Human iPSC-Derived Cardiomyocyte Networks on Multiwell Micro-electrode Arrays for Recurrent Action Potential Recordings
Summary
This article contains a set of protocols for the development of human induced pluripotent stem cell-derived cardiomyocyte (hiPSC-CM) networks cultured on multiwell MEA plates to reversibly electroporate the cell membrane for action potential measurements. High-throughput recordings are obtained from the same cell sites repeatedly over days.
Abstract
Cardiac safety screening is of paramount importance for drug discovery and therapeutics. Therefore, the development of novel high-throughput electrophysiological approaches for hiPSC-derived cardiomyocyte (hiPSC-CM) preparations is much needed for efficient drug testing. Although multielectrode arrays (MEAs) are frequently employed for field potential measurements of excitable cells, a recent publication by Joshi-Mukherjee and colleagues described and validated its application for recurrent action potential (AP) recordings from the same hiPSC-CM preparation over days. The aim here is to provide detailed step-by-step methods for seeding CMs and for measuring AP waveforms via electroporation with high precision and a temporal resolution of 1 µs. This approach addresses the lack of easy-to-use methodology to gain intracellular access for high-throughput AP measurements for reliable electrophysiological investigations. A detailed work flow and methods for plating of hiPSC-CMs on multiwell MEA plates are discussed emphasizing critical steps wherever relevant. In addition, a custom-built MATLAB script for rapid data handling, extraction and analysis is reported for comprehensive investigation of the waveform analysis to quantify subtle differences in morphology for various AP duration parameters implicated in arrhythmia and cardiotoxicity.
Introduction
Human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) are the gold standard for an increasing number of laboratories1,2,3,4,5,6,7,8,9,10. Beating embryoid bodies11,12,13 and monolayer3,7,10,11,12,13,14,15,16,17 differentiation are the preferred methods for cardiomyocyte production and the multielectrode array (MEA) has become a common modality for monitoring the electrodynamics of these networks18,19,20. While parameters that can be extracted from field potentials (FPs) such as beating rate, amplitude, duration and RR intervals are baseline electrophysiological responses of spontaneously beating monolayers18,21,22,23, the action potential (AP) components underlying these extracellular FP signals are difficult to extrapolate24. Our recent publication on the discovery of an application of MEAs for direct recurrent AP measurements provides proof of methodology for exemplary intracellular AP readouts with an extensive waveform analysis at various repolarization phases across multiple batches of hiPSC-derived cardiomyocyte networks3. In the study we demonstrated that the delivery of electroporating pulses to networks of hiPSC-derived cardiomyocytes enables intracellular access for AP recordings. These transient AP recordings are dependent on transmembrane potential recoveries observed through the injury site3,25,26. Waveforms recorded via MEA and patch- clamp in our study showed similar AP morphologies thus validating the reliability of the approach3.
A few laboratories have reported measuring APs from various electrogenic cells using custom-built MEAs18,21,26,27,28,29,30, but the reliability of using MEAs for consistent and recurrent AP measurements was not assessed. Currently, the gold standard patch-clamp technique is limited to terminal recordings7,31 whereas, MEA-based AP measurements are transient and therefore can be conducted multiple times on the same cell. We also show that one can easily record high-quality AP signals in the millivolt range requiring minimal filtering. Researchers can therefore conduct not only acute but also chronic drug studies in the same preparations using MEAs. Additionally, this technology permits simultaneous FP/AP measurement generating electro-biome libraries in a short period of time. Given the growing emphasis on arrhythmia prediction and drug associated cardiotoxicity24,32,33,34,35, integration of AP measurement approaches will enhance drug safety and efficacy assessments.
Here, we present protocols for 1) pre-plating of cryopreserved hiPSC-CMs for maturation, 2) dissociating and plating of hiPSC-CMs on multiwell MEAs, 3) recording of FPs and APs from hiPSC-CM networks, 4) segmenting and extracting the data for analysis, and 5) restoring the arrays for multiple reuse. Each step has been optimized emphasizing critical steps wherever relevant. Requirements for cell attachment to ensure a beating syncytial monolayer are discussed and procedures for multiwell MEA restoration for repetitive electrophysiological studies are explained. Finally, a custom GUI developed in the laboratory is presented for AP signal extraction, quality assurance, and segmentation workflow to quantify and analyze AP parameters.
Protocol
1. Preparation of solutions and materials (see Table of Materials)
- 6-well tissue-culture plate substrate-coating
- Thaw the coating substrate on ice or at 4 °C.
- Prepare a 1:100 coating substrate dilution in cold DMEM/F12 medium. Mix the solution by slow pipetting.
- Transfer 2 mL of the coating substrate solution (step 1.1.2) per well of a 6-well tissue culture plate.
- Immediately place the coated tissue culture plate in the cell culture incubator at 37 °C and 5% CO2 for at least 7 h and use within 7 days.
- hiPSC-CM culture medium
- Add 10 mL of CM media supplement thawed at 4 °C to 500 mL of CM base medium. Store at 4 °C for up to 2 weeks.
- Aliquot required amount of media for the day and bring to room temperature before use.
- hiPSC-CM thawing medium: Prepare fresh by mixing hiPSC-CM culture medium (step 1.2) with 10% fetal bovine serum (FBS). Bring the media to room temperature prior to thawing of CMs for suspension.
- Fibronectin 1 mg/mL stock solution: Aliquot 200 µL into 1.5 mL sterile microfuge tubes and stored at 4 °C for later use. Prepare working solution of 50 µg/mL concentration freshly on ice.
- Multiwell cleaning solution: Combine 0.5 g of enzymatic detergent with 50 mL of sterile double distilled water (ddH2O). Vortex to mix the contents. Filter and store at 4 °C for up to a week.
2. Pre-plating of cryopreserved hiPSC-CM for maturation (Figure 1)
NOTE: This section is intended for thawing and culturing hiPSC-CMs that were differentiated using the feeder-free monolayer method3,16 and cryopreserved in liquid nitrogen 10 days post-differentiation at 1-2 million cells/vial. Cells from one vial are plated into two substrate-coated wells of a 6-well tissue culture plate. Cardiomyocytes tend to settle at the bottom of the tube so gentle mixing at the time of pre-plating is important for achieving even cell density across wells.
- Aliquot 2 mL of FBS per vial of hiPSC-CMs being thawed into a 15 mL conical tube and bring to room temperature.
- Thaw vials of cryopreserved hiPSC-CMs by placing them in a 37 °C water bath and swirl gently for even thawing for no more than 3 min.
- Immediately transfer the vial contents to the tube containing FBS (see step 2.1), mix by swirling and centrifuge at 200 x g for 5 min.
- Aspirate the supernatant and resuspend the cell pellet in 1 mL of hiPSC-CM thawing medium (see step 1.3) per vial thawed. Use a transfer pipette to resuspend the pellet by gentle trituration. Assess cell viability.
- Add additional 3 mL of thawing medium per vial of hiPSC-CM thawed in step 2.4 and suspend gently using a transfer pipette to further dissociate cell clumps.
- Dispense 2 mL of cell suspension gently into each well of substrate-coated 6-well plates (see step 1.1). Place in cell culture incubator at 37 °C and 5% CO2.
- Replace with fresh hiPSC-CM culturing medium after 24 h and 3 times weekly thereafter for 20 days.
NOTE: Cells should adhere to the substrate coating by 24 h and beat spontaneously at 48 h post-plating (See Video 1 and Video 2).
3. Multiwell MEA plate sterilization and coating (Figure 2 and Figure 3)
NOTE: The protocol described here is for preparing 24-well MEA plates with 12 micro gold PEDOT-coated electrodes on glass for hiPSC-CM plating. Avoid touching the bottom of the plate as this may damage the electrodes.
- Two days prior to cell plating, add 0.5 mL of hiPSC-CM culture medium (see step 1.2) to each well and perform a baseline recording to verify signal-to-noise ratio for quality check of the MEAs.
- Aspirate the media, rinse with sterile ddH2O and sterilize under UV light inside a laminar flow hood overnight.
- The day prior to cell plating, add 0.1 mL of FBS to each well for hydrophilic treatment of MEA surfaces. Incubate for 30 min at room temperature. This step is necessary for cell attachment.
- Aspirate the FBS and rinse with 0.5 mL of sterile ddH2O per well. Repeat one more time.
- Leave the plate to dry in the laminar flow hood overnight.
- Prepare a working dilution of 50 µg/mL fibronectin in cold DMEM/F12 medium from the stock (see step 1.4). Keep the solution on ice.
- Pipette 5 µL of the working fibronectin dilution (see 3.6) and carefully dispense the droplet to the center of each well to cover all 12 electrodes. It is important to work swiftly across the 24 wells to prevent the droplet from drying.
- Immediately place the fibronectin-coated multiwell MEA plate on a raised surface inside a humidifying chamber containing sterile ddH2O to cover the entire dish surface. Place the chamber with the multiwell MEA plate for 3 h in the cell culture incubator.
NOTE: Placing the 24 well MEA plate in a humidifying chamber is critical to prevent fibronectin droplets from drying out during the incubation period.
4. hiPSC-CM dissociation and plating on Multiwell MEA plate (Figure 3)
NOTE: Start this step about 1 h prior to MEA fibronectin incubation is complete. Ensure that the cell dissociation solution is at 37 °C and the iPSC-CM thawing medium is at room temperature. Dissociation methods have been optimized for 30 days post-differentiated hiPSC-CMs cultured on substrate-coated 6-well plates (see step 2) to obtain about 90% viable CMs for MEA plating. Care should be taken not to introduce air bubbles while trituration to prevent cell death.
- Aspirate the culture medium from each well of the 6-well tissue culture dish with 30 day post-differentiated hiPSC-CM culture (see step 2.7) and wash with 2 mL of sterile D-PBS per well.
- Add 1 mL of pre-warmed cell dissociation solution (see Table of Materials) per well and incubate for 4 min at 37 °C. Use a transfer pipette to gently triturate to loosen cells for dissociation. If most of the cells are still adherent, incubate for another 3 min at 37 °C and triturate again. This additional incubation should help for maximal cell recovery. Do not incubate for more than 7 min as this might result in low cell viability.
- Using a transfer pipette, pool all the dissociated cells into a conical tube containing hiPSC-CM thawing medium (see step 1.3). It is recommended to suspend the cells in at least twice the volume of media (2 mL per well being harvested) to block the activity of the cell dissociation solution.
- Centrifuge at 200 x g for 5 min.
- Aspirate the solution carefully as the pellet is loosely attached to the surface and resuspend in 0.1 mL of hiPSC-CM thawing medium. Use a transfer pipette to resuspend the pellet a few times by gentle trituration.
- Aliquot 2 µL of dissociated cells and dilute with 18 µL of media in a 1.5 mL microfuge tube. Add 20 µL of Trypan Blue and mix for cell count and viability assessment.
- Adjust the cell density to 6,000 cells/µL by adding the appropriate volume of hiPSC-CM thawing medium. Suspend the pellet by gentle flicking a few times. The hiPSC-CMs easily detach from glass surfaces especially when plated at high densities. We have optimized the seeding density and plating conditions to achieve 15 days of electrical recordings.
- When ready, bring the multiwell MEA plate into the laminar flow hood for cell seeding. It is critical to perform the following two steps one well at a time to prevent the fibronectin from drying.
- Carefully remove the fibronectin droplet using a P10 pipette without touching the electrodes.
- Immediately dispense a 5 µL cell droplet (30,000 cells) to the center of the well of the MEA plate covering all 12 electrodes. Repeat the step until all the 24 wells are plated. Intermittent mixing of cell suspension by flicking is recommended.
- Place the multiwell MEA plate back in the loosely covered humidifying chamber and return to the cell culture incubator at 37 °C and 5% CO2 for 3 h for cell attachment.
- Carefully add 200 µL of hiPSC-CM thawing medium to each well without disturbing the cells using a P200 pipette. Add the media dropwise to the side of the well.
- Place the multiwell MEA plate back in the cell culture incubator.
- Replace with fresh hiPSC-CM culture medium at 24 h post plating. Spontaneous beating of cardiomyocytes can be observed at this point (See Video 3).
- Change the media every 2 days until the end of experiments.
5. hiPSC-CM electroporation and signal acquisition (Figures 4 – 6)
NOTE: This protocol is for simultaneous recording of high-throughput electrode signals (12 sites for each of the 24 wells). The 24-well multiwell MEA system is used with the acquisition software (see Table of Materials). All MEA recordings are conducted at 37 °C.
- Turn on the interface board and initiate the acquisition software. Allow enough time for the multiwell MEA headstage to reach 37 °C temperature (see arrow no. 1 in Figure 4).
- Insert multiwell MEA plate from step 4 into the multiwell MEA headstage by placing it covered over the recording platform and click the Insert button (see arrow no. 2 in Figure 4). Allow for temperature to stabilize before starting recordings.
- Adjust acquisition and electroporation settings
- Click on the Define Experimental Flow icon (see arrow no. 3 in Figure 4) and set recording time to 2 min or as desired.
- Click on the Data Acquisition Setup icon (see arrow no. 4 in Figure 4) and set sampling rate to 20 kHz, high-pass filter to 0.1 Hz and low-pass filter to 3500 Hz.
- Click on the Stimulator Settings icon (see Figure 5). Under the Stimulus Definition tab, define stimulation as biphasic symmetric voltage pulses of 1 mV, 1 ms and 1 Hz. Under the Stimulation Electrodes tab, select electroporation sites by highlighting all relevant electrodes.
- Click the Explore button to visualize signals in all wells. Verify signal quality and steady state conditions. Take notes on electrodes with FP signals in the mV range. Click the same button to stop the exploration. No data has been recorded up to this point.
- Start recording by clicking on the Go! button. The electrodes in each well will show FP signals in the raw data window (Figure 6). After 30 s of recording, click on the Stimulate button and allow electroporation to take place on the selected sites for 30 s; then, click on the same button to stop stimulation and continue recording for the remaining 60 s.
NOTE: The recorded file can be played back by switching to ‘Replayer Mode’ in the ‘Application’ dropdown menu.
6. Multiwell MEA plate cleaning for reuse
- After final experiments, clean all the wells in the multiwell plate by aspirating all media contents from each well and carefully avoiding touching the electrode surface.
- Add 1 mL of sterile ddH2O per well. Aspirate and repeat once.
- Add 0.3 mL of multiwell cleaning solution (see step 1.5) per well. Incubate overnight at room temperature to dislodge cells and debris.
- The following morning, aspirate the solution and rinse with 1 mL of sterile ddH2O. Incubate for 5 – 7 min and aspirate. Repeat 5 times.
- Add 0.5 mL of sterile ddH2O per well. Record the baseline of the cleaned multiwell plate for quality check of the cleaned MEAs (Figure 7).
- Store at 4 °C until ready to use.
7. Data file conversion and export
NOTE: Four data files will be generated for every recording: MWR, MWC, MWD, and MWS files. Using the converter software, the MWD file can be converted to H5 file for subsequent analysis using custom-built script (see Supplementary File 1).
- Initiate the converter software (see Table of Materials).
- Select Set Input Path from the File menu. Select folder containing data files of interest.
- Select Set Output Path from the File menu. Select folder where converted files are to be saved.
- Highlight MWD file of interest.
- Click the EXPORT to HDF5 button.
8. Data segmentation and analysis (Figures 8-10)
NOTE: Matlab-based custom software is used to segment and extract various FP and AP data parameters. Software is available on demand.
- Run the Waveform Analysis code using Matlab (see Figure 8 for a view of the GUI’s main window).
- Click on File and select Process .h5.
- Find and select the mwd.h5 file created according to step 7 above.
- Click on the Save Directory button to change the storage location of the output files.
- Create a signal processing queue by selecting electrode/well combinations of interest and then clicking on the Queue button. Repeat this step to append more electrode/well combinations to be processed to the queue.
- Edit the queue by clicking directly on Med Name / Med Concentration if cells were treated with drugs (Figure 8).
- Once the queue is final, click the Initialize Waveforms button. This will start the preliminary processing in which signals are identified and extracted for segmentation.
- Click on the Zoom In button and select the action potential area of interest with the cursor (Figure 9).
- Click the Keep button and review the panels. Peaks (red ‘x’) and troughs (yellow circles) are detected for every waveform and the normalized action potentials are superimposed. Click the Keep button and move on to the next trace in the queue (Figure 10).
- Repeat steps 8.8-8.9 for the rest of the electrode/well combination signals in the queue.
NOTE: A .csv file will be generated with APD parameters measured for every waveform. A .mat file for each .h5 file is also saved to allow additional processing of segmented data.
Representative Results
The viability and plating density of post-thawed hiPSC-CMs is critical for multiwell MEA culture. Pre-plating of 1-2 million hiPSC-CMs/vial into two wells of a 6-well tissue culture plate with 50% or greater viability will produce a healthy monolayer culture with spontaneous beating at 48 h. Poor viability of CMs will result in cultures with a high percentage of non-myocyte populations. These monolayers when dissociated for multiwell MEA plating generally produce inconsistent results and bad quality signals and therefore should be discarded. Figure 1 shows examples of optimal vs. sub-optimal hiPSC-CMs cultures at 48 h post plating. Thawing the CMs on substrate-coated tissue culture plates rather than directly on multiwell MEAs, allows for cell recovery and maturation3. Direct plating of cryopreserved CMs on the array is not recommended as it produced inconsistent results.
In addition to the quality of the dissociated CMs, cell attachment on multiwell MEA is highly dependent on cell density and the fibronectin coating technique. The fibronectin droplet size is critical as the CMs will conform to the boundaries of the fibronectin-coated area. For this reason, only 5 µL of the fibronectin solution are dispensed directly over the electrode array area. To ensure that the droplet does not disperse, the well surface must be completely dry at the time of coating. Figure 2 shows the layout of the multiwell MEA plate with schematics of step-by-step pretreatment for optimal preparation. Additionally, to prevent the fibronectin from drying the multiwell MEA plates must be placed inside a humidifying chamber during the incubation period lasting no more than 3 h (see step 3.8). Once the incubation period is complete, it is important to remove the fibronectin droplet from each well just before CM plating and only then proceed to next well plating. Working swiftly and carefully dispensing of the CMs is the key to successful cell attachment.
hiPSC-CM cultures at 30 days post-differentiation are dissociated for multiwell MEA plating using the enzymatic cell dissociation method (see step 4). CMs will attach to the fibronectin-coated MEA surfaces by 3 h and a monolayer covering the arrays will be visible after 24 h post-plating (Figure 3). Synchronous beating of the monolayer will be observed at 24-48 h. Cell droplet dispersion will affect the culture density or even lead to drying and cell death. Precise cell placement directly on the array is of utmost importance and therefore the technique must be practiced for optimal plating. Cell adhesion to the reference electrode will hinder electrical signal production. See Figure 3 for images of optimal CM placement, and culture after 24 h.
The CMs cultured on multiwell MEAs are subjected to quality check for electrical activity at 48 h post-plating. Typically, FP signal amplitude increases from the µV range to mV in approximately 4 days3. If 50% of the electrodes within a network and 70% of the total networks do not produce FP signals, then the network or the culture are suboptimal and should be discarded. Only cultures that pass the quality check are processed for FP and AP analysis. Figure 6 shows examples of good and sub-standard FP signals.
Electroporation-mediated AP recordings can be obtained multiple times from cultures 48 h post-MEA plating. Employing electroporation, we gained intracellular access to record high-resolution APs from multiple hiPSC-derived cardiomyocyte networks. Low-voltage pulses (1 V, 1 ms, 1 Hz) for 30 s were delivered for transient, reversible transformation of FP to AP. The electroporation allows successful intracellular access for AP measurement in approximately 75% of the electrodes. Electrical signals are recorded for 2 min that include 30 s pre-electroporation, 30 s during and 1 min post-electroporation. A train of 10 s AP waveforms 10 s post-electroporation are evaluated across all sites for signal quality and analysis. Any trace not conforming to pure AP signal are discarded. To investigate if AP amplitudes correlate to FP signal we electroporated all 288 sites to simultaneously record waveforms. Representative FP and AP signals recorded from the same cell site from two different electrodes are shown in Figure 11A. We observed no correlation between FP amplitudes and post electroporation AP amplitudes recorded from the same cell site. Additionally, multiple electroporations of the same cell site at 0, 24, 48, 72 and 96 h had no significant effect on the AP shape over time (Figure 11B).
Given the high-throughput nature of the system, a manual technique to extract and quantify parameters of interest such as RR interval, instantaneous frequency and differential action potential duration is inefficient and time consuming. A custom-built MATLAB script available to the research community upon request is employed to perform waveform measurements with 1 µs resolution. Electroporation time points are overlaid with the extracted signal to identify 10 s of AP post-electroporation to conduct signal extraction, quality assurance, and segmentation workflow (Figure 8, Figure 9, Figure 10). The user interface allows for selection of the desired segment using the overlaid electroporation indicators as a guide. The segmented waveform is processed by subroutines to further identify individual AP waveforms. This is completed through peak detection, where the highest and lowest voltage is identified for each cycle. Once this process is completed, the amplitudes are normalized, and the associating time vectors are shifted to define time zero at a peak value of 1. Interpolation of intersection points along the individual cycles was used to determine APD measurements. Thus, partial automation workflow for AP waveform segmentation allows efficient data analysis for various APD parameters across multiple batches of cultures in a short period of time. Further automation of inclusion and exclusion criteria for FPs and APs is ongoing for real-time data analysis.
A significant advantage of the multiwell MEA plate is that it can be reused multiple times. This restoration enables repetitive electrophysiological studies for cost-effective and consistent data collection. Recordings of APs from the same array after 6 restorations are shown in Figure 12. Signal-to-noise ratio is similar across multiple reuses. To demonstrate the reliability of the array for repetitive electrophysiological studies, a total of 3815 AP waveforms are pooled from three restoration batches and AP duration data is extracted to examine the repeatability of the results. Distribution plots for individual waveform APD30, APD80, triangulation (APD80—APD30) and fractional shortening ((APD80—APD30)/(APD80)) are displayed (Figure 13).
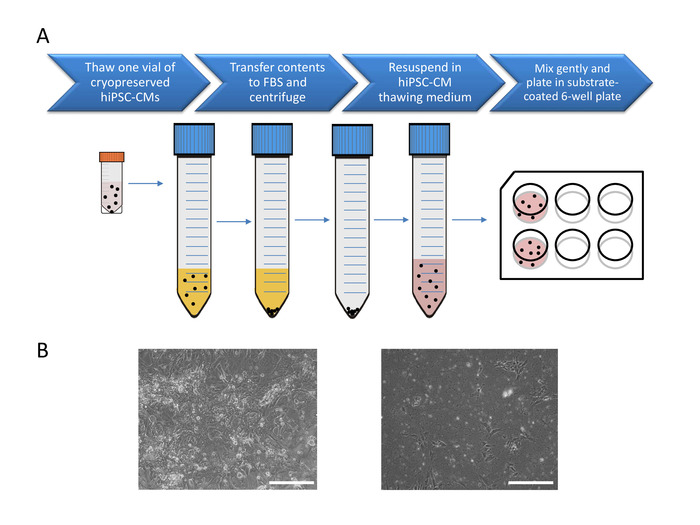
Figure 1: Pre-plating of cryopreserved hiPSC-CM for maturation. (A) Cell processing for pre-plating 1 vial of 10 days post-differentiation cryopreserved hiPSC-CMs. (B) Phase contrast images of successful (left) and unsuccessful (right) hiPSC cultures. Scale bar: 275 µm. See Video 1 and Video 2 for successful 14 and 24 days post-differentiation culture examples. Please click here to view a larger version of this figure.
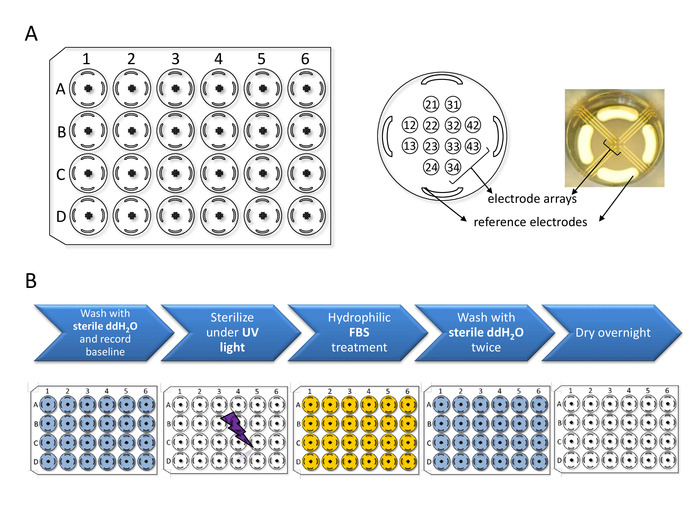
Figure 2: Multiwell MEA plate setup and preparation. (A) Multiwell MEA plate schematics: The plate consists of 24 wells (A1 through D6) each containing 12 microelectrode arrays and 4 peripheral reference electrodes. Electrode diameter: 30 µm / Inter-electrode distance: 300 µm. Recordings can be obtained from the 288 electrodes simultaneously. (B) Sterilization and hydrophilic treatment steps to be conducted prior to hiPSC-CM plating. Please click here to view a larger version of this figure.

Figure 3: hiPSC-CM dissociation and plating on Multiwell MEA plate. (A) Schematics of hiPSC-CM MEA plating steps for each well. (B) Microscopic image illustrating correct cell droplet placement covering all 12 electrodes without spreading to the 4 reference electrodes. (C) Phase contrast microscopic images of an exemplary (left) and suboptimal (right) hiPSC-CM platting on MEA at 24 h post-plating. Scale bar= 275 µm. See Video 3 for successful MEA plating example. Please click here to view a larger version of this figure.
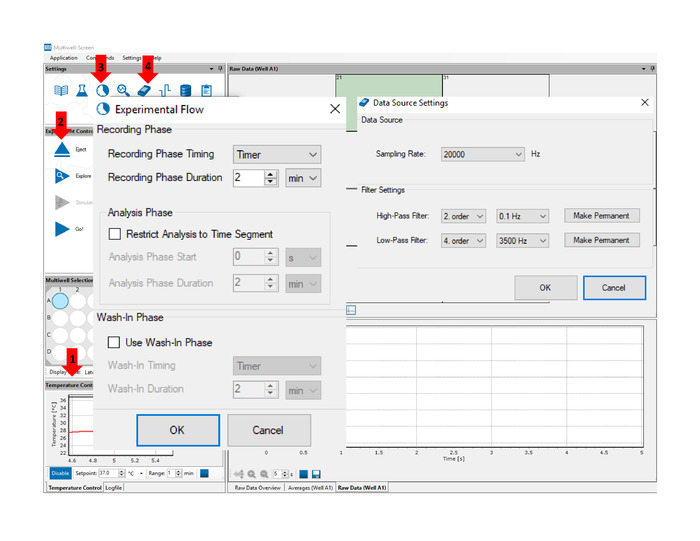
Figure 4: Multiwell-screen acquisition software. Arrows indicate the location of key features and functions referenced in the text: Temperature Control (1) panel allows for real time temperature monitoring throughout the experiment. Insert/Eject (2) button engage and release the Multiwell MEA Plate. Define Experimental Flow (3) function allows the user to set the duration of the recording. Data Acquisition Setup (4) function allows the user to set the sampling rate and acquisition filter settings. Please click here to view a larger version of this figure.
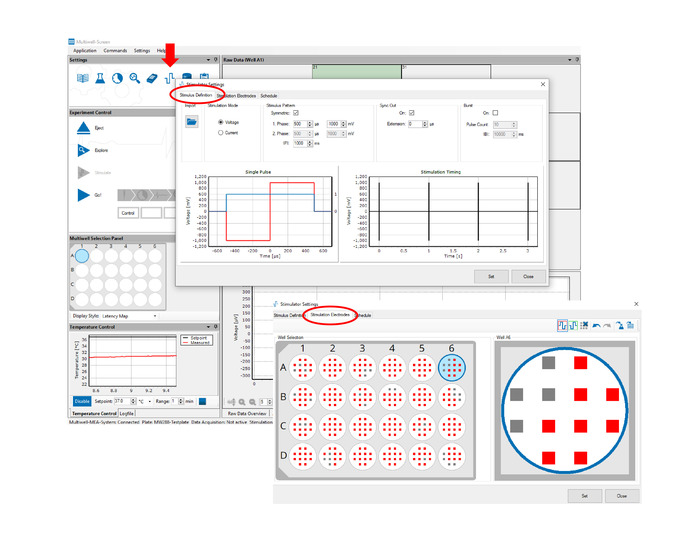
Figure 5: hiPSC-CM electroporation and signal acquisition. Stimulus Definition tab allows the user to define the electroporating pulse parameters. Stimulation Electrodes tab allows the user to select the electroporating electrodes. Any combination of the 288 electrodes can be selected. Please click here to view a larger version of this figure.
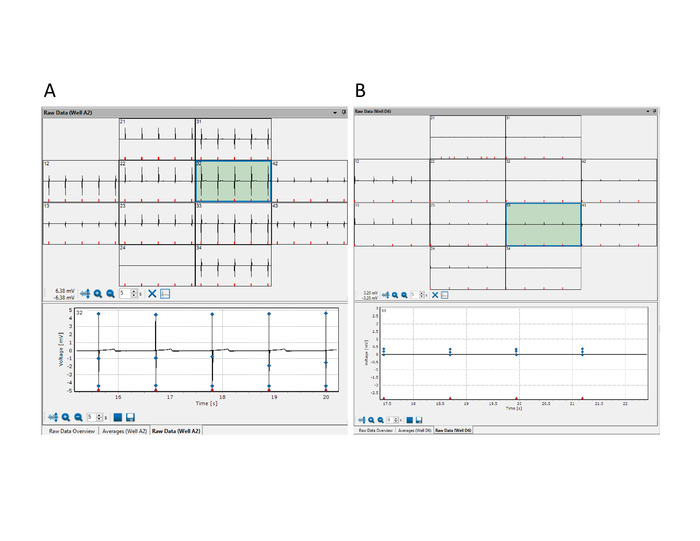
Figure 6: Quality check of Multiwell MEAs for electrical activity. Multiwell–Screen acquisition software showing raw data windows with representative examples of optimal (A) and sub-standard (B) FP signals. Please click here to view a larger version of this figure.
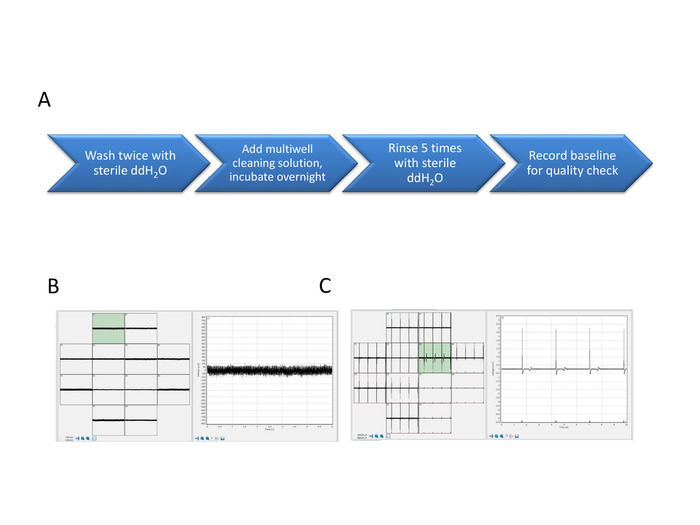
Figure 7: FP and AP signals from the new and restored array. Multiwell MEA enzymatic cleaning steps (A). The baseline signal of the new array shows minimal signal to noise ratio (B) and FP signals show the electrical activity of the network (C). Please click here to view a larger version of this figure.
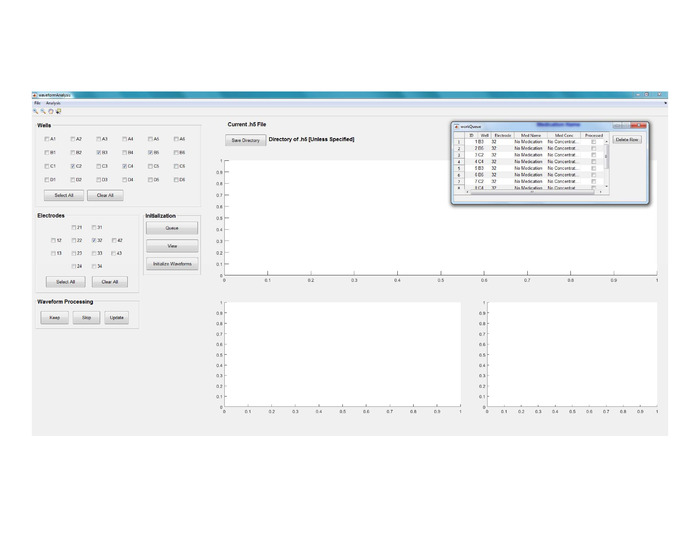
Figure 8: Data segmentation and analysis. View of GUI’s main window for Waveform Analysis. Please click here to view a larger version of this figure.
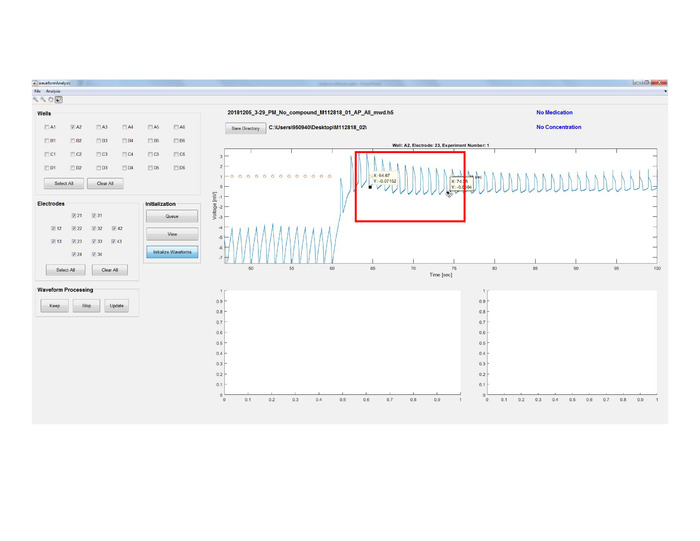
Figure 9: Data segmentation and analysis. Initialize Waveforms button to identify and extract AP waveforms for segmentation and to start the preliminary processing by zooming in and selecting the action potential area of interest. Red circles are the electroporation indicators. Please click here to view a larger version of this figure.
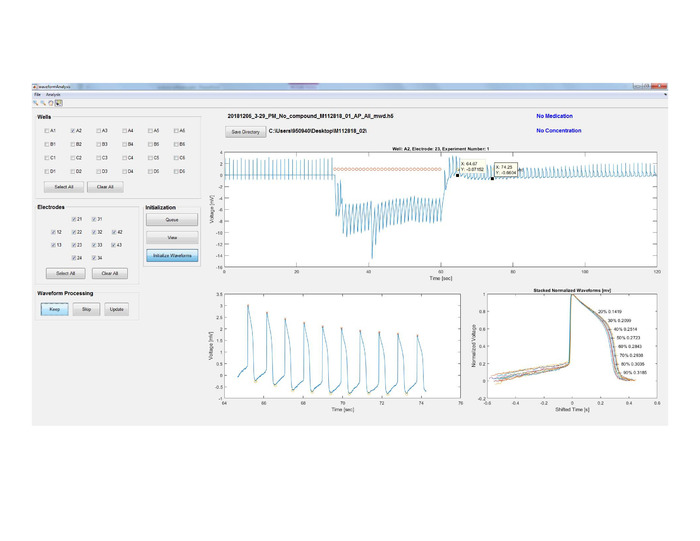
Figure 10: Data segmentation and analysis. Peaks (red ‘x’) and troughs (yellow circles) are detected for every waveform and the normalized APs are superimposed for a quality check of the waveforms. Please click here to view a larger version of this figure.
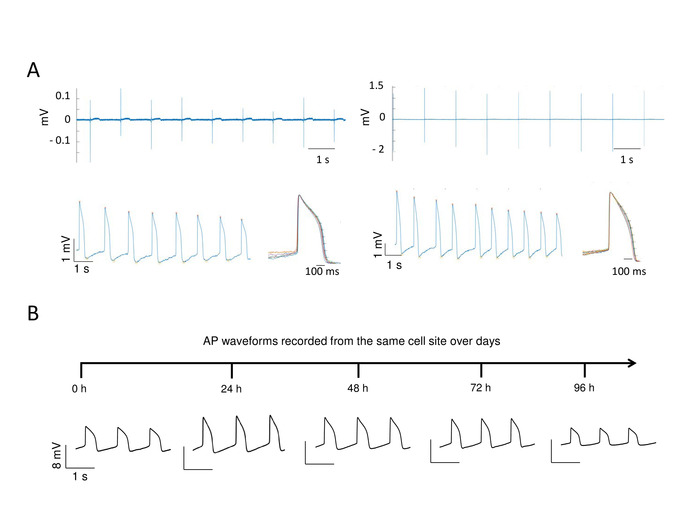
Figure 11: AP Amplitude dependence on the FP signal for multiple recordings from the same cell site. FP amplitude in µV ranges (A, top left panel) or mV ranges (A, top right panel) recorded from two independent electrodes produce AP amplitude in mV range (A, bottom left and right panels) showing no correlation between FP amplitudes and post-electroporation AP amplitudes. The normalized AP waveforms for each recording are superimposed as shown for each recording. Multiple electroporations of the same cell site at 0 to 96 h produced high quality AP waveforms allowing tracking of membrane electrodynamics (B). Please click here to view a larger version of this figure.
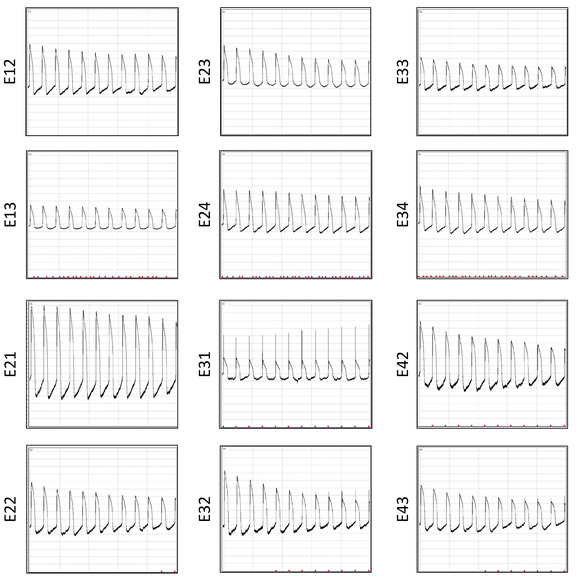
Figure 12: AP recordings after six restorations. AP waveforms recorded simultaneously 10 s post-electroporation across 12 electrodes from the same well are displayed. Please click here to view a larger version of this figure.
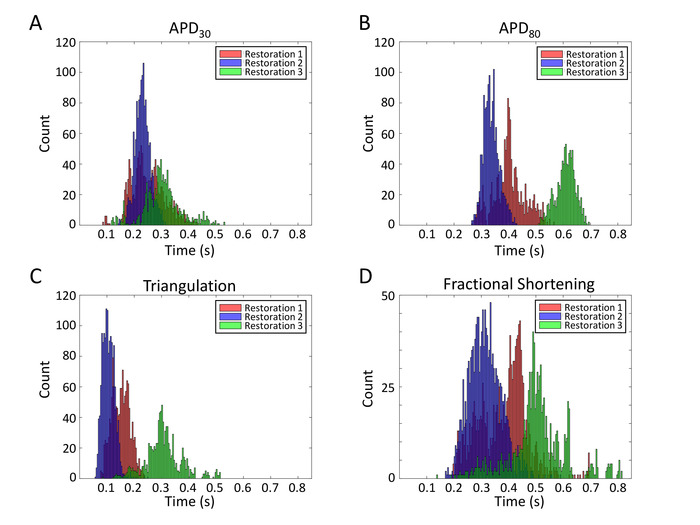
Figure 13: APD parameter histograms from multiple restorations. Distribution plots for individual waveform APD30 (A), APD80 (B), triangulation (APD80—APD30) (C) and fractional shortening ((APD80—APD30)/(APD80)) (D) are displayed. Please click here to view a larger version of this figure.
Supplementary files. Videos 1-3. Please click here to download this file.
Discussion
Over the years, the application of MEAs has been limited to carrying out FP measurements of excitable cells to study their electrophysiological properties36,37,38,39. Only a few groups have reported AP traces from electrogenic cells using custom MEA based technology18,29,30. However, these approaches have not been investigated for repeated recordings from the same preparations. We developed an innovative and accurate methodology for studying APs from the same cell site over days in multiple hiPSC-CM networks simultaneously3. In our published study, a multiwell micro-gold MEA platform was employed to generate AP waveform libraries from multiple batches of hiPSC-CM cultures with high precision and with a temporal resolution of 1 µs. The protocol described here explains seeding of hiPSC-CMs on the array for efficient development of syncytial CM networks for high-throughput AP recordings. Several critical steps in the protocol are: 1) production of multiple high-purity batches of quality-controlled CMs for cryopreservation banking, 2) highly viable post-thaw CMs for pre-plating and maturation, 3) treatment of the multiwell MEA plate for CM seeding, 4) hiPSC-CM culture dissociation at 30 days post-differentiation for MEA plating, and 5) restoration of the MEAs for multiple reuse.
It is important to note that batch-to-batch variation in hiPSC differentiation might affect experimental outcomes. The monolayer method of differentiation was optimized in-house for high percent cardiomyocyte production3,40. The FACS analysis of MLC2v and TNNT2 markers of our cultures demonstrate a ≥90% ventricular-like phenotype3. These quality-controlled cultures are cryopreserved for experimental studies. The current differentiation approaches yield a heterogeneous mix of nodal-, atrial- and ventricle-like cells3,16,17,41. Therefore, strategies employed for CM subtype population enrichment can further improve the specificity of the cultures. Additionally, tissue engineering approaches can be employed to enhance their maturation. The methods proposed here can easily be implemented for other CM sources.
The AP waveforms recorded using MEA were similar to those recorded from networks of cardiomyocytes by optical mapping42,43, complementary metal oxide semiconductor-based MEA18,21, and simulated AP using FP recordings20. To address the mechanism of AP measurements via MEA Hai and Spira25 demonstrated that electropore-electrode interface mimic the established sharp glass microelectrode technique. However, the resting membrane potential and true amplitude values in our study cannot be established given that the electropore-electrode interface in MEA systems is not calibrated, and that the amplitude is a function of the sensitivity and resolution of the technique. Our approach shares similar limitations to optical mapping when it comes to AP amplitude.
The multiwell MEA-based FP/AP readouts reported here open new possibilities for drug safety assessment. Although spontaneous, these hiPSC-CM monolayers beat at constant rates. Analysis of APD parameters across multiple networks provide insight on electrical heterogeneity (Figure 13). However,comprehensive APD restitution analyses must incorporate preceding diastolic intervals. Furthermore, high-quality AP waveforms recorded from the same cell site over 96 h (Figure 11B) is the first report to track membrane electrodynamics over time which will be of value in development and in disease.
The protocol described here for quantifying AP parameters can be used to generate dose-response curves to test compounds. As recently reported by Edwards et al.3, dose response of norepinephrine, isoproterenol and E 4031 are plotted for APD at various repolarization phases. The published study demonstrated the accuracy and reliability of the approach for identification of the dose-dependent subtle changes in the AP waveforms in real time. This technique could easily be extended for other compounds or small molecule libraries for understanding various electrophysiological responses.
The MEA based approach for AP measurements presented in this study will be of interest not only to electrophysiologists but also to cell biologists and in-silico modelers. Furthermore, FP/AP recordings from the same cell site on hiPSC-CMs will enable researchers to generate bioelectric data libraries of wide array of excitable cellular networks within a short period of time. Availability of these resources will be valuable for drug discoveries and disease modeling.
Disclosures
The authors have nothing to disclose.
Acknowledgements
None
Materials
| Accutase | Sigma Aldrich | A6964-100ML | cell dissociation solution |
| Acquisition software | Multichannel Systems | Multiwell-Screen v 1.9.2.0 | |
| B27 Supplement | ThermoFisher | 17504-044 | CM media supplement |
| Converter software | Multichannel Systems | MultiChannel DataManager | |
| DMEM/F12 | ThermoFisher | 11330-032 | |
| D-PBS | ThermoFisher | 14190-250 | |
| FBS | Fisher Scientific | SH3007103HI | |
| Fibronectin | Sigma Aldrich | F1141-5MG | |
| Geltrex | ThermoFisher | A1413202 | coating substrate |
| Interface board | Multichannel Systems | MCS-IFB 3.0 Multiboot Interface Board | |
| Multiwell MEA Plate | Multichannel Systems | 24W300/30G-288 | |
| RPMI 1640 | ThermoFisher | 11875-093 | CM base medium |
| Terg-a-zyme | Sigma Aldrich | Z273287-1EA | enzymatic detergent |
| Transfer pipettes, individually wrapped | Fisher Scientific | 1371148 | |
| Trypan Blue | Sigma Aldrich | T8154-100ML | |
| Ultrapure sterile water | ThermoFisher | 10977-023 | |
| 6-well tissue-culture treated plates | Fisher Scientific | 08-772-1B |
References
- Dambrot, C., Passier, R., Atsma, D., Mummery, C. L. Cardiomyocyte differentiation of pluripotent stem cells and their use as cardiac disease models. Biochemical Journal. 434 (1), 25-35 (2011).
- Dunn, K. K., Palecek, S. P. Engineering Scalable Manufacturing of High-Quality Stem Cell-Derived Cardiomyocytes for Cardiac Tissue Repair. Frontiers in medicine. 5, (2018).
- Edwards, S. L., et al. A Multiwell Cardiac muGMEA Platform for Action Potential Recordings from Human iPSC-Derived Cardiomyocyte Constructs. Stem Cell Reports. 11 (2), 522-536 (2018).
- Herron, T. J., Lee, P., Jalife, J. Optical imaging of voltage and calcium in cardiac cells & tissues. Circulation Research. 110 (4), 609-623 (2012).
- Huebsch, N., et al. Miniaturized iPS-Cell-Derived Cardiac Muscles for Physiologically Relevant Drug Response Analyses. Scientific Reports. 6, 24726 (2016).
- Lundy, S. D., Zhu, W. Z., Regnier, M., Laflamme, M. A. Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cells and Development. 22 (14), 1991-2002 (2013).
- Ma, J., et al. High purity human-induced pluripotent stem cell-derived cardiomyocytes: electrophysiological properties of action potentials and ionic currents. American Journal of Physiology-Heart and Circulatory Physiology. 301 (5), H2006-H2017 (2011).
- Sharma, A., et al. Use of human induced pluripotent stem cell-derived cardiomyocytes to assess drug cardiotoxicity. Nature Protocols. , (2018).
- Zhang, D., et al. Tissue-engineered cardiac patch for advanced functional maturation of human ESC-derived cardiomyocytes. Biomaterials. 34 (23), 5813-5820 (2013).
- Zhang, J., et al. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method. Circulation Research. 111 (9), 1125-1136 (2012).
- Burridge, P. W., Holmstrom, A., Wu, J. C. Chemically Defined Culture and Cardiomyocyte Differentiation of Human Pluripotent Stem Cells. Current Protocols in Human Genetics. 87, (2015).
- Burridge, P. W., et al. Chemically defined generation of human cardiomyocytes. Nature Methods. 11 (8), 855-860 (2014).
- Burridge, P. W., Zambidis, E. T. Highly efficient directed differentiation of human induced pluripotent stem cells into cardiomyocytes. Methods in Molecular Biology. , 149-161 (2013).
- Laflamme, M. A., et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nature Biotechnology. 25 (9), 1015-1024 (2007).
- Sharma, A., et al. Derivation of highly purified cardiomyocytes from human induced pluripotent stem cells using small molecule-modulated differentiation and subsequent glucose starvation. Journal of Visualized Experiments. (97), (2015).
- Bhattacharya, S., et al. High efficiency differentiation of human pluripotent stem cells to cardiomyocytes and characterization by flow cytometry. Journal of Visualized Experiments. (91), (2014).
- Zhang, J., et al. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circulation Research. 104 (4), e30-e41 (2009).
- Jans, D., et al. Action potential-based MEA platform for in vitro screening of drug-induced cardiotoxicity using human iPSCs and rat neonatal myocytes. Journal of Pharmacological and Toxicological Methods. 87, 48-52 (2017).
- Strauss, D. G., Blinova, K. Clinical Trials in a Dish. Trends in Pharmacological Science. 38 (1), 4-7 (2017).
- Tertoolen, L. G. J., Braam, S. R., van Meer, B. J., Passier, R., Mummery, C. L. Interpretation of field potentials measured on a multi electrode array in pharmacological toxicity screening on primary and human pluripotent stem cell-derived cardiomyocytes. Biochemical and Biophysical Research Communications. 497 (4), 1135-1141 (2018).
- Braeken, D., et al. Open-cell recording of action potentials using active electrode arrays. Lab Chip. 12 (21), 4397-4402 (2012).
- Otsuji, T. G., et al. Progressive maturation in contracting cardiomyocytes derived from human embryonic stem cells: Qualitative effects on electrophysiological responses to drugs. Stem Cell Research. 4 (3), 201-213 (2010).
- Shinozawa, T., Imahashi, K., Sawada, H., Furukawa, H., Takami, K. Determination of appropriate stage of human-induced pluripotent stem cell-derived cardiomyocytes for drug screening and pharmacological evaluation in vitro. Journal of Biomolecular Screening. 17 (9), 1192-1203 (2012).
- Raphel, F., et al. Identification of Ion Currents Components Generating Field Potential Recorded in MEA From hiPSC-CM. IEEE Transactions on Biomedical Engineering. 65 (6), 1311-1319 (2018).
- Hai, A., Spira, M. E. On-chip electroporation, membrane repair dynamics and transient in-cell recordings by arrays of gold mushroom-shaped microelectrodes. Lab Chip. 12 (16), 2865-2873 (2012).
- Xie, C., Lin, Z., Hanson, L., Cui, Y., Cui, B. Intracellular recording of action potentials by nanopillar electroporation. Nature Nanotechnology. 7 (3), 185-190 (2012).
- Cohen, A., Shappir, J., Yitzchaik, S., Spira, M. E. Reversible transition of extracellular field potential recordings to intracellular recordings of action potentials generated by neurons grown on transistors. Biosensors and Bioelectronics. 23 (6), 811-819 (2008).
- Ojovan, S. M., et al. A feasibility study of multi-site,intracellular recordings from mammalian neurons by extracellular gold mushroom-shaped microelectrodes. Scientific Reports. 5, 14100 (2015).
- Shmoel, N., et al. Multisite electrophysiological recordings by self-assembled loose-patch-like junctions between cultured hippocampal neurons and mushroom-shaped microelectrodes. Scientific Reports. 6, 27110 (2016).
- Lin, Z. C., Xie, C., Osakada, Y., Cui, Y., Cui, B. Iridium oxide nanotube electrodes for sensitive and prolonged intracellular measurement of action potentials. Nature Communications. 5, 3206 (2014).
- Stett, A., Burkhardt, C., Weber, U., van Stiphout, P., Knott, T. CYTOCENTERING: a novel technique enabling automated cell-by-cell patch clamping with the CYTOPATCH chip. Receptors Channels. 9 (1), 59-66 (2003).
- Kanda, Y., Yamazaki, D., Osada, T., Yoshinaga, T., Sawada, K. Development of torsadogenic risk assessment using human induced pluripotent stem cell-derived cardiomyocytes: Japan iPS Cardiac Safety Assessment (JiCSA) update. Journal of Pharmacological Sciences. 138 (4), 233-239 (2018).
- Yang, X., Papoian, T. Moving beyond the comprehensive in vitro proarrhythmia assay: Use of human-induced pluripotent stem cell-derived cardiomyocytes to assess contractile effects associated with drug-induced structural cardiotoxicity. Journal of Applied Toxicology. 38 (9), 1166-1176 (2018).
- Sala, L., Bellin, M., Mummery, C. L. Integrating cardiomyocytes from human pluripotent stem cells in safety pharmacology: has the time come. British Journal of Pharmacology. 174 (21), 3749-3765 (2017).
- Zlochiver, V., Edwards, S., Joshi-Mukherjee, R. Longitudinal Cardiotoxic Effect of Doxorubicin in a Multicellular Cardiac Model. Biophysical Journal. 116 (3), (2019).
- Del Alamo, J. C., et al. High throughput physiological screening of iPSC-derived cardiomyocytes for drug development. Biochimica et Biophysica Acta. 1863 (7 Pt B), 1717-1727 (2016).
- Clements, M. Multielectrode Array (MEA) Assay for Profiling Electrophysiological Drug Effects in Human Stem Cell-Derived Cardiomyocytes. Current Protocols in Toxicology. 68, 21-22 (2016).
- Navarrete, E. G., et al. Screening drug-induced arrhythmia [corrected] using human induced pluripotent stem cell-derived cardiomyocytes and low-impedance microelectrode arrays. Circulation. 128 (11 Suppl 1), S3-S13 (2013).
- Harris, K., et al. Comparison of electrophysiological data from human-induced pluripotent stem cell-derived cardiomyocytes to functional preclinical safety assays. Toxicological Sciences. 134 (2), 412-426 (2013).
- Zlochiver, V., Edwards, S. L., Joshi-Mukherjee, R. Longitudinal Cardiotoxic Effect of Doxorubicin in a Multicellular Cardiac Model. Biophysical Journal. 116 (3), (2019).
- Waas, M., et al. Are These Cardiomyocytes? Protocol Development Reveals Impact of Sample Preparation on the Accuracy of Identifying Cardiomyocytes by Flow Cytometry. Stem Cell Reports. , (2019).
- Gorospe, G., et al. Automated grouping of action potentials of human embryonic stem cell-derived cardiomyocytes. IEEE Transactions on Biomedical Engineering. 61 (9), 2389-2395 (2014).
- Zhu, W., Varga, Z., Silva, J. R. Molecular motions that shape the cardiac action potential: Insights from voltage clamp fluorometry. Progress in Biophysics & Molecular Biology. 120 (1-3), 3-17 (2016).

