Implementation of a Nonlinear Microscope Based on Stimulated Raman Scattering
Summary
In this manuscript, the implementation of a stimulated Raman scattering (SRS) microscope, obtained by the integration of an SRS experimental set-up with a laser scanning microscope, is described. The SRS microscope is based on two femtosecond (fs) laser sources, a Ti-Sapphire (Ti:Sa) and synchronized optical parametric oscillator (OPO).
Abstract
Stimulated Raman scattering (SRS) microscopy uses near-infrared excitation light; therefore, it shares many multi-photon microscopic imaging properties. SRS imaging modality can be obtained using commercial laser-scanning microscopes by equipping with a non-descanned forward detector with proper bandpass filters and lock-in amplifier (LIA) detection scheme. A schematic layout of a typical SRS microscope includes the following: two pulsed laser beams, (i.e., the pump and probe directed in a scanning microscope), which must be overlapped in both space and time at the image plane, then focused by a microscope objective into the sample through two scanning mirrors (SMs), which raster the focal spot across an x-y plane. After interaction with the sample, transmitted output pulses are collected by an upper objective and measured by a forward detection system inserted in an inverted microscope. Pump pulses are removed by a stack of optical filters, whereas the probe pulses that are the result of the SRS process occurring in the focal volume of the specimen are measured by a photodiode (PD). The readout of the PD is demodulated by the LIA to extract the modulation depth. A two-dimensional (2D) image is obtained by synchronizing the forward detection unit with the microscope scanning unit. In this paper, the implementation of an SRS microscope is described and successfully demonstrated, as well as the reporting of label-free images of polystyrene beads with diameters of 3 µm. It is worth noting that SRS microscopes are not commercially available, so in order to take advantage of these characteristics, the homemade construction is the only option. Since SRS microscopy is becoming popular in many fields, it is believed that this careful description of the SRS microscope implementation can be very useful for the scientific community.
Introduction
In life science applications, SRS microscopy has emerged as powerful tool for label-free imaging. The basic idea of SRS microscopy is to combine the strength of vibrational contrast and its ability to acquire images in a few seconds.
SRS is a process in which the frequency difference between two laser beams frequencies (pump signal and stokes signal at different frequencies) matches the molecular vibration of an investigated sample, causing stimulated Raman scattering and a significant increase in the Stokes signal. Unlike linear Raman spectroscopy, SRS exhibits a nonlinear dependence on the incoming light fields and produces coherent radiation. SRS has two fundamental advantages: 1) speed, which makes images less sensitive to artefacts arising from sample movement or degradation, and 2) an excellent signal-to-noise ratio (SNR). In addition, SRS exhibits a spectrum identical to the spontaneous Raman, and the SRS signal is linearly proportional to the concentration of the chemical bond excited1,2,3,4,5.
In our microscope, a femtosecond (fs) SRS experimental set-up is integrated with an inverted optical microscope equipped with a fast mirrors scanning unit (Figure 1)6,7,8. Two pulsed laser sources are used to implement this microscope. The first is a fs-Ti:Sa with a pulse duration of approximately 140 fs, repetition rate of 80 MHz, and emission wavelengths in the range of 680-1080 nm. The second, used as probe beam and pumped by Ti:Sa, is a femtosecond synchronized optical parametric oscillator (SOPO), with a pulse duration of approximately 200 fs, repetition rate of 80 MHz, and emission wavelengths in the range of 1000-1600 nm. It should be noted that the minimum photon energy difference between the Ti:Sa and SOPO beam is 2500 cm-1. Therefore, using this combination of laser systems, only the high frequency C-H region (2800-3200 cm-1) of Raman spectra can be explored6,7,8.
In order to set up a SRS microscope, there are three crucial issues to consider, which are described in the successive paragraphs. The first is the implementation of a high-frequency modulation transfer method (see Figure 2 and step 2.1 of the protocol for a description). In a SRS experimental investigation, a crucial parameter is the sensitivity of the system. A SRS signal is detected as a small change in the intensity of excitation beams; therefore, it can be corrupted by laser intensity noise and shot noise. This issue can be overcome by integrating this system with a high-frequency modulation transfer method (see Figure 2 and step 2.1 of the protocol for details). In this method, an electro-optic modulator (EOM) is used to modulate the pump. The modulation transferred to the probe beam can then be detected by a PD after blocking the pump beam with a stack of optical filters [stimulated Raman gain (SRG) detection mode]. The PD output is connected by a low pass filter to a lock-in amplifier (LIA), which demodulates the measured signal. By increasing the modulation frequency of the beam to frequencies above 1 MHz, the intrinsic limit of PDs can be obtained.
The second issue to consider is the installation of a mechanical mount which permits to carry out forward detection and at the same time to preserve microscope observation in brightfield. In addition, it has to reduce the noise due to mechanical vibration during the generation of images and to allow the precise repositioning of detection system (see Figure 3 and step 2.2 of the protocol).
The third is the synchronization of the signal acquired by the phase-sensitive detection scheme, with the beam positioned onto the sample monitored by the scan head of the microscope. In order to realize images, the SMs require three TTL signals that are made available by the microscope controller connected to the scan head unit: pixel clock, line sync, and frame sync. The synchronization is achieved by controlling using a PCI card, the three TTL signals, and the acquisition of a voltage signal at the output channel of LIA6,7,8. A homemade software has been developed and described previously6,7,8, while the hardware of the synchronization system is reported in Figure 4.
A fundamental procedure when carrying out SRS imaging is microscope alignment. It is realized over the course of four steps, which are described in the successive paragraphs. The first is the spatial overlap of two beams (see step 3.1 of the protocol). In this experimental set-up, the two beams were spatially collinearly combined by a dichroic mirror. The preliminary step is the alignment of OPO and Ti:Sa so that each reaches the microscope. Then, considering OPO as a reference beam and taking advantage of a position sensitive detector, the Ti:Sa is spatially overlapped to OPO.
The second crucial aspect is the temporal overlap of two beams (see step 3.2 of the protocol). Even if the pump and OPO beams are perfectly synchronized9, since they follow slightly different beam paths inside the OPO housing, at the OPO exit they have a time delay of about 5 ns and spatial difference of 5 cm. Therefore, Ti:Sa and OPO require being re-timed optically to ensure temporal overlap at the sample. This is typically accomplished with a finely tunable optical delay line, which in this case is inserted between the Ti:Sa and microscope (see Figure 1). In order to obtain the temporal overlap of two beams, two techniques are used. The first is carried out using a fast PD and oscilloscope, while the second is based on auto- and cross-optical correlations. Using the first technique, a rough overlap of two beams is obtained (uncertainty of 10 ps), while an accurate temporal overlap of two beams is obtained using a cross-correlator (resolution of 1 fs).
The third crucial aspect is alignment of the two beams inside the microscope (see step 3.3 of the protocol). A preliminary white light observation of sample allows to individuate the desired field of view (FOV). Afterwards, laser beams, entering the microscope by a side port of microscope, are aligned in order to reach the PD mounted on the upper part (Figure 3). However, for a correct image acquisition, setting a number of parameters is required (for example, pixel dimension and pixel dwell time). The sampling frequency must respect the constraint imposed by Nyquist’s theorem in order to preserve all information in an image, while for a correct correspondence between the spatial coordinates of pixels and SRS value measured in each pixel, the integration time of LIA should be equal or comparable to the pixel dwell time.
In the final step of microscope alignment, numerous tests are carried out to optimize the spatial and temporal alignment (see step 3.4 of the protocol). A number of transmission images (TI) for both Ti:Sa and OPO are acquired in order to optimize spatial overlap. In a TI, a single beam is used, and the transmitted beam intensity from the sample is measured by a PD. In the case of TI realized by OPO, the PD output signal is directly connected to PCI card, while in the case of TI realized by Ti:Sa, the PD output signal is connected to LIA and analog output of LIA is connected to PCI card. The transmission images are very useful to optimize the FOV, the illumination, the focal position of microscope objectives and to check if the two beams are spatially overlapped6,7,8.
The optimization of the pump and probe beam’s temporal overlap is obtained by scanning the delay line with steps of 0.001 mm corresponding to a 3.3 fs time-shift and carrying out a SRS measurement in a single point of a polystyrene bead sample 3 µm in diameter. The amplitude of an SRS signal measures values from LIA, as a function of the probe-pump delay, and provides a maximum corresponding with exact temporal overlap of the two beams6,7,8. Before concluding, it should be noted that all discussed steps are mandatory to obtain a high quality image.
Protocol
1. Starting up the laser system
- Check if the temperature of chillers is maintained at or below 20 °C.
- Check if the humidity control unit is working properly and humidity is maintained at a value around 40%.
- Turn on the Ti:Sa laser, strictly following the instructions in the manual.
- Set the wavelength to 810 nm.
- Turn on the OPO and the connected mini-computer. Run the application that controls the OPO laser.
- Select bypass if 100% of the Ti:Sa laser output is required at the exit of the OPO box.
- Deselect bypass if 20% of the Ti:Sa laser output and the OPO laser output are required at the exit of the OPO box.
- Open the shutter of Ti:Sa and release the Ti:Sa beam to the OPO input.
- Release the two laser beams at the OPO exit by clicking signal-out and pump-out.
- Wait until both lasers Ti:Sa and OPO are stabilized (around 45–60 min).
- Verify the beam spot at the exit of the OPO box for both Ti:Sa and OPO using a paper detector card and check the power using a power meter.
- Tune the OPO laser wavelength to 1076 nm.
- Reduce the power to ~10 mW for each laser beam to perform alignment.
2. Setting up the microscope
- Implementation of high frequency modulation transfer method
- Carry out the optical alignment procedure of the modulator such that the Ti:Sa beam enters and exits without any distortion.
- Turn on the function generator and generate a TTL signal (square wave with amplitude = 5 V, offset = 2.5 V, frequency = 5 MHz).
- Divide the TTL signal into two parts using a T junction; one for EOM and the other for the lock-in amplifier (LIA) (see Figure 2).
- Check all signal levels with an oscilloscope.
- Turn on the LIA and connect the generator output channel to the reference channel of the LIA.
- Connect the generator output channel to the high voltage power amplifier of EOM.
- Turn on the amplifier and set the voltage to almost the maximum level. Monitor the beam power at the exit of EOM.
- Integration of mechanical mounting to fix PD and assign x and y relative motion
NOTE: With the microscope, an external mount is introduced, equipped with a micrometer that has motion control in x and y directions.- Mount two travel translation stages to allow movements along the x and y directions (see Figure 3).
- Fix the stage on a Ø1.5" post of appropriate height.
- Mount the PD to an external mount.
- Maximize the beam power at PD, adjusting the PD positions (x and y coordinates) using the micrometers attached to the mount (see Figure 3).
3. Alignment of microscope
- Spatial overlap of the beams
NOTE: Considering the OPO beam as a reference and taking advantage of a position sensitive detector, the Ti:Sa should be overlapped to OPO according to the following procedure:- Align the OPO and the Ti:Sa laser beams so that they both reach the microscope.
- Place the laser beam position sensors detectors in two positions in between dichroic mirror 1 and mirror 6, the first position is located close to dichroic mirror 1 and the second one is close to mirror 6. For each position, use the sensors to detect the x and y coordinates of the OPO beam (follow Figure 1).
- Verify that the x and y coordinates of the Ti:Sa laser beam are the same OPO in both positions of the sensors detectors. If in some positions the coordinates of Ti:Sa and OPO do not coincide, tune the tilt of the adjacent mirror to compensate the difference (follow Figure 1).
- Follow the same procedure to align the Ti:Sa beam positions with respect to OPO for the path in between M6-M7 (follow Figure 1).
- Temporal synchronization of the beams
- Use of the fast photodiode plus oscilloscope:
- Stop propagation of the Ti:Sa and OPO beams and place a fast detector in front of the OPO beam (in between M6 and M7).
- Connect the trigger signal provided by the Ti:Sa laser box with an oscilloscope in channel 2.
- Connect the detector cable with the oscilloscope in channel 1 and visualize the OPO temporal profile.
- Record the time (abscissa) measured by the oscilloscope corresponding to its maximum value, namely t1.
- Stop the OPO beam and release the Ti:Sa beam.
- Visualize the Ti:Sa temporal profile and record the time (abscissa) corresponding to its maximum value, namely t2.
- Minimize the difference between t1-t2 using the delay line to overlap the two beams. In our case, the minimum measurable difference is 10 ps.
- Remove the fast detector in between M6 and M7.
- Use of the autocorrelator:
NOTE: In the schematic diagram shown in Figure 1, an autocorrelator is installed without interfering with optical paths of the beams. In addition, an additional mirror is introduced and mounted on a flip-flop mount (referred to as FFM/AM) in between M6 and M7 to divert the beam into the autocorrelator.- Flip the AM to direct the beam into the autocorrelator.
- Stop the Ti:Sa and release the OPO.
- Set the beam distance adjustment screw micrometer of the autocorrelator to Normal position (8.35 mm).
- Power on the autocorrelator controller and start the software application on the personal computer controlling it.
- Project the OPO beam from FFM/AM to the input mirror into the autocorrelator.
- Control the reflection spot (using a paper detector card) of the beam on the alignment window of the autocorrelator.
- In the case of no-beam or low-beam intensity, adjust the position and orientation of FFM/AM to the optimum extent, and try to adjust the input mirror (mounted on the autocorrelator) to maximize the laser pulse signal. The autocorrelator signal is obtained as shown in Figure 5a.
- Stop the OPO and project of the Ti:Sa beam from FFM/AM to input mirror into the autocorrelator. Repeat steps 3.2.2.6 and 3.2.2.7. The autocorrelator signal is obtained as shown in the Figure 5b.
- Set the beam distance adjustment screw micrometer to Cross position (7.30 mm).
- Release both beams.
- Scan the delay line to obtain the two beams OPO and Ti:Sa overlapped. The cross-correlator signal is obtained as shown in the Figure 6.
- Flip the mirror FFM/AM so that the beams can reach M7 and scan head of the microscope.
- Use of the fast photodiode plus oscilloscope:
- Microscope alignment
- Perform white light microscopic observation:
NOTE: Prior to microscopic observation, ensure that the microscope is properly aligned.- Prepare the test sample, which consists of a phosphate buffer solution in which polystyrene beads with diameters of 3 μm are dispersed. The solution is placed inside a sandwich of two glass slides.
- Turn on the microscope and power supply of white light. Follow the manual for observation under white light.
- Use a condenser to illuminate the sample. Use objective of 60x to collect light. Place the sample onto the stage. Optimize the focal position of the 60x microscope objective.
- Select the FOV of interest. Take a CCD image of the sample (Figure 7).
- Turn off the power supply of white light.
- Microscope alignment with femtosecond laser beams: OPO and Ti:Sa
- Remove the condenser using the escape button to temporarily retract the 60x microscope objective lens. Move the 60x microscope objective lens off the optical path, rotating the nosepiece.
- Mount the detector to the upper part of microscope using the external mechanical mount. Connect the detector output through a low-pass filter of 50Ω to oscilloscope and monitor the OPO signal.
- Turn on the processor that controls the scanner head. Project the OPO beam into the scanner head of the microscope.
- Check the position of the beam inside the microscope, make sure that the location of the beam is in the center or near the center.
- Check that the position of the beam inside the head of PD is in the center.
- Maximize the power measured by the detector using an x-y translator.
- Switch the beam from OPO to Ti:Sa and verify that a maximum signal is also obtained for the titanium-sapphire laser. THis indicates that both beams are well-aligned.
- Finalize the beam alignment, introducing the 60x microscope objective lens and rotating back the nosepiece.
- Use the refocus button on the microscope to regain the finalized focus to the 60x microscope objective lens.
- Place the objective with magnification 40x in place of the condenser without touching or disturbing the sample.
- Perform white light microscopic observation:
- Optimization of spatial and temporal synchronizations of beams
- Temporal synchronization
- Set the power of Ti:Sa and OPO measured before the microscope to 30 mW for both beams. Set the wavelength of OPO to a different value with respect to the previous one so that the pump and probe are not in resonance with the vibrational frequency of the beads.
- Release both beams (Ti:Sa and OPO) so that they enter the microscope.
- Run the scanning delay line computerized translator and record the measured intensity by LIA for each position of the delay line. Wait until the delay line scanning is complete. The obtained temporal profile is visualized in Figure 8a.
- Set the wavelength of OPO to 1076 nm again so that pump and probe are in resonance with the vibrational frequency of the beads. Repeat step 3.4.1.3 (the obtained temporal profile is visualized in Figure 8b).
- Set the obtained overlap beam position in the delay line to acquire SRS images.
- Spatial synchronization of the beams
NOTE: The transmission images are useful to optimize the FOV, illumination, and focal position of the microscope objectives, and to check if the two beams are spatially overlapped.- Transmission image acquisition of OPO
- Stop the Ti:Sa beam and reduce the OPO power to 8 mW.
- Connect the detector readout to the data acquisition card.
- Run the data acquisition program along with microscope scanning console.
- Save the file and process the data to get the image. The raw image appears as shown in Figure 9a.
- Transmission image acquisition of Ti:Sa
- Stop the OPO beam and reduce the Ti:Sa power to 2.5–4.5 mW.
- Connect the detector with LIA and LIA readouts with the data acquisition card.
- Repeat steps 3.4.2.1.3 and 3.4.2.1.4. The raw image appears as shown in Figure 9b.
- Transmission image acquisition of OPO
- Temporal synchronization
4. SRS image acquisition
NOTE: A dedicated algorithm has been realized in order to store data. It supports the following image formats: 512 px x 512 px and 256 px x 256 px, with acquisition times of 16 s, 8 s, 4 s, and 2 s.
- Introduce a stack of filters in between the 40x objective and PD to remove the pump pulses (Ti:Sa) and acquire only the Stokes signal (OPO).
- Set the pump signal to 810 nm with a focused power of 8 mW and the probe signal to 1076 nm with a focused power of 8 mW to investigate a typical C-H bond of polystyrene (Raman shift of 3054 cm−1).
- Connect the detector with the LIA and LIA readout to the data acquisition card.
- Set the image acquisition pixel format as per requirement and set the acquisition time.
- Run the program that controls the microscope controller.
- Run the dedicated algorithm program that acts as synchronization between the microscope controller, detection system, and DAQ (see Figure 4).
- Save the matrix file once the acquisition is completed.
- Import the raw data file and save the image in the required format (typically saved in .tif format) using ImageJ software. The image is shown in the Figure 10.
Representative Results
An example of SRS measurement (i.e., SRS measurement in a single point of the sample) is reported in Figure 7. When the beams are not overlapped in time or space, the obtained result is reported in Figure 8a. In off-resonance, the amplitude of signal measured by LIA is zero, while the phase of signal measured by LIA jumps between negative and positive values. Whereas, when the beams are overlapped in space, moving the delay line in an appropriate range, the obtained results are reported in Figure 8b. The signal measured by LIA increases and reaches its maximum when the beams are perfectly overlapped in time, while the phase starts to achieve a fixed value during the time at which the beams are overlapped in time.
The absorption images obtained using a single beam (Ti:Sa or OPO) of the same polystyrene beads are represented in Figure 9a,b with scale bars of 6 µm. In order to acquire the SRS images, the delay line is set to the position achieved in Figure 7b, a typical SRG image is shown in Figure 10 with a scale bar of 6 µm.

Figure 1: Schematic layout of the f-SRS microscope system. OPO = optical parametric oscillator; Ti:Sa = Titanium-Sapphire laser; M1-M7= femtosecond broadband mirrors; FFM/AM = Flip-Flop Mirror/ Autocorrelator Mirror; DM1, DM2 = dichroic mirrors; DL = Delay Line; AC = autocorrelator ; EOM = electro-optic modulator; FG = function generator; GM = Galvo mirror; Obj1, Obj2 = microscope objectives; PD = photodiode; DAQ = data acquisition system; PC = personal computer. Please click here to view a larger version of this figure.
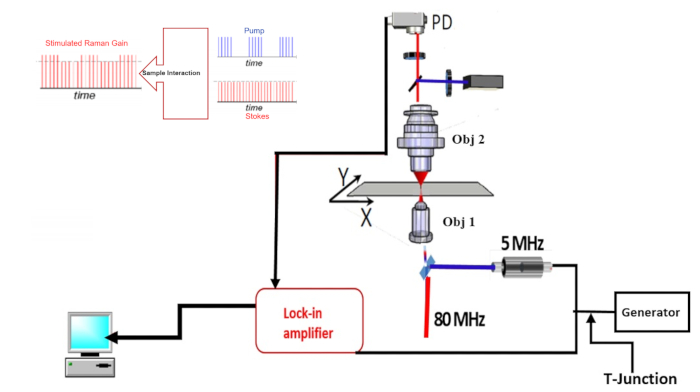
Figure 2: Scheme of high frequency modulation transfer method. In the inset figure, the two lasers beams before interaction inside the sample and modified probe due to interactions of the probe and pump inside the sample are represented. Time is represented in ns. Please click here to view a larger version of this figure.

Figure 3: Representation of photodiode mount with mechanical mounting system. Please click here to view a larger version of this figure.
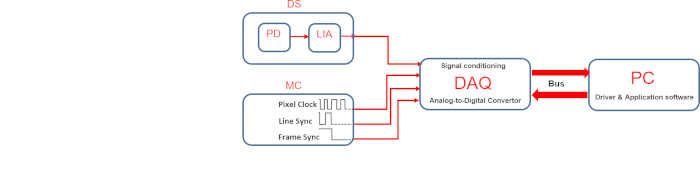
Figure 4: Schematic of data acquisition system. PD = photodiode, LIA = lock-in amplifier, DS = detection system, MC = microscope control, DAQ= Data acquisition system, PC = personal computer. Please click here to view a larger version of this figure.

Figure 5: Autocorrelator function of OPO (a) and Ti:SA (b). Please click here to view a larger version of this figure.

Figure 6: Cross correlation function of OPO and Ti:Sa. Please click here to view a larger version of this figure.

Figure 7: CCD image of polystyrene beads.

Figure 8: Amplitude and phase of SRS signal measured by lock-in amplifier: off resonance (on the left) and in resonance (on the right). Please click here to view a larger version of this figure.
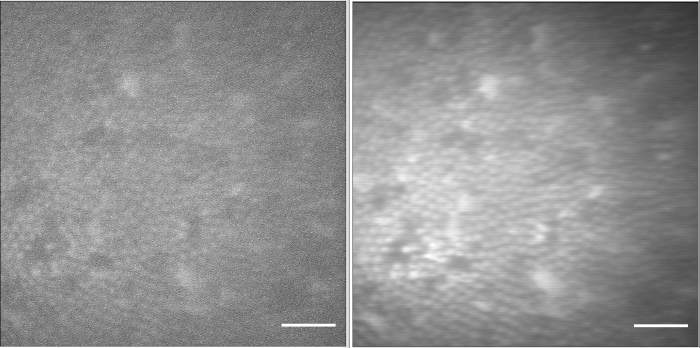
Figure 9: Transmission images of polystyrene beads achieved by OPO (a) and Ti:Sa (b). Scale bar = 16 µm. Please click here to view a larger version of this figure.

Figure 10: SRS image of polystyrene beads. Scale bar = 12 µm.
Discussion
SRS microscopy has taken label-free imaging to new heights, especially in studies of complex biological structures such as lipids, which are fundamental to cells and cellular architecture. Lipids are involved in multiple physiological pathways such as production of biological membranes, and they serve as biosynthetic precursors and signal transducers10. Lipids are packaged into specialized intracellular organelles, also called lipid droplets (LDs). Their diameters vary from few tens of nanometers to tens of micrometers11,12. LDs not only participate abundantly in adipose- and steroid-producing cells but are also present in other cell lines. LDs cooperate in a several physiological processes such as lipid storage. They are featured prominently in common pathologies (e.g., altered cholesterol metabolism)13,14.
Traditionally, visualization of lipids is achieved using fluorescence microscopy and neutral lipid-specific dye-labeled fixed cells10. It should be noted that as lipids are smaller sized in comparison to proteins and DNA, structural and functional changes and unwanted artifacts can occur when adding fluorophores15,16. SRS has been shown to be powerful for studying lipid-rich structures. Lipids are abundant in C-H2 groups. Therefore, the relatively isolated peaks associated with C-H bond vibrational states at 2845 cm-1 in their Raman spectra provide a unique signature for lipids inside a cell. Unfortunately, since the differentiable vibrational signatures are finite, it is rather difficult to distinguish a target biomolecule from the other related species inside cells that share similar chemical bonds. However, it is possible to add tiny Raman-active vibrational probes (e.g., alkynes and stable isotopes) to obtain specificity for imaging of small biomolecules17.
For biological and biomedical in vivo applications, simultaneous mapping of various chemical species in a given sample is necessary for investing the co-distribution and dynamic correlations between pairs of biomolecules18,19. Therefore, many efforts have been made to obtain multiple chemical contrasts. In the simplest option of multicolor imaging, to image different Raman modes of a sample, the frequency of the pump beam or Stokes beam are tuned in sequential scans18. However, using the wavelength tuning approach may cause loss of co-localization information of different Raman modes, especially when the sample is in a dynamic environment18.
As a consequence of nonlinear excitation, SRS offers intrinsic 3D resolving capabilities of the selected chemical bond within biological samples20. Volume reconstruction of the selected chemical bond and its spatial distributions can be simply achieved by collecting SRS images at different focal plane along the z-axis. Since the images are acquired with high spatial and temporal resolutions, other pieces of key information (i.e., 3D structure, chemical composition, etc.) about the biological sample can be obtained.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We appreciate V. Tufano from IMM CNR for his valuable technical assistance and Giacomo Cozzi, product specialist from Nikon Instruments, for useful discussions and continuous support. This work was partially supported by Italian National Operative Programs PONa3 00025 (BIOforIU) and by Euro-Bioimaging large-scale panEuropean research infrastructure project.
Materials
| Acquisation tool | Nikon | Nikon C2Tool | Acquisation supported tool |
| APE Pulse link control software | APE- | APE Pulse link control software | software control |
| Autocorrelator | APE | APE PulseCheck USB 50 | Autocorrelator |
| Detector | Thorlabs | Thorlabs DET10A | Photodiode |
| Detector card | Thorlabs | Thorlabs VRC | IR detector Card |
| Dichroic mirror | Semrock | Semrock FF875-Di01-25X36 | Dichroic mirror |
| Dichroic mirror | Semrock | FF875-Di01-25×36 | Dichroic mirror |
| EOM | Conoptics | (EOM CONOPTICS 3350-160 KD*P). | Pockels cell |
| Fast detector | Thorlabs | Thorlabs DET025AL/M | Photodiode |
| Fast mirror scanning unit | Nikon | C2 | Microscpe scanning head |
| Femtosecond laser Ti:SA | Coherent | Coherent Chameleon Ultra II | Chameleon Ultra II |
| Function generator | TTi | TG5011 AIM – TTi | Function generator |
| Inverted optical microscope | Nikon | Eclipse TE-2000-E, Nikon | Eclipse TE-2000-E, Nikon |
| Lock-in Amplifier | Standford Research System | SR844-200 MHz dual phase | A lock-in amplifier from Stanford Research Systems |
| Notch filter, | Semrock | NF03-808E-25 | Notch filter |
| Optical delay line | Newport | Newport M-ILS200CC | Tunable optical delay line |
| Optical Parametric Oscillator | Coherent | Coherent Compact OPO | Coherent Compact OPO |
| Oscilloscope | WaveRunner | 640Zi 4GHz OSC/LeCroy | Digital Oscilloscope |
| PCI Card | National instrument | NI PCIe 6363 | Data acquisation card |
| Position Sensors Detectors | Newport | Newport Conex PSD9 | Position detector sensor |
| Power meter head | Coherent | PowerMax PM10, | Laser power detector |
| Translation Stages | Thorlabs | Thorlabs PT1/M | Meachnical Translation Stage with Standard Micrometer |
References
- Saar, B. G., et al. Video-Rate Molecular Imaging in Vivo with Stimulated Raman Scattering. Science. 330 (6009), 1368-1370 (2010).
- Zhang, D., Wang, P., Slipchenko, M. N., Cheng, J. X. Fast Vibrational Imaging of Single Cells and Tissues by Stimulated Raman Scattering Microscopy. Accounts of Chemical Research. 47 (8), 2282-2290 (2014).
- Alfonso-García, A., Mittal, R., Lee, E. S., Potma, E. O. Biological imaging with coherent Raman scattering microscopy: a tutorial. Journal of Biomedical Optics. 19 (7), 071407 (2014).
- Cheng, J. X., Xie, X. S. Vibrational spectroscopic imaging of living systems: An emerging platform for biology and medicine. Science. 350 (6264), aaa8870 (2015).
- Camp, C. H., Cicerone, M. T. Chemically sensitive bioimaging with coherent Raman scattering. Nature Photonics. 9 (5), 295-305 (2015).
- D’Arco, A., et al. Subcellular chemical and morphological analysis by stimulated Raman scattering microscopy and image analysis techniques. Biomedical Optics Express. 7 (5), 1853 (2016).
- D’Arco, A., et al. Label-free imaging of small lipid droplets by femtosecond-stimulated Raman scattering microscopy. Journal of Nonlinear Optical Physics & Materials. 26 (4), (2017).
- Ranjan, R., et al. Integration of stimulated Raman gain and stimulated Raman losses detection modes in a single nonlinear microscope. Optics Express. 26 (20), 26317 (2018).
- Reid, D. T., Sun, J., Lamour, T. P., Ferreiro, T. I. Advances in ultrafast optical parametric oscillators. Laser Physics Letters. 8 (1), 8-15 (2011).
- Zumbusch, A., Langbein, W., Borri, P. Nonlinear vibrational microscopy applied to lipid biology. Progress in Lipid Research. 52 (4), 615-632 (2013).
- Suzuki, M., Shinohara, Y., Ohsaki, Y., Fujimoto, T. Lipid droplets: Size matters. Journal of Electron Microscopy. 60 (1), S101-S116 (2011).
- Rizzatti, V., et al. Lipid droplets characterization in adipocyte differentiated 3T3-L1 cells: size and optical density distribution. European Journal of Histochemistry. 57 (3), 159-162 (2013).
- Alfonso Garcia, A., et al. D38-cholesterol as a Raman active probe for imaging intracellular cholesterol storage. Journal of Biomedical Optics. 21 (6), (2016).
- Mukherjee, S., Zha, X., Tabas, I., Maxfield, F. R. Cholesterol distribution in living cells: fluorescence imaging using dehydroergosterol as a fluorescent cholesterol analog. Biophysical Journal. 75 (4), 1915-1925 (1998).
- Fukumoto, S., Fujimoto, T. Deformation of lipid droplets in fixed samples. Histochemistry and Cell Biology. 118 (5), 423-428 (2002).
- Kinkel, A., et al. Oil red-O stains non-adipogenic cells: A precautionary note. Cytotechnology. 46 (1), 49-56 (2004).
- Wei, L., et al. Live-Cell Bioorthogonal Chemical Imaging: Stimulated Raman Scattering Microscopy of Vibrational Probes. Accounts of Chemical Research. 49 (8), 1494-1502 (2016).
- Ozeki, Y., Asai, T., Shou, J., Yoshimi, H. Multicolor Stimulated Raman Scattering Microscopy with Fast Wavelength-Tunable Yb Fiber Laser. IEEE Journal of Selected Topics in Quantum Electronics. 25 (1), 1-11 (2019).
- Saar, B. G., Contreras-Rojas, L. R., Xie, X. S., Guy, R. H. Imaging Drug Delivery to Skin with Stimulated Raman Scattering Microscopy. Molecular Pharmaceutics. 8 (3), 969-975 (2011).
- Chen, X., et al. Volumetric chemical imaging by stimulated Raman projection microscopy and tomography. Nature Communications. 8, 15117 (2017).
- Ferrara, M. A., Filograna, A., Ranjan, R., Corda, D., Valente, C., Sirleto, L., et al. Threedimensional label-free imaging throughout adipocyte differentiation by stimulated Raman microscopy. PLoS ONE. 14 (5), e0216811 (2019).

