100 µL on a LB ampicillin antibiotic agar plate. Incubate overnight at 37 °C.
- Extract the plasmid DNA from the combined colonies using a maxi-prep column as detailed in the manufacturer's protocol.
- Scrape all the colonies from the LB agar plate and inoculate a starter culture of 2 mL of LB ampicillin antibiotic medium and incubate overnight at 37 °C with vigorous shaking (~200 x g).
- Dilute the starter culture 1:500 into 100 mL of LB ampicillin medium and incubate at 37 °C for 12-16 h with vigorous shaking (~200 x g).
- Harvest the bacterial cell pellet by centrifugation at 6,000 x g for 15 min at 4 °C.
- Re-suspend the bacterial pellet in 10 mL of suspension buffer.
- Lyse the suspended pellet with 10 mL of the lysis buffer, and vigorously invert 4-6 times. Incubate the lysate for 5 min at room temperature.
- Neutralize the lysate with 10 mL of chilled Neutralization Buffer. Mix by gently inverting the tubes 4-6 times and incubate it for 20 min on ice.
- Spin down at 13,500 x g for 30 min at 4 °C. Promptly transfer the supernatant containing the plasmid DNA to a new tube.
- Repeat step 2.3.7, and promptly transfer the supernatant containing the plasmid DNA to a new tube.
- Equilibrate the column by applying 10 mL of equilibration buffer and allow the column to empty by gravity flow.
- Add the supernatant to the column and allow it to enter the resin by gravity flow.
- Wash the column with 2x 30 mL of washing buffer.
- Elute the DNA with 15 mL of elution buffer.
- Precipitate the DNA with 10.5 mL of room-temperature isopropanol to the eluted DNA. Mix and spin down immediately at 15,000 x g for 30 min at 4 °C, and gently decant the supernatant.
- Wash the DNA pellet with 5 mL of 70% ethanol, centrifuge DNA pellet at 15,000 x g for 10 min and discard the clear supernatant.
- Repeat step 2.5.14 twice more.
- Centrifuge DNA pellet at 15,000 x g for 10 min, and gently decant the supernatant without disturbing the DNA pellet.
- Air-dry the pellet for 5-10 min, and dissolve the DNA in a required volume of buffer (TE buffer, pH 8.0).
3. The High Titer sgRNA Library Lentivirus Generation
- Cell preparation: Culture HEK293T cells in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% (vol/vol) fetal bovine serum (FBS) and 1% (vol/vol) penicillin-streptomycin (PS) antibiotic in T-25 flasks. Place them in the incubator at 37 °C and 5% CO2.
- Package lentivirus: Co-transfect HEK293T cells with 20 µg of purified library vectors from step 2, 15 µg of the package plasmid (psPAX2) and 10 µg of the envelope plasmid (pMD2.G) for 48 h before harvesting the viruses.
- Virus collection: After 48 h, collectthe virus supernatant and filter the virus supernatant through a 0.45 µm low protein binding PVDF membrane.
- Virus concentration: Concentrate the lentiviral supernatant by 50-fold using the concentrator and test the virus MOI in step 5.
- Virus storage: Aliquot the concentrated viruses and store in a -80 °C freezer.
4. Optimized Puromycin Concentration
- Leukemia cell culture: Culture MOLM13 AML cells in RPMI 1640 supplemented with 10% (vol/vol) fetal bovine serum (FBS) and 1x penicillin-streptomycin (PS) antibiotics in a T-125 flask. Place them in an incubator at 37 °C and 5% CO2.
NOTE: Cells are typically passed every 4-5 d at a split ratio of 1:4 or 1:6, never allowing cells to reach more than 70% confluency. - Set up MOLM13 cells in a 12-well plate with a density of 1.0 x 104 cell/mL, at a total volume of 2 mL per well (2.0 x 104 cells).
- Time-course assay: Treat MOLM13 cells with puromycin for 7 days in increasing concentrations (0.1 µg/mL, 0.2 µg/mL, 0.5 µg/mL, 1.0 µg/mL and 2.0 µg/mL)
- Set up MOLM13 cells without puromycin treatment on day 0 and set up 3 replicate wells without puromycin treatment as a control from day 0 to day 7.
- Treat MOLM13 cells with 0.1 µg/mL, 0.2 µg/mL, 0.5 µg/mL, 1.0 µg/mL and 2.0 µg/mL, separately, with each experimental condition containing 3 replicate wells.
- Count the live cell ratio and make a survival curve from day 0 to day 7 containing all conditions.
- Survival curve: Stain cells with Trypan blue and count viability daily to obtain the survival curves for each puromycin concentration.
- Optimizing minimal puromycin concentration: Determine the minimal puromycin concentration through Trypan blue staining, in which all MOLM13 cells are killed between 5-7 days.
5. Titration of Lentiviral Library in MOLM13 Leukemia Cells
- AML cells preparation: Collect MOLM13 AML cells with the transduction medium (RPMI 1640, 10% FBS, 1% PS, and 8.0 µg/mL coating medium) at a density of 1.5 x 106 cells /mL.
- Place MOLM13 cells in the 12-well plate with 1.5 x 106 cells in each well.
- Thaw the lentivirus: Remove the concentrated lentivirus from the -80 °C freezer and thaw it on ice.
- Mix MOLM13 cells with a different dose of the concentrated lentivirus in separate wells, including 0, 1, 2.5, 5, 7.5 and 10 µL (total 6 groups).
- Immediately centrifuge these mixtures at 1,000 x g for 2 h at 33 °C and transfer the 12-well plates back to the incubator at 37 °C and 5% CO2 for 4 h.
- After 4 h, spin down the infected cells at 400 x g for 5 min at room temperature.
- Gently aspirate the supernatant without disturbing the cell pellet, and re-suspend the transduced cells with fresh media (RPMI 1640, 10% FBS and 1% PS), and then transfer them to T-25 flasks and incubate at 37 °C for 48 h without puromycin.
- After 48 h, split these cells into 2 flasks (2 groups): an experimental group treated with 1 µg/mL puromycin for 5 days, and a control group without puromycin treatment for 5 days.
- Carry out puromycin selection for 5 days with 1 µg/mL puromycin according to the step 4 until all the non-transduced control cells are dead. Exchange for fresh media every 2 days.
- Measure the optimized MOI value for transduction by dividing the number of live cells treated with puromycin with the number of cells without puromycin treatment.
6. Transduction of the Pooled CRISPR-Cas9 KO Library
- Transduction with lentivirus: Infect 1.5 x 106 MOLM13 cells with 0.3 MOI of sgRNA pooled lentivirus in medium (RPMI 1640, 10% FBS, 1% PS, and 8 µg/mL coating medium) in 6-well plate and use the cells without the lentivirus infection as a control.
- Immediately centrifuge the 6-well plate at 1,000 x g for 2 h at 33 °C to spinfect the cells and transfer the plates back to the incubator at 37 °C and 5% CO2 for 4 h.
- Spin down the infected cells at 400 x g for 5 min at room temperature.
- Gently aspirate supernatant without disturbing the cell pellet, and re-suspend the transduced cells with fresh media (RPMI 1640, 10% FBS and 1% PS), and then transfer them to T-25 flasks and incubate at 37 °C for 48 h without puromycin.
- After 48 h, treat cells with 1 µg/mL puromycin for 5 days. Exchange for fresh media after 2 days and keep at an optimal cell density.
- Seed the single clone in 96-well plates with limiting dilution methods and incubate these single clones at 37 °C and 5% CO2. Culture them for 3-4 weeks.
- After a single cell grows up into a population, transfer half of the cells into 24-well plates for further culture under puromycin selection and verify these clones in the next step. Keep the rest of the cells.
7. Screening of the Pooled CRISPR-Cas9 KO Library with One-step RT-qPCR
- Determine the effectiveness of the sgRNA integrated clone screening by evaluating the expression of the marker gene HOXA9 with one step reverse-transcriptase polymerase chain reaction (one-step RT-qPCR).
NOTE: HOXA9 are highly expressed in MOLM13 AML cells in leukemogenesis22,23. - Count the sgRNA integrated MOLM13 cell and transfer 1 x 104 cells per well to a 96-well PCR plate.
- Centrifuge the tube at 1,000 x g for 5 min, and then thoroughly remove and discard the supernatant with a pipet without disturbing the cell pellet.
- Wash cells with 125 µL of PBS buffer, and centrifuge the tube at 1,000 x g for 5 min. Then remove 120 µL of the supernatant using a pipette and retain approximately 5 µL of PBS in each well.
- Add 50 µL of the cell lysis master mix containing 48 µL of cell lysis buffer, 1 µL of proteinase K solution (10 mg/mL) and 1 µL of DNase solution (1 mg/mL) to each well. Then pipet up and down 5 times to re-suspend the cell pellet.
- Incubate the mix for 10 min at room temperature, followed by 5 min at 37 °C, and then 75 °C for 5 min.
- Store the cell lysate at -80 °C freezer.
- The preparation of one-step RT-qPCR reaction: Thaw the one-step reaction mix and other reaction components to 4 °C. Then spin down briefly to collect solutions at the bottom of tubes, and place on ice without light. Mix and spin gently.
- Add 1 µL of cell lysate to the PCR wells with the RT-qPCR reaction mix, including 1 µL of the marker gene's forward primer (300 nM) and reverse primer (300 nM), 0.125 µL of reverse transcriptase (10 U/µL), and 5 µL of one-step reaction mix (2x).
- Seal wells with optically transparent film, and gently vortex and mix the reaction components.
- Place the 96-well PCR plate on a real-time PCR instrument.
- Run the reverse transcription reaction for 10 min at 50 °C, followed by polymerase inactivation and DNA denaturation for 1 min at 95 °C.
- Perform RT-PCR with 40 cycles of PCR reaction: denaturation for 15 s at 95 °C, annealing/extension and plate fluorescence reading for 20 s at 60 °C, and then melt curve analysis at 65-95 °C via 0.5 °C increments at 2-5 s/step.
- Set up upregulated, downregulated and no change groups according to the expression levels of HOXA9 gene by comparison to the control, separately. Use β-actin gene as a housekeeping gene control.
8. Verification of Integrated sgRNAs Positive Clones through Genotyping and Sanger Sequence
- Verify the HOXA9 decreased expression clones through Sanger sequencing and perform PCR with 50-100 ng MOLM13 genome DNA, 5 µL polymerase reaction buffer (10x), 1 µL forward primer (10 µM) (AATGGACTATCATATGCTTACCGTAACTTGAAAGTATTTCG) and 1 µL reverse primer (10 µM) (TCTACTATTCTTTCCCCTGCACTGTTGTGGGCGATGTGCGCTCTG), 1 µL dNTP (10mM), 1 unit polymerase (5 U /µL). Perform the PCR reaction with the initial denaturation at 94 °C for 30 s, and then more denaturation at 94 °C for 20 s, annealing at 56 °C for 20 s, extension at 68 °C for 20 s (total 30 cycles), final extension at 68 °C for 10 min, and then holding at 4 °C.
- Extract and purify the PCR products (size 285 bp) with a PCR Purification Kit.
- Ligate the purified PCR products into the T vector with 2 µL T4 ligation buffer (10x), 50 ng T vector DNA (50 ng/µL), 25 ng purified PCR DNA (285 bp), 1 µL T4 ligase (3 units/µL), and place the ligation mix into an incubator at 16 °C overnight.
- Transfer the ligation mix into DH5α competent cell, grow on a LB ampicillin antibiotic agar plate, and incubate overnight at 37 °C.
- Pick the single clones from the LB plate and verify them by genotyping and Sanger sequencing.
9. Detection of sgRNAs Induced Indel Mutation by Nuclease Digestion Assay
- Detect the sgRNA integrated single clone induced Indel rates by a nuclease test assay.
- Separately prepare PCR amplicons with 50-100 ng Indel mutant (test) and wild-type (WT, reference) DNA as PCR template, 5 µL polymerase reaction buffer (10x), 1 µL dNTP (10mM), 1 unit polymerase (5 U/µL), 1 µL forward primer (10 µM)( 5'-GAGATGGCGGCGCGGAAG-3'), and 1 µL reverse primer (10 µM) (5'-AAATATAGGGCGGCTGTTCACT-3'). The PCR reaction was performed with initial denaturation at 98 °C for 30 s, and then denaturation at 98 °C for 20 s, annealing at 56 °C for 20 s, extension at 72 °C for 30 s (total 30 cycles), and final extension at 72 °C for 10 min, and holding at 4 °C.
- Set up the heteroduplex mixture group with 200 ng of the "reference" (20 ng/µL) and 200 ng of "test" (20 ng/µL) PCR amplicons in 0.2 mL PCR tube, and the homoduplex mixture group with only 400 ng of "reference" PCR amplicons as a control.
- Separately incubate the heteroduplex and homoduplex mixture at 95 °C for 5 min in a 1 L beaker filled with 800 mL of water and then cool down gradually to room temperature to anneal and form heteroduplex or homoduplexes.
- Separately digest 400 ng of the annealed heteroduplex and homoduplex mixture with 1 µL indel mutation detection nuclease (2.5 U/µL) and 2 µL nuclease reaction buffer (10x) at 42 °C for 60 min.
- Analyze the digested samples with agarose gel electrophoresis, the heteroduplex mixture DNA should be cut into small fragments (70-250 bp), and the homoduplex DNA (320 bp) should not be cut.
Representative Results
CRISPR-Cas9 technology is a powerful research tool for functional genomic studies. It is rapidly replacing conventional gene editing techniques and has high utility for both genome-wide and individual gene-focused applications. Here, the first individually cloned loci-specific CRISPR-Cas9-arrayed sgRNA library contains 1,070 sgRNAs consisting of sgRNAs targeting 303 random targeting genes, 60 positive controls, 500 non-Human-targeting controls, and 207 CTCF elements or lncRNA targeting genes in four HOX loci (Figure 1, Table 1). This library targets all CTCF core binding motifs, HOX gene associated lncRNAs, known regulatory elements, and several HOX genes as positive controls in the HOX loci. It also contains sgRNAs targeting random non-HOX genes, non-human genes and intergenic regions as negative controls. To enhance efficiency and specificity of CTCF site knock-out (KO) by lentiCRISPR transduction, each targeting site contains 5-10 sgRNAs (Table 1). In the protocol described here, sgRNA libraries are designed according to CTCF binding sites at the HOXA/B/C/D loci and lncRNAs in these loci, which is based on the Broad Institute sgRNA tools (Figure 1, Figure 2). After transduction at a low multiplicity of infection with a MOI of 0.3 in MOLM13 cells carrying the MLL-AF9 fusion, the infection rate is less than one sgRNA/cell followed by puromycin selection, and then the resistant clones grown from seeded single cell were screened for impairment of HOXA9 gene expression.
The workflow for sgRNA library screening was briefly described (Figure 3). First, the virus containing sgRNA library were generated in HEK293T cells with the help of two vectors (psPAX2 and pMD2.G). sgRNA pooled library lentiviruses were concentrated and transduced into MOLM13 AML cells with polybrane (8.0 µg/mL). After a 48 h transduction, cells were treated with the optimal concentration of puromycin. After 5 days, the cells were seeded one cell/well into 96-well plates and the single clones were generated in the presence of puromycin. Finally, sgRNA single clones integrated into genome were identified by one-step RT-PCR, Sanger sequencing and Indel mutation detection (Figure 3). The puromycin resistant single clones are identified through one-step droplet digital RT-qPCR (RT-ddqPCR) according to altering expression of HOXA9 oncogene (Figure 4). Genotyping and Sanger sequence were performed for sgRNA library construction and verification (Figure 2, Figure 4).
sgRNA targeting MOLM13 positive clones in a 96-well PCR plate were further confirmed with the RT-qPCR method based on theexpression levels of HOXA9 genes through comparison with the control cells. Out of the 528 surviving clones screened, 10 clones exhibited more than 50% reduction in HOXA9 levels (Figure 4A). sgRNAs integrated into the HOXA9-reduced, HOXA9-unchanged, and HOXA9-increased clones were further confirmed by PCR amplification of the sgRNA sequences using flanking vector primers. The purified PCR products were ligated into the T vector system through T4 ligase and sent out for identification by Sanger sequence (see step 8). The sequence data indicated that out of 30 clones sequenced, 21 clones included single sgRNA (Table 2). The categories of sgRNA were identified and analyzed according to the HOXA9 expression levels. Six of ten clones showing a reduction in HOXA9 levels contained sgRNAs targeting the CBS7/9 site, but not in the non-human genes, random human genes, and other CTCF site controls (Figure 4 and Table 2).
sgRNA integrated positive single clone-induced Indel mutations are determined by PCR-based genotyping and nuclease digestion based on the nuclease assay (Figure 5). The nuclease digestion assay has been performed to identify Indel mutations occurred in the CBS7/9 boundary in the representative HOXA9-reduced, HOXA9-unchanged, and HOXA9-increased clones. The results revealed that the CBS7/9 mutation has been found in 4 out of the 6 HOXA9-reduced clones: clones #5, 6, 28, and 121, but not in clones #15 and #31 (Figure 5). However, clone #15 contained the sgRNA targeting HOTTIP lncRNA site, while clone #31 contained several sgRNAs targeting HOAIRM1 lncRNA, HOTAIR lncRNA, and HOXD9/10 CTCF binding site (Figures 4B, 5 and Table 2).
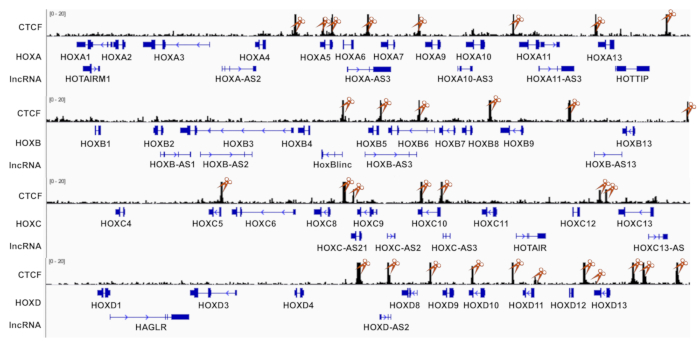
Figure 1: Schematic diagram showing CTCF binding sites and lncRNAs in four HOX gene loci. Each targeting DNA element contains 5-10 different sgRNAs. CTCF ChIP-seq dataset was downloaded from GEO (GSM1335528) and visualized with Integrated Genomic Viewer (IGV). SgRNA targeting CTCF sites in HOX loci were labelled with orange scissors. Please click here to view a larger version of this figure.
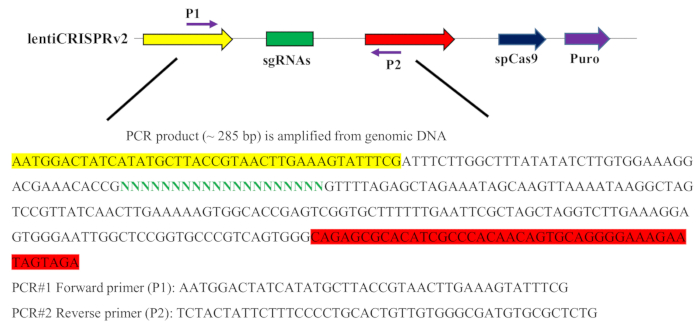
Figure 2: Schematic diagram representing the part of integrating sgRNA vector sequence and PCR amplification primers. The PCR amplification primers were designed according to the blank sequence of the sgRNA lentiviral vector. The forward primer (P1) was highlighted in yellow, the reverse primer (P2) was highlighted in red, and the sgRNA was highlighted in green in the sgRNA lentiviral vector. Please click here to view a larger version of this figure.
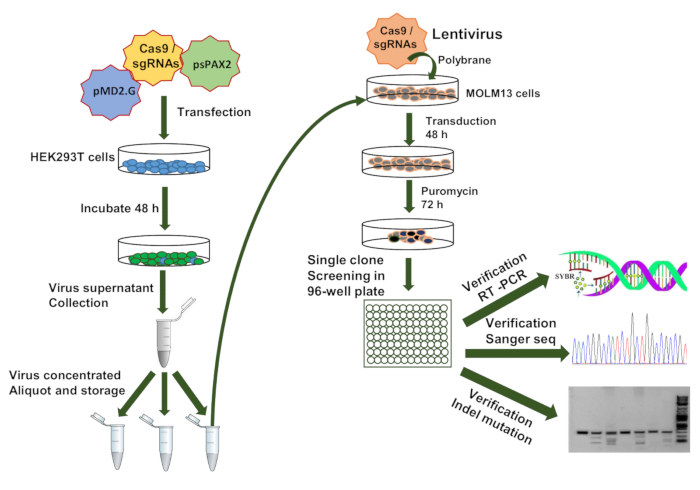
Figure 3: Schematic diagram representing the workflow for sgRNAs library design, construction and verification. This workflow is as follows. First, the sgRNA library was designed and cloned into a lentiviral CRISPR vector, and then the lentivirus was packaged with the sgRNA library lentiviral vector, psPAX2 and pMD2.G vectors in the HEK293T cells. Next, MOLM13 cells were infected with a low MOI (0.3) virus and these cells underwent puromycin selection. Then, the single clone was seeded in a 96-well plate. Finally, the sgRNA single clones integrated into a genome were identified by one-step RT-qPCR, Sanger sequence and Indel mutation detection. Please click here to view a larger version of this figure.
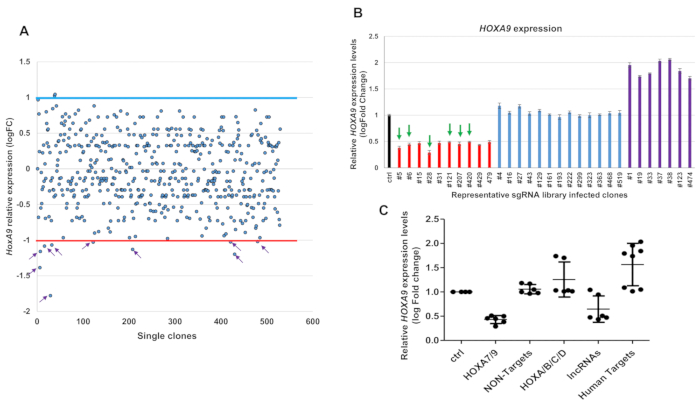
Figure 4: Pooled CRISPR-Cas9 KO library screening identified with one-step RT-qPCR and Sanger sequence. (A) One step RT-droplet digital PCR screening of the HOXA9 expression in single clones infected with lentivirus containing the sgRNA library. The screening of 528 sgRNA library infected clones for HOXA9 expression levels is shown (528 dots). Ten of 528 clones exhibited more than 50% reduction in HOXA9 levels (purple arrows). The red line signifies the boundary of a 2-fold decrease change by comparing with the control cells; the blue line signifies the boundary of a 2-fold increase change. (B) The six clones #5, 6, 28, 121, 207 and 420 were targeted by the CBS7/9 specific sgRNA through Sanger sequence (green arrows). (C) The RT-ddqPCR analysis of HOXA9 levels in WT MOLM13 and the 21 clones containing single targeted sgRNA. The HOXA9 expression data were grouped into five groups in accordance with the categories of sgRNA sequences: HOXA7/9 CTCF site, non-human targets, other CTCF sites in the HOX loci, HOX associated lncRNAs, and other human targets. This figure has been modified from Luo et al.12. For statistics, this data was represented as the mean ± SD from three independent experiments with the Student's t-test. Please click here to view a larger version of this figure.
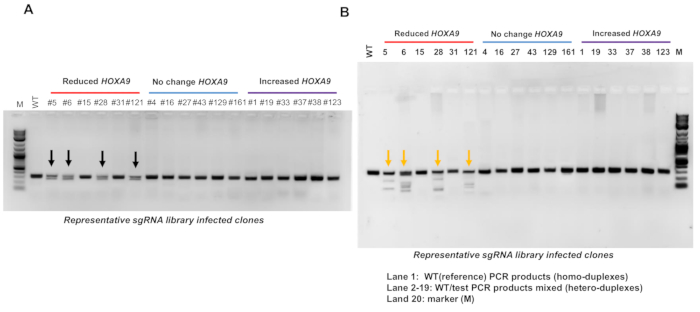
Figure 5: Indel mutations of integrated sgRNAs positive clone confirmed with the PCR-based genotyping and nuclease assay. (A) Genomic DNA was isolated from the representative CRISPR-Cas9 KO library screened clones that exhibited reduced, unchanged, or increased HOXA9 expression levels. The heterozygous deletion of the CTCF site located between HOXA7 and HOXA9 genes (CBS7/9 boundary) was identified by PCR-based genotyping. The HOXA9-decreased clones #5, 6, 28, and 121 exhibited deletion in the CBS7/9 boundary location (black arrows). (B) The Indel mutations in the CBS7/9 site were analyzed by the nuclease digestion assay from the representative clones that exhibited reduced (red line), unchanged (blue line), or increased (purple line) HOXA9 expression levels. The HOXA9-decreased clones #5, 6, 28, and 121 exhibited mutations in the CBS7/9 boundary location (orange arrows). This figure has been modified from Luo et al.12. Please click here to view a larger version of this figure.
Table 1: sgRNAs pool library targeting information. This data is from Luo et al.12. Please click here to download this table.
Table 2: Sangersequencing results of sgRNAs presented in the selected HOXA9-decreased, HOXA9-unchanged, and HOXA9-increased clones. HOXA9-decreased, unchanged and increased clones are highlighted in red, blue and purple, separately. This data is from Luo et al.12. Please click here to download this table.
Discussion
Protein-coding gene related sgRNA libraries have been applied in a functional screening system to identifying genes and networks regulating specific cellular functions through sgRNA enrichment24,25,26,27,28. Several non-coding region related sgRNA libraries were also shown in gene-specific functional screens for distal and proximal regulating elements, including BCL11A, Tdgf1a and drug-resistance regulating genes28,29,30. These sgRNA libraries were all generated by a detailed bioinformatics design, oligonucleotide synthesis, and sub-cloning the oligonucleotide pool(s) into vectors. The whole genome-wide screening approach is very powerful and useful but requires computational expertise for genome-wide sgRNA design and consistent funding for the expensive synthesis; thus, it is still challenging for most laboratories. However, our loci-specific sgRNA library screening approach is both convenient and efficient to identify the specific DNA element such as the CTCF binding site involved in chromatin organization and transcriptional regulation (Figure 1 and Figure 2). By targeting the CTCF boundaries, we applied a one-step RT-PCR to evaluate sgRNA targeted clones according to the expression level of a specific marker gene, HOXA9. In addition, we performed Sanger sequencing to confirm these positive integrated sgRNAs clones (Figure 2 and Figure 4). To functionally confirm these positive sgRNAs targeted clones, we carried out a PCR-based genotyping and mutation detection assay in order to determine whether the sgRNA induces the target site insert or deletion mutations (Figure 5). This gives us a promising method to target specific non-coding DNA elements and evaluate their biological function in mammalian cells.
In our protocol, we mentioned that a specific oligonucleotide design will ensure more efficient sub-cloning into lentiCRIPSRV2 vectors and more reliably generate an accurate sgRNA library (steps 1–2). In order to obtain the high titer sgRNA library lentivirus, the lentiviral supernatant should be concentrated 50-fold using the concentrator following the protocol (step 3), and stored in a -80 °C freezer in multiple aliquots (steps 3.1–3.5). An additional concern is finding the optimal MOI value for transduction. If the MOI is too low, the number of infected cells will decrease and lead to sgRNA screening failure. If the MOI is too high, it will integrate more than one sgRNA into a single cell, and it will interfere with the sgRNA library screening through the one-step RT-PCR and Indel mutation detection. Therefore, before screening, finding the optimal MOI for each group of cells through titration of the lentiviral library is an important step. Titration of the lentiviral library in MOLM13 leukemia cells and evaluation of the MOI will be carried out in the protocol (steps 5.1–5.10). Moreover, a thorough lysing of cells for reverse transcription can ensure successful one-step RT-PCR. This can be done by increasing the incubation time for lysis at all temperature stages in the protocol (steps 7.5–7.6). Therefore, in order to enhance the efficient for screening of the pooled CRISPR-Cas9 KO library, thorough cell lysis and reverse transcription play a critical role in determining the one-step RT-qPCR (steps 7.1–7.14). Additionally, increasing the quality of PCR products can ensure successful indel mutation detection, because low quality PCR products will affect the heteroduplex/homoduplex generation process (steps 9.1–.6).
In addition, the method can be used to identify the role of CTCF in HOX gene regulation in early embryonic development and certain leukemia with aberrant HOX gene signature. For example, HOX genes play critical roles during embryonic development and all four clusters of HOX gens are temporally and spatially restricted in their expression patterns in embryonic development. Furthermore, NPM1 mutations are among the most common genetic abnormality in AML and account for 30% of AML patients with normal cytogenetic karyotype31. This subset of AML exhibits an aberrant HOXA and HOXB gene signature, which becomes a dominant mechanism of leukemic transformation17. It is critical to elucidate how HOX gene are regulated in normal development and dysregulated during leukemogenesis. We and others have shown that CTCF plays an essential role in chromatin organization and gene transcription in HOX loci9,12. Thus, the HOX loci focused sgRNA library screening provides a convenient means to entangle the specific function of the CTCF binding site in HOX gene regulation during development and hematopoietic malignancies. However, a limitation of the approach is the difficulty of finding a useful marker for the high-throughput next-generation sequencing. One of the future research goals will be to find a highly selective marker and carry out genome-wide next generation sequencing in order to see the marker's effects. Therefore, using a specific fluorescent marker-tagged gene as the tracking reporter will become a crucial tool in future research plans.
Enhancers play a multitude of critical roles in the regulation of promoter function and gene expression. However, it can also activate promoter activity from long distance in a position and orientation independent manner, and enhancers often regulate gene expression in a trans orientation. Thus, it is challenging to pinpoint the enhancer(s) for specific genes, especially in the post-genomic era. Traditional reporter assays and correlative functional analyses (e.g., chromatin immunoprecipitation and DNaseI hypersensitive assays) have been used to examine enhancer function32,33. Similarly, small scaled locus-focused screenings were also applied to explore the activities of distal and proximal regulatory elements for specific genes34. Recently, the pooled sgRNA-KO library strategy that targets non-coding regulatory elements in the HOX gene loci successfully identified a CTCF binding site located between HOXA7 and HOXA9 genes, as well as a HOTTIP lncRNA that is critical for controlling posterior HOXA chromatin domain organization, which drives ectopic HOXA gene expression in acute myeloid leukemia (AML)12. These studies demonstrated that the pooled sgRNA-KO library screening is also a powerful genetics approach to identify and evaluate biological function of non-coding elements in our genome in situ.
CTCF, as a chromatin insulator protein, plays an important role in genome organization by defining chromatin neighborhoods for specific gene expression patterns in specific cell type11,35. Alteration of topologically associated domain (TAD) structure changes the enhancer/promoter interactions, resulting in a diseased state5,11. CTCF is highly conserved in metazoan and is enriched at the TAD boundaries. However, it remains unclear whether and how CTCF contributes to maintain chromatin boundary structure and TAD formation. Although the pooled CTCF sgRNA-knockout library screening was focused on the four HOX loci, it proved to be a powerful method to identify and dissect CTCF boundaries, and define TAD domain as well as enhancer/promoter interaction and transcription within the TAD domain12. Additionally, this method can be efficiently applied to identify the lncRNA elements and transcription factors that mediate chromatin conformation and accessibility activity in HOX loci. We are also trying to explore the CRISPR/sgRNA library containing the genome-wide CTCF sites through next generation sequencing identification according to the CTCF ChIP-seq and ChIA-PET data in future research. Thus, this strategy can be extended to a whole chromosome or even the whole genome.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors also thank Nicholas Cesari for editing the manuscript. The work was supported by grants from National Institute of Health (S.H., R01DK110108, R01CA204044).
Materials
| Lipofectamine 3000 reagent | Thermo Fisher Scientific | L3000-008 | |
| Proteinase K | Thermo Fisher Scientific | 25530049 | |
| Puromycin | Thermo Fisher Scientific | A1113802 | |
| Stbl3 cells | Life Technologies | C737303 | |
| HEK293T | ATCC | CRL-3216 | |
| MOLM-13 | DSMZ | ACC 554 | |
| lentiCRISPRv2 | Addgene | 52961 | |
| pMD2.G | Addgene | 12259 | |
| psPAX2 | Addgene | 12260 | |
| pGEM®-T Easy Vector Systems | Promega | A137A | |
| T4 ligase | New England Biolabs | M0202S | |
| QIAquick Gel Extract kit | QIAGEN | 28706 | |
| QIAuick PCR purification kit | QIAGEN | 28106 | |
| SingleShot™ SYBR® Green One-Step Kit | Bio-Rad Laboratories | 1725095 | |
| QIAGEN Plasmid Maxi Kit | QIAGEN | 12163 | |
| Dulbecco’s Modified Eagle Medium | Thermo Fisher Scientific | 11965084 | |
| RPMI 1640 | Thermo Fisher Scientific | 11875093 | |
| Fetal bovine serum (FBS) | Thermo Fisher Scientific | 10-082-147 | |
| Penicillin/streptomycin/L-glutamine | Life Technologies | 10378016 | |
| Lenti-X Concentrator | Clontech | 631232 | |
| Trypan Blue Solution | Thermo Fisher Scientific | 15250061 | |
| Polybrene | Santa Cruz Biotechnology | sc-134220 | |
| Phosphate Buffered Saline (PBS) | Genessee Scientific | 25-507 | |
| TAE buffer | Thermo Fisher Scientific | FERB49 | |
| Surveyor® Mutation Detection Kits | Integrated DNA Technologies | 706020 | |
| Biorad Universal Hood II Gel Doc System | Bio-Rad | 170-8126 | |
| Centrifuge 5424 R | Eppendorf | 5404000138 | |
| Digital Dry Baths/Block Heaters | Thermo Fisher Scientific | 88870002 | |
| TSX Series Ultra-Low Freezers | Thermo Fisher Scientific | TSX40086V | |
| Forma™ Steri-Cult™ CO2 Incubators | Thermo Fisher Scientific | 3308 | |
| Herasafe™ KS, Class II Biological Safety Cabinet | Thermo Fisher Scientific | 51022484 | |
| Sorvall™ Legend™ XT/XF Centrifuge Series | Thermo Fisher Scientific | 75004506 | |
| Fisherbrand™ Isotemp™ Water Baths | Thermo Fisher Scientific | FSGPD02 | |
| Thermo Scientific™ Locator™ Plus Rack and Box Systems | Thermo Fisher Scientific | 13-762-353 | |
| CFX96 Touch Real-Time PCR Detection System | Bio-Rad | 1855195 | |
| MiniAmp™ Thermal Cycler | Applied Biosystems technology | A37834 | |
| Thermo Scientific™ Owl™ EC300XL2 Compact Power Supply | Thermo Fisher Scientific | 7217581 | |
| Thermo Scientific™ Owl™ EasyCast™ B1 Mini Gel Electrophoresis Systems | Thermo Fisher Scientific | 09-528-178 | |
| VWR® Tube Rotator and Rotisseries | VWR International | 10136-084 | |
| VWR® Incubating Mini Shaker | VWR International | 12620-942 | |
| Analytical Balance MS104TS/00 | METTLER TOLEDO | 30133522 | |
| DS-11 FX and DS-11 FX+ Spectrophotometer | DeNovix Inc. | DS-11 FX |
References
- Dixon, J. R., et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 485 (7398), 376-380 (2012).
- Cuddapah, S., et al. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome research. 19 (1), 24-32 (2009).
- Phillips, J. E., Corces, V. G. CTCF: master weaver of the genome. Cell. 137 (7), 1194-1211 (2009).
- Tang, Z., et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell. 163 (7), 1611-1627 (2015).
- Lupianez, D. G., et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 161 (5), 1012-1025 (2015).
- Dowen, J. M., et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 159 (2), 374-387 (2014).
- Phillips-Cremins, J. E., et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 153 (6), 1281-1295 (2013).
- Narendra, V., Bulajic, M., Dekker, J., Mazzoni, E. O., Reinberg, D. CTCF-mediated topological boundaries during development foster appropriate gene regulation. Genes & Development. 30 (24), 2657-2662 (2016).
- Narendra, V., et al. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 347 (6225), 1017-1021 (2015).
- Deng, C., et al. HoxBlinc RNA Recruits Set1/MLL Complexes to Activate Hox Gene Expression Patterns and Mesoderm Lineage Development. Cell Reports. 14 (1), 103-114 (2016).
- Patel, B., et al. Aberrant TAL1 activation is mediated by an interchromosomal interaction in human T-cell acute lymphoblastic leukemia. Leukemia. 28 (2), 349-361 (2014).
- Luo, H., et al. CTCF boundary remodels chromatin domain and drives aberrant HOX gene transcription in acute myeloid leukemia. Blood. 132 (8), 837-848 (2018).
- Dou, D. R., et al. Medial HOXA genes demarcate haematopoietic stem cell fate during human development. Nature Cell Biology. 18 (6), 595-606 (2016).
- Lawrence, H. J., et al. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. 106 (12), 3988-3994 (2005).
- Deng, C., et al. USF1 and hSET1A mediated epigenetic modifications regulate lineage differentiation and HoxB4 transcription. PLOS Genetics. 9 (6), e1003524 (2013).
- Rawat, V. P., Humphries, R. K., Buske, C. Beyond Hox: the role of ParaHox genes in normal and malignant hematopoiesis. Blood. 120 (3), 519-527 (2012).
- Alharbi, R. A., Pettengell, R., Pandha, H. S., Morgan, R. The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia. 27 (5), 1000-1008 (2013).
- Rice, K. L., Licht, J. D. HOX deregulation in acute myeloid leukemia. Journal of Clinical Investigation. 117 (4), 865-868 (2007).
- Bassett, A. R., Kong, L., Liu, J. L. A genome-wide CRISPR library for high-throughput genetic screening in Drosophila cells. Journal of Genetics and Genomics. 42 (6), 301-309 (2015).
- Zhu, S., et al. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nature Biotechnology. 34 (12), 1279-1286 (2016).
- Kurata, J. S., Lin, R. J. MicroRNA-focused CRISPR-Cas9 library screen reveals fitness-associated miRNAs. RNA. 24 (7), 966-981 (2018).
- Collins, C. T., Hess, J. L. Role of HOXA9 in leukemia: dysregulation, cofactors and essential targets. Oncogene. 35 (9), 1090-1098 (2016).
- Kroon, E., Thorsteinsdottir, U., Mayotte, N., Nakamura, T., Sauvageau, G. NUP98-HOXA9 expression in hemopoietic stem cells induces chronic and acute myeloid leukemias in mice. The EMBO Journal. 20 (3), 350-361 (2001).
- Koike-Yusa, H., Li, Y., Tan, E. P., Velasco-Herrera Mdel, C., Yusa, K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nature Biotechnology. 32 (3), 267-273 (2014).
- Shalem, O., et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 343 (6166), 84-87 (2014).
- Wang, T., Wei, J. J., Sabatini, D. M., Lander, E. S. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 343 (6166), 80-84 (2014).
- Zhou, J., et al. Dual sgRNAs facilitate CRISPR/Cas9-mediated mouse genome targeting. The FEBS Journal. 281 (7), 1717-1725 (2014).
- Sanjana, N. E., et al. High-resolution interrogation of functional elements in the noncoding genome. Science. 353 (6307), 1545-1549 (2016).
- Rajagopal, N., et al. High-throughput mapping of regulatory DNA. Nature Biotechnology. 34 (2), 167-174 (2016).
- Korkmaz, G., et al. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nature Biotechnology. 34 (2), 192-198 (2016).
- Rezaei, N., et al. FMS-Like Tyrosine Kinase 3 (FLT3) and Nucleophosmin 1 (NPM1) in Iranian Adult Acute Myeloid Leukemia Patients with Normal Karyotypes: Mutation Status and Clinical and Laboratory Characteristics. Turkish Journal of Haematology. 34 (4), 300-306 (2017).
- Yaragatti, M., Basilico, C., Dailey, L. Identification of active transcriptional regulatory modules by the functional assay of DNA from nucleosome-free regions. Genome Research. 18 (6), 930-938 (2008).
- Wilken, M. S., et al. DNase I hypersensitivity analysis of the mouse brain and retina identifies region-specific regulatory elements. Epigenetics Chromatin. 8, 8 (2015).
- Narlikar, L., Ovcharenko, I. Identifying regulatory elements in eukaryotic genomes. Briefings in Functional Genomics and Proteomics. 8 (4), 215-230 (2009).
- Hnisz, D., et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 351 (6280), 1454-1458 (2016).

