Successful In vivo Calcium Imaging with a Head-Mount Miniaturized Microscope in the Amygdala of Freely Behaving Mouse
Summary
In vivo microendoscopic calcium imaging is an invaluable tool that enables real-time monitoring of neuronal activities in freely behaving animals. However, applying this technique to the amygdala has been difficult. This protocol aims to provide a useful guideline for successfully targeting amygdala cells with a miniaturized microscope in mice.
Abstract
In vivo real-time monitoring of neuronal activities in freely moving animals is one of key approaches to link neuronal activity to behavior. For this purpose, an in vivo imaging technique that detects calcium transients in neurons using genetically encoded calcium indicators (GECIs), a miniaturized fluorescence microscope, and a gradient refractive index (GRIN) lens has been developed and successfully applied to many brain structures1,2,3,4,5,6. This imaging technique is particularly powerful because it enables chronic simultaneous imaging of genetically defined cell populations for a long-term period up to several weeks. Although useful, this imaging technique has not been easily applied to brain structures that locate deep within the brain such as amygdala, an essential brain structure for emotional processing and associative fear memory7. There are several factors that make it difficult to apply the imaging technique to the amygdala. For instance, motion artifacts usually occur more frequently during the imaging conducted in the deeper brain regions because a head-mount microscope implanted deep in the brain is relatively unstable. Another problem is that the lateral ventricle is positioned close to the implanted GRIN lens and its movement during respiration may cause highly irregular motion artifacts that cannot be easily corrected, which makes it difficult to form a stable imaging view. Furthermore, because cells in the amygdala are usually quiet at a resting or anesthetized state, it is hard to find and focus the target cells expressing GECI in the amygdala during baseplating procedure for later imaging. This protocol provides a helpful guideline for how to efficiently target cells expressing GECI in the amygdala with head-mount miniaturized microscope for successful in vivo calcium imaging in such a deeper brain region. It is noted that this protocol is based on a particular system (e.g., Inscopix) but not restricted to it.
Introduction
Calcium is a ubiquitous second messenger, playing a crucial role in almost every cellular functions8. In neurons, action potential firing and synaptic input cause rapid change of intracellular free [Ca2+]9,10. Therefore, tracking calcium transients provides an opportunity to monitor neuronal activity. GECIs are powerful tools that allow for monitoring [Ca2+] in defined cell populations and intra-cellular compartments11,12. Among many different types of protein based calcium indicator, GCaMP, a Ca2+ probe based on a single GFP molecule13, is the most optimized and thus widely used GECI. Through multiple rounds of engineering, a number of variants of GCaMP has been developed12,14,15,16. We use one of the recently developed GCaMPs, GCaMP7b, in this protocol16. GCaMP sensors have greatly contributed to the study of neural circuit functions in a number of model organisms such as imaging of Ca2+ transients during development17, in vivo imaging in a specific cortical layer18, measurement of circuit dynamics in motor task learning19 and imaging of cell ensemble activity related with associative fear memory in the hippocampus and amygdala20,21.
Optical imaging of GECIs has several advantages22. Genetic encoding enables GECIs to be stably expressed for a long-term period of time in a specific subset of cells that are defined by genetic profile or specific patterns of anatomical connectivity. Optical imaging enables in vivo chronic simultaneous monitoring of hundreds to thousands of neurons in live animals. A few optical imaging systems have been developed for in vivo imaging and analysis of GECIs within the brain of freely behaving mice with head-mount miniaturized fluorescence microscopes21,23,24,25. Despite the in vivo optical imaging technique based on GECIs, GRIN lens, and a head-mount miniature microscope being a powerful tool to study the link between neural circuit activity and behavior, applying this technology to the amygdala has been difficult due to several technical issues related with targeting the GRIN lens to cells expressing GECIs in the amygdala without causing motion artifacts that severely reduce the quality of image acquisition and finding cells expressing GECIs. This protocol aims to provide a helpful guideline for surgical procedures of baseplate attachment and GRIN lens implantation that are critical steps for successful in vivo optical calcium imaging in the amygdala. Although this protocol targets the amygdala, most procedures described here are commonly applicable to other deeper brain regions. Although this protocol is based on a particular system (e.g., Inscopix), the same purpose may be easily achieved with other alternative systems.
Protocol
All procedures were approved by the Animal Ethics Committee at the Korea Advanced Institute of Science and Technology. All experiments were performed in accordance with the guideline of the Institutional Animal Care and Use Committee.
NOTE: This protocol consists of six major steps: virus injection surgery, GRIN lens implant surgery, validation of GRIN lens implantation, baseplate attachment, optical recording of GCaMP signal during a behavior test, and data processing (Figure 1A). Except for surgery, the commercial software package (Inscopix) is used.
1. Stereotaxic surgery – AAV Virus injection
NOTE: The mouse strain used in this surgical procedure is C57BL6/J. The animal’s body is covered with a sterile drape during surgery and all steps in protocol are performed with wearing sterile gloves. Multiple surgeries are usually not performed in the same day. However, if multiple mice have to have the same surgery in the same day, use a separate set of autoclaved surgical tools for each mouse and 70% ethanol to disinfect the surgical instruments between mice. To keep mouse warm during the surgical procedure, the mouse is covered with a custom-made surgical blanket after fixed to the stereotaxic frame.
- Disinfect all required surgical tools with 70% ethanol, and prepare a Hamilton syringe and a glass pipette. Fill the glass pipette with distilled water.
NOTE: All surgical tools used in the protocol are autoclaved. Because the GRIN lens and skull screws cannot be autoclaved for sterilization, 70 % ethanol is used as a disinfectant to prevent inflammation. Although not tried, other forms of sterilization may be used e.g. gas or chemical sterilization. - Anesthetize the mouse with pentobarbital (83 mg∙kg-1 of body weight) by intraperitoneal injection. After the mouse becomes unconscious, shave the fur on the skull.
- Clean the skin above the skull with 70% ethanol and fix the mouse to the stereotaxic frame. Carefully remove the skin and wipe the skull surface with a 35% hydrogen peroxide solution using a cotton-tip applicator. Apply lubricant eye ointment to prevent corneal drying during the surgery.
NOTE: Although usually there is no problem with using 70% ethanol only for aseptic skin preparation, it is recommended using 3 alternating rounds of disinfectant e.g. iodophors or chlorhexidine solution, and 70% ethanol. Higher concentration of hydrogen peroxide is used in this protocol to make sure that connective tissue on the mouse skull is removed as completely as possible, which is critical for reducing motion artifact during later imaging because the connective tissue on the skull interferes with a firm attachment of the baseplate to the skull. It should be noted that 35% hydrogen peroxide is a lot higher than what is normally used (3 %). - Fix the pin with a stereotaxic probe holder. Align the height of the bregma and lambda with the pin.
- Find the specific location on the mouse skull for craniotomy.
NOTE: Coordinates of the lateral amygdala (LA) for virus injection are (AP, ML, DV) = (-1.6 mm, -3.5 mm, -4.3 mm) from bregma in this protocol (AP; Anterior-Posterior, ML; Medial-Lateral, DV; Dorsal-Ventral, each abbreviation indicates relative axial distance from the bregma)26. The coordinates should be optimized for each experimental condition. - Drill the skull surface (Figure 1B). Wash skull fractions with phosphate buffered saline (PBS). Remove the remaining layer by using forceps or a 27 G needle.
- When drilling the skull, try not to touch the last dura layer to avoid damage to the brain tissue surface. A damaged brain surface causes inflammation around the lens, which induces severe motion artifacts and high autofluorescence during recording. The size of the craniotomy is 1.6 mm. The drill speed is 18,000 rpm, and the bit size is 0.6 mm.
- For lowering the GRIN lens from the brain tissue surface of the drilled site to the target amygdala site, calculate the distance named as “Value A” by subtracting the DV difference between the bregma and the exposed brain surface from DV coordinate of the lens implantation, which is set based on the bregma (4.2 mm in this case, Figure 1C).
- Load 3 µL of mineral oil and 0.7 µL of the AAV virus solution in the glass pipette.
NOTE: Mineral oil helps to separate virus solution and water. 0.7 µL is an optimized volume for the LA virus injection that can fully cover the LA. - Remove the pin from stereotaxic probe holder and fix the glass pipette to it.
- Locate the glass pipette at the injection site mentioned in step 1.5. Insert the glass pipette into the brain tissue after bleeding is completely stopped.
- Deliver the virus through a glass pipette using an injection pump.
NOTE: The titer of the virus should higher than 1 x 1013 vg/mL to obtain a sufficient number of GCaMP expressing cells. Injection speed is 0.1 µL/min and diffuse for 10 min. If there is bleeding, wash the brain surface with PBS. Keep the brain surface wet in this step. - After finishing injection, remove the glass pipette slowly.
2. Stereotaxic surgery – GRIN lens implantation
NOTE: GRIN lens implant surgery is the most critical step in this protocol. Since a consistent slow speed of the GRIN lens movement is critical for successful GRIN lens implantation, a motorized surgery arm can be useful. The motorized surgery arm is a stereotaxic manipulator that is controlled by computer software. Although the motorized arm is used in this protocol, other ways can also be used as long as the lens moves consistently and slowly during implantation. Detailed information about the device is in the Table of Materials.
- Drill the skull to implant two skull screws. Wash all skull fractions with PBS.
NOTE: To reduce the motion artifacts in later optical imaging, at least two screws are required. The two skull screws and the GRIN lens implant position form an equilateral triangle (Figure 1B). The size of the craniotomy should tightly fit with the diameter of the screw. If there is bleeding, wash the bleeding surface with PBS. The drill speed is 18,000 rpm, and the bit size is 0.6 mm. - Disinfect the screws with 70% ethanol to prevent inflammation. Implant the screws as deep as possible.
NOTE: Screws should be tightly fixed without additional adhesion such as cement. - Install a motorized surgery arm on the stereotaxic frame. Connect the required hardware to the laptop computer in which controlling software is installed (Table of Materials).
- Before lowering the GRIN lens, prepare to make a needle track with a 26 G needle.
NOTE: The needle track is a sort of guide track that helps implanting the relatively thick GRIN lens into deep brain regions such as amygdala without causing a severe distortion of brain structure along the injection path. This guide track is called a needle track because it is made by a lowering and elevating procedure of relatively thin 26 G needle. - Attach the cannula holder to stably hold the needle on the stereotaxic frame.
- Grip the 26 G needle with the cannula holder and calculate the coordinates of the needle insertion site.
NOTE: In this protocol, the coordinates of the needle insertion site are 50 µm lateral from the virus injection site (AP, ML, DV) = (-1.6 mm, -3.55 mm, -3.7 mm). - Access the controlling software and complete preparation for manipulating needle position.
- Run the controlling software and select the Start new project button to start controlling session.
NOTE: In the controlling session, there are several blank boxes and menu buttons. The blank boxes are input space for coordinates. After entering coordinates in the blank boxes, the motorized stereotaxic arm moves by clicking the Go To button. The Go To button changes into a STOP button when the stereotaxic arm is moving. Among several menu buttons, only the Tool button is required in this protocol. - Click the Tool button. Calibrate the controlling software by clicking the Calibrate the frame menu button. Calibration is the process by which the actual position of manipulator is set as values on the Stereodrive software.
- Run the controlling software and select the Start new project button to start controlling session.
- Locate the tip of the needle at the exposed brain surface on the skull surface using stereotaxic manipulator.
NOTE: The speed of the needle is set using the Microdrive speed menu in the Tool menu. The default speed is 3.0 mm/s. However, a 0.5 mm/s speed is recommended in this step. The movement of the needle is controlled by the arrow keys. - Add 2-3 drops of PBS to keep the brain surface wet. Fill in the blank box on the computer screen with appropriate DV coordinates of the LA. If the brain surface is dry, it hinders penetration of the needle into the brain tissue.
- Lower the 26 G needle by clicking the Go To button. It automatically stops when the needle reaches the target site set by DV coordinates. The speed of the needle is 100 µm/min. If there is bleeding, stop the needle by clicking the STOP button and wash the brain surface with PBS. Keep the brain surface wet in this step. After bleeding is stopped, start lowering again.
- After the needle completely stops moving, enter 45.00 mm in the blank box on the computer screen.
NOTE: 45.00 mm is a value of DV coordinate that makes needle return to the start position. - Elevate the needle by clicking the Go To button. The speed of the needle movement is 100-200 µm/min.
- After the needle stops moving, remove the PBS on the brain surface. Uninstall the needle and cannula holder from the stereotaxic frame. The experimenter can manually stop the needle by clicking the STOP button when necessary.
- Equip the stereotaxic rod with the lens holder. Install the GRIN lens to the lens holder.
NOTE: The GRIN lens optimized for targeting the LA (0.6 mm in diameter, 7.3 mm in length) is used in this protocol. If the lens holder is not prepared, an alternative way is to use a bulldog clip to grip the GRIN lens. - Disinfect the lens with 70% ethanol and attach the stereotaxic rod to the stereotaxic frames.
- Locate the tip of the GRIN lens at the designated site on the skull surface (same method described in step 2.8).
NOTE: The coordinate of GRIN lens is (AP, ML, DV) = (-1.6 mm, -3.55 mm, -4.2 mm) (Figure 1C). The lens should not touch the skull to avoid damage on its tip. - Calculate the DV coordinate by subtracting the absolute value of “Value A” from the DV coordinate of the lens implantation described in the step 2.16.
- Drop 2-3 drops of PBS on the brain surface. Enter 1000 µm for lowering and 300 µm for raising the lens in the blank box on the computer screen.
- Repeat lowering (1000 µm downward) and raising (300 µm upward) of the GRIN lens until it reaches the target site. This up and down procedure is used to help release the pressure generated inside the brain tissue due to the lowering procedure of lens that otherwise can cause distortion of brain structures along the implantation path.
NOTE: The speed of the GRIN lens movement is 100 µm/min. Even if there is bleeding, do not stop moving the GRIN lens and wash the brain surface with PBS. Keep the brain surface wet in this step. If the brain surface is dry, brain tissue sticks to the GRIN lens surface and it causes distortion of brain structure. - If the GRIN lens reaches the target site, remove PBS and carefully apply the resin dental cement around the GRIN lens, the screws, and their sidewall (Figure 1D).
NOTE: Applying the resin cement all over the skull may reduce motion artifacts. On the contrary, if the resin cement accidentally covers the muscles attached to the skull, it increases motion artifacts. - After the resin dental cement hardens, disassemble the GRIN lens from the lens holder and apply acrylic cement all over the skull (Figure 1D).
- To attach the head plate, disassemble the lens holder from the stereotaxic rod and attach the head plate on the tip of the rod by using tapes. Confirm whether the head plate is horizontal.
NOTE: A tilted head plate causes a tilted view. A head plate is required for fixing the mouse head in the mobile home cage system (in the baseplate attachment section). If the experimenter does not use the system, skip this step (step 2.22) and the following step 2.23. - Slowly lower the rod until the head plate locates at the top of the lens cuff. Lower the head plate 1000 µm more. Move the head plate for the implanted GRIN lens to locate at the right edge of the internal ring of the headplate.
NOTE: The head plate should be positioned lower than the implanted GRIN lens surface; otherwise, the microendoscope cannot approach close enough to the implanted GRIN lens for adjusting focal plane due to the acryl cement. - Apply acrylic cement to attach the head plate to the cement layers (Figure 1D).
- Attach paraffin film on the implanted GRIN lens surface to protect the lens surface from dust. Then apply removable epoxy bond on the paraffin film to keep paraffin film remain on the lens surface.
- Inject Carprofen, an analgesic (0.5 mg/mL in PBS), and Dexamethasone, an anti-inflammatory drug (0.02 mg/mL in PBS) solution, in the peritoneal cavity of the mouse.
NOTE: The dose of the drug is dependent on the body weight: Carprofen (5 mg∙kg-1 of body weight) and Dexamethasone (0.2 mg∙kg-1 of body weight). Both Carprofen and Dexamethasone is administered after surgery and once a day for 7 days post-surgery. - Mix 0.3 mg/mL amoxicillin, an antibiotic, in home cage water to administer the drug for 1 week.
- Return the mouse to the home cage and allow 5-8 weeks for recovery and virus transduction.
NOTE: The mouse is recovered in a pre-warmed cage on an electric blanket until the mouse awakes from the anesthetic.
3. Validation of the GRIN lens implantation
NOTE: Validation of the GRIN lens implantation provides information on whether the implanted GRIN lens is on-target to GCaMP expressing cells. Based on this information, the experimenter saves their time from time-consuming processes such as the behavior test and data processing by excluding animals with off-targeted GRIN lens implantation. During the validation procedure, cells with GCaMP expression should be focused in the recording field of view. A mobile home cage is used in this step. A mobile home cage is a specialized round-shaped apparatus that allows head-fixed mice to freely move their legs during the validation of the GRIN lens implantation and baseplate attachment. An airlifting table in this apparatus on which the legs of head fixed mice are placed enables such free movement of legs although the head is fixed. The cells in the lateral nucleus of the amygdala is usually quiet in a resting or anesthetized state so GCaMP fluorescence signals are rarely detected in these conditions, which makes it very difficult, sometimes impossible, to find cells expressing GCaMP during the validation of the GRIN lens implantation and baseplate attachment. However, the movement of legs often produces GCaMP fluorescence signals in the cells of lateral nucleus of amygdala and thereby can help to locate the microscope. Thus, the mobile home cage is used in this protocol.
- Assemble the stereotaxic manipulator to the mobile home cage system.
- Anesthetize the mouse with 1.5% isoflurane gas. After the mouse is completely unconscious, place the mouse on to the head bar of the mobile home cage system.
- Locate the carbon cage under the mouse. Close the carbon cage and turn on the airflow to make the cage freely move without friction. Wait a few minutes until the mouse becomes active.
NOTE: The speed of airflow should be optimized for each experimental condition (here, 100 L/min). - Remove paraffin film and epoxy bond on the surface of the implanted GRIN lens and wipe the lens surface with 70% ethanol using lens paper.
- Prepare the data acquisition (DAQ) software (e.g., Inscopix). Set the imaging conditions.
NOTE: The DAQ software is used for controlling the microscope (LED power, lens focus, etc.) and data storage.- Turn on the DAQ box and plug a microscope to it. Then, connect it to a laptop computer either directly using a cable or via wireless internet.
NOTE: The DAQ software is installed in the extra hardware called DAQ box (e.g., Inscopix). By connecting the DAQ box to the laptop computer, DAQ software becomes accessible in a laptop computer. - Access the DAQ software through an internet browser (Firefox is recommended).
- Set all the necessary details (Date, mouse ID, etc.) in the Defined Session. Then click Save and Continue. After saving, access the Run session.
- In the Run Session, set the imaging conditions such as exposure time, gain, electronic focus, and exposure-LED power.
NOTE: Gain and exposure-LED power are changeable depending on laboratory conditions. The recommended value for electrical focus is 500 and the exposure time is 50 ms. There is no restriction for changing LED-power and gain. However, a higher intensity of light can cause more bleaching. The exposure time of recording determines a frame rate of video recording. Higher frame rates will reduce signal intensity. Frame rates should be optimized for the GECIs used for the imaging.
- Turn on the DAQ box and plug a microscope to it. Then, connect it to a laptop computer either directly using a cable or via wireless internet.
- To hold the microscope on a stereotaxic manipulator, attach the stereotaxic rod with microscope gripper to stereotaxic manipulator.
NOTE: The gripper is a small accessory that can be attached to the stereotaxic apparatus to hold a microscope body for placing it to the target brain location. - Grip the microscope with the microscope gripper. Turn on the LED light connected to the microscope and align the microscope vertically. Lower the microscope while observing image until the surface of implanted GRIN lens shows up.
- Align the center of the implanted GRIN lens with the center of the view.
NOTE: Validation of the GRIN lens implantation can be conducted in this step (mentioned in the result section, Figure 2B, Figure 2D, Figure 2F). - Capture image of the implanted GRIN lens surface (Click Prt Sc button on keyboard). The image of the implanted GRIN lens surface is required for deselecting non-neuronal components during later data processing (step 6.8).
- Find the appropriate position of the microscope. Slowly elevate the objective lens of microscope while observing image to search for cells with GCaMP7b expression.
NOTE: It may be necessary to adjust the gain of the image acquisition or the exposure-LED power. Validation of the GRIN lens implantation can be conducted in this step (mentioned in result section, Figure 2A, Figure 2C, Figure 2E).
4. Baseplate attachment
NOTE: The baseplate attachment step follows the validation of GRIN lens implantation. The baseplate is a platform for mounting the microscope to the head of mice. As mentioned in the previous section, a mobile home cage is used in this protocol.
- After validating GRIN lens implantation, disassemble a miniature microscope from the microscope gripper.
- Attach baseplate to the microscope. Tighten the screw using hex-key for stable attachment of baseplate to the microscope.
- Grip the microscope with the microscope gripper. Turn on the LED light connected to the microscope and align the microscope vertically. Lower the microscope until the surface of implanted GRIN lens is observed. Align the center of the implanted GRIN lens with the center of the view.
- Find the appropriate position of the microscope. Slowly elevate the objective lens of the microscope until finding a focal plane that gives best images of cells with GCaMP7b expression (Figure 2A).
NOTE: It may be necessary to adjust the gain of image acquisition or exposure-LED power during this step. - Apply acrylic cement to stably attach the baseplate to the head plate (Figure 2G). Build up the sidewalls with acrylic cement between the baseplate and head plate.
NOTE: Acrylic cement shrinks when it hardens. For this reason, do not remove the microscope until the cement completely hardens (this step takes at least 30 min). Cementing should be done very carefully to avoid unwanted cementing of the microscope and its objective lens with acrylic cement. - After acrylic cement completely hardens, loosen the screw and elevate the microscope body slowly.
- If there is a gap on the sidewall, redo additional cementing to protect the implanted GRIN lens from dust.
- Cover the baseplate to protect the implanted GRIN lens from dust and tighten the screw of the baseplate.
- Release the head plate from the head bar. Return mouse to home cage until future optical imaging.
5. Optical recording of GCaMP signal during a behavior test
NOTE: The procedure of GCaMP signal recording during behavior can be very different depending on the systems used for optical imaging, experimental design and laboratory environment. Therefore, it is described in a simple way in this section.
- Conduct several days of handling before the behavior test.
- Prepare the behavior apparatus (e.g., fear conditioning chamber, behavior software, and the Coulbourn Instrument) and laptop computer.
- Plug the microscope into the DAQ box and turn on the DAQ box.
- Connect the DAQ box to laptop computer via wireless internet connection.
- Start the DAQ software (Firefox browser is recommended) on the laptop computer. Click File manager button and check the remaining storage capacity of the DAQ box for saving recorded data.
NOTE: Data size is approximately 80 GB for 30 min recording. - Enter the necessary information (Date, mouse ID, etc.) in the Defined session and enter into the Run session.
- Gently grab the mouse and remove baseplate cover on its head. Placing the microscope on the baseplate platform.
- Tighten the baseplate screw and place the animal with a head-mount microendoscope into the behavior chamber.
NOTE: The mouse tries to escape in this step if handling is not enough. - Connect the NI board to the TRIG port of the DAQ box using a BNC cable. NI board receives TTL signals from behavior software (e.g., FreezeFrame) and coordinates simultaneous recording procedure of GCaMP signal and animal’s behavior.
- Adjust the focal plane.
NOTE: The focal plane is determined by the distance between the objective lens of microscope and the GRIN lens implanted in the brain. To adjust the focal plane, the objective lens position is manipulated while observing images. The distance between those two lenses that shows clear morphology of cells and blood vessels is set as the focal plane for imaging. In this protocol, the DAQ software is used to control the movement of the objective lens during adjustment. - Set the optimized exposure time, gain, LED power in Run session.
NOTE: Generally, 50 ms is used for exposure time. Gain and LED power are set based on image histograms. Histograms indicate the distribution of the fluorescence intensity of the pixels. Based on visual inspection, the right edge of the histogram should have a value between 40% and 60% in the case of GCaMP7b. It changes depending on the GECIs used for imaging. - Change recording option to Triggered recording. Click Record and start the behavior.
NOTE: The triggered recording option provides a matched timeline of the recording session and the behavior session. If the TTL signal is received, a red light above the TRIG port turns on. - After finishing the behavior test, detach the microscope and attach baseplate cover.
- Return the mouse to the home cage. Then, select File manager and export data from the DAQ box.
- After the behavior test, sacrifice the mouse for postmortem histological verification.
6. Data processing
NOTE: Data processing procedure is very different depending on the data processing software and the GECIs used for the imaging experiment. Therefore, it is described in a simple way in this section. This protocol uses commercial data processing software (see Table of Materials). Alternatively, other open source software mentioned in the discussion section can also be used with no problem. The variables used in this protocol are listed in Table 1.
- Import the recording video data file (1280 pixels x 800 pixels) from the DAQ box into desktop computer for data processing. Launch the data processing software.
NOTE: Data processing software is different from DAQ software in previous steps. In data processing software, there are 6 steps: down-sampling, cropping, spatial filter, motion correction, ΔF/F calculation, event detection which are necessary for extracting single cell calcium transient data from recorded video. - Downsample and crop the video.
NOTE: Downsampling and cropping are necessary to boost up data processing speed. Crop the area except region of interest where the GCaMP signal is observed. - Apply spatial filter. After applying spatial filter, create a mean intensity projection image.
NOTE: Spatial filter distinguishes each pixel in recorded video image and provide a high contrast image. There are two types of cut-off filter, low and high cut-off filter. The higher value of low cut-off filters increases contrast of image but at the same overall gray noise also increases. The lower value of high cut- off filter makes the image become blur. - Apply motion correction. Use the mean intensity projection image as a reference image for motion correction.
- Apply motion correction again using the image of first frame as a reference image.
NOTE: Two steps of motion correction significantly decrease motion artifact. - Apply ΔF/F calculation to visualize pixels that show a change in brightness.
NOTE: The brightness change of pixels is caused by fluorescence intensity change of GCaMP, and it indicates calcium transients. - Apply the PCA/ICA algorithm to visualize calcium transient curve of each cell.
NOTE: PCA/ICA is an algorithm for sorting a group of data into a set of single component data. The variables should be optimized for each laboratory condition. In this protocol, all variables are default except ICA temporal weight (0.4 is used in this protocol)27. - Deselect manually non-neuronal components among identified components based on the criteria described in the following Note.
NOTE: Two major criteria are used for this purpose. First, the GCaMP signal originated from a neuron looks like a clear spherical shape. Thus, only the signal with a clear spherical shape is selected as a neuron. Second, given the diameter of the cell body of amygdala pyramidal cell is known to be approximately 20 µm28,29,30, only the signal roughly matches with this size is selected as a neuron. To determine the size of the GCaMP signal, the actual size of a single pixel is calculated based on the captured image of the lens surface (step 3.9) and the maximum width of single spherical shape is calculated. For example, in this protocol, the diameter of the implanted GRIN lens is 600 µm. Therefore, if the diameter of the lens is 800 pixels, the actual size of the single pixel is 1.5 µm (20 µm is about 7 pixels). - Apply the event detection function to detect a significant increase of calcium transient (calcium event).
NOTE: In this step, the variable called event smallest decay time (τ) is required. In this protocol, 1.18s is used as an optimized variable for GCaMP7b. The average half decay time (t1/2=0.82s) is calculated based on ΔF/F curve during spontaneous activity of cells. Since GCaMP follows exponential decay, τ =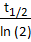 .
.
Representative Results
Validation of GRIN lens implantation
Before chronically attaching the baseplate to the brain by cementing, the GRIN lens implantation needs to be validated. In animals with successful lens implantation, both GCaMP expressing cells and blood vessels were clearly observed within a focal plane range determined by the distance between objective lens of microscope and implanted GRIN lens (Figure 2A and B). In contrast, in animals with off-target implantation, a clear image of GCaMP expressing cells was not observed within the focal plane range (either out-of-focus or out-of-view). In the case of out-of-focus, well-focused blood vessels could be observed but only blurry images for cells within the focal plane range (Figure 2C and D). In the case of out-of-view, blood vessels were not observed in most cases and no fluorescence signal was detected within the focal plane range (Figure 2E). Moreover, unlike other cases, the implanted GRIN lens showed a bright edge (Figure 2F). The baseplate attachment is conducted only on validated animals with on target GRIN lens implantation.
Recording of GCaMP signals in the LA neuron in response to auditory stimuli
In this protocol, 4 instances of pseudorandomized tone (2.8 kHz, 200 ms duration, 25 pulses) were presented to a mouse, and GCaMP signals were optically recorded in the LA cells of mice with head-mount microendoscope. Mice injected with AAV1-Syn-GCaMP7b-WPRE in the unilateral lateral amygdala were used for the imaging. In animals with successful GRIN lens implantation, virus expressing cells and blood vessels were clearly observed within the focal plane range (Figure 3A). In the behavior condition, approximately 50-150 cells usually displayed a significant fluorescence change in the field of view in the LA even without tone as shown in ΔF/F image, likely spontaneously active cells (Figure 3B). Upon tone presentation, only a few cells displayed a tone-specific change of GCaMP signal as determined by ΔF/F image analysis (6 cells in the Figure 3C and Figure 3D). The same procedures were conducted on mice injected with AAV2/1-CaMKIIα-GFP as a control. Although GFP expressing cells were detected within the focal plane range, no cells displayed a significant fluorescence change with or without tone as determined by ΔF/F image analysis (Figure 3E and Figure 3F).
For histological verification of GCaMP expression and targeting of GRIN lens, coronal sections of the postmortem brain are examined under the fluorescence microscope (Figure 4A and Figure 4B). DAPI staining is used to confirm intact cells and no sign of tissue damage due to inflammation of the brain tissue around the GRIN lens (Figure 4C).
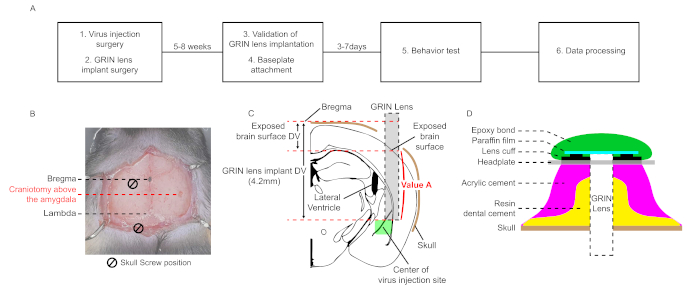
Figure 1: Schematic workflow and diagrams for stereotaxic surgery for in vivo microendoscopic calcium imaging in the LA. (A) A schematic workflow of in vivo microendoscopic calcium imaging in the LA. (B) A microscopic picture showing two-dimensional coordinates of craniotomy sites for the virus injection and the GRIN lens implant surgery. Two skull screws and the lens form an equilateral triangle. (C) A diagram for the relative position between virus injection site and the implanted GRIN lens, and schematic explanation for calculating “Value A”. (D) Cross-section of the cement layer from the skull surface to the headplate in the final steps of surgery. Resin dental cement should cover the wall of the screws and the implanted GRIN lens. Acrylic cement is applied on the resin dental cement. Headplate is attached on the cement layer. Paraffin film covers the implanted GRIN lens surface and protect it from dust. Removable epoxy bond prevents detaching of paraffin film. Please click here to view a larger version of this figure.
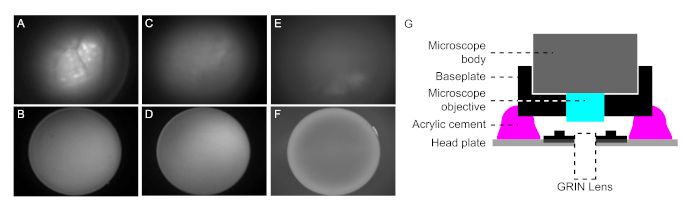
Figure 2: Validation of the GRIN lens implantation and configuration of cement layers after baseplate attachment. (A) A representative endoscopic snapshot image from animals with successful GRIN lens implantation. Both GCaMP expressing cells and blood vessels are clearly observed. (B) Snapshot image of the implanted GRIN lens surface from animals with successful GRIN lens implantation. (C) A representative endoscopic snapshot image from animals with out-of-focus GRIN lens implantation. (D) Snapshot image of the implanted GRIN lens surface from animals with out-of-focus GRIN lens implantation. In the case of out-of-focus, blood vessels are sometimes clearly observed but only blurry images for cells within the focal plane range. (E) A representative endoscopic snapshot image from animals with out-of-view GRIN lens implantation. In the case of out-of-view, blood vessels are not observed in most cases and no fluorescence signal is detected within the focal plane range. (F) A snapshot image of the implanted GRIN lens surface from animals with out-of-view GRIN lens implantation. Unlike other cases, the bright edge is observed in this case. (G) The configuration of cement layers for baseplate attachment. Baseplate and headplate are attached by acrylic cement. There should a space unfilled with acrylic cement between the baseplate and GRIN lens. Please click here to view a larger version of this figure.
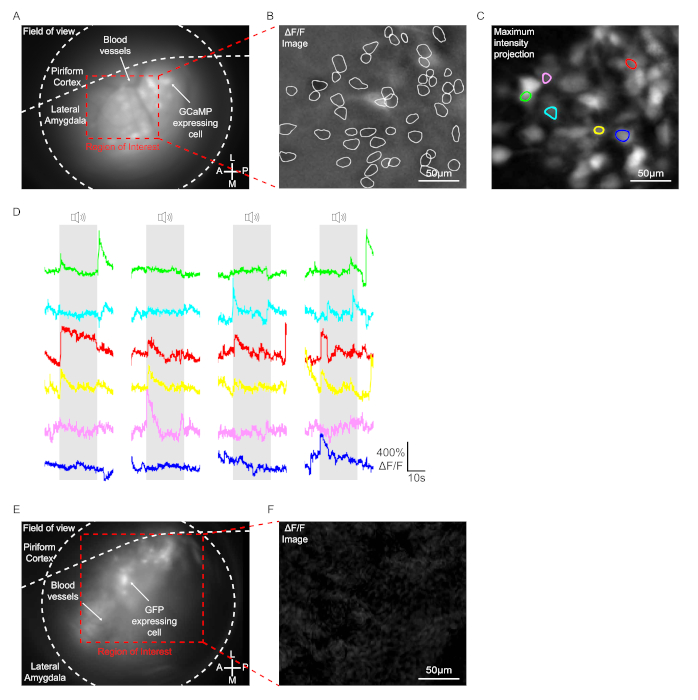
Figure 3: Recording of GCaMP signals in the LA neurons. (A to C) Data obtained from a mouse injected with AAV1-Syn-GCaMP7b-WPRE. (A) A representative endoscopic snapshot image during a behavior test. GCaMP7b expressing cells and blood vessels were clearly observed within the focal plane range. (B) A representative snapshot image showing ΔF/F signal. White line indicates cells that displayed a significant fluorescence change in the region of interest during the behavior test. Scale bar = 50 µm. (C) Maximum intensity projection image. Total 6 cells displayed a tone-specific change of GCaMP signal. Scale bar = 50 µm. (D) Representative ΔF/F traces of tone responsive cells. Each color corresponds to each individual cell with the same color in panel (C). (E and F) Data obtained from a mouse injected with AAV2/1-CaMKIIα-GFP. (E) A representative endoscopic snapshot image during a behavior test. GFP expressing cells were clearly detected within the focal plane range. (F) A representative snapshot image showing ΔF/F signal. Scale bar = 50µm. Please click here to view a larger version of this figure.
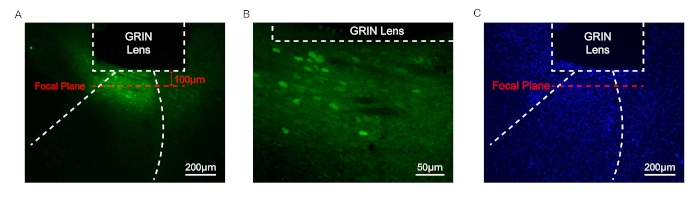
Figure 4: Histological verification of the GRIN lens position and GCaMP7b expression. (A) A representative coronal section image showing GCaMP7b expression in the LA. Scale bar = 200µm. (B) Magnified image. Scale bar = 50 µm. (C) Fluorescence microscopic image showing DAPI staining signal. Scale bar = 200 µm. Please click here to view a larger version of this figure.
| Factor | value |
| Spatial down sampling factor | 2 |
| Temporal down sampling factor | 2 |
| Spatial filter: High cut-off | 0.5 |
| Spatial filter: Low cut-off | 0.005 |
| Motion correction: High cut-off | 0.016 |
| Motion correction: Low cut-off | 0.004 |
| Motion correction: Maxtranslation | 20 |
| ΔF/F reference frame | Mean frame |
| PCA/ICA: blockSize | 1000 |
| PCA/ICA: convergenceThreshold | 1×10^-5 |
| PCA/ICA: icaTemporalWeight | 0.4 |
| PCA/ICA: numICs | 120 |
| PCA/ICA: numPCs | 150 |
| PCA/ICA: unmixType | Temporal |
| Event Detection: Decay Constant | 1.18 |
| Event Detection: Threshold | 4 |
Table 1: List of variables for Data processing
Discussion
Skillful surgery techniques are essential for achieving successful in vivo optical calcium imaging with head-mount miniature microscopy in deeper brain regions such as the amygdala as we described here. Therefore, although this protocol provides a guideline for optimized surgical processes of baseplate attachment and GRIN lens implantation, additional optimization processes might be necessary for critical steps. As mentioned in the protocol section, amygdala coordinates in surgery, airflow speed in baseplate attachment step, image acquisition settings (frame rate, LED power, etc.) in calcium recording and variables (ICA temporal weight, event smallest decay time, etc.) in data processing need to be optimized.
The baseplate attachment step can be modified. The head plate is necessary because it helps fix the head of awake mice during the baseplate attachment conducted in mobile home cage. However, if the mobile home cage is not prepared in the laboratory, the isoflurane gas anesthesia system is an alternative option. For this alternative way, the concentration of isoflurane gas may be critical. We observed that GCaMP signal is rarely detected in the amygdala of mice under 1.5% isoflurane. On the contrary, the GCaMP signal is detected under an 0.8% isoflurane condition and mice stay in an almost awake state but without substantial head movement in this condition. This anesthetization condition thus allows for conducting the baseplate attachment without using additional devices such as head plate and mobile home cage.
Unstable attachment of the skull screw, cement, baseplate, and microscope can cause motion artifacts that can be corrected by using motion correction software algorithm. The movement of the lateral ventricle is thought to cause an irregular type of motion artifacts that cannot be easily corrected with currently available motion correction software. Such irregular motion artifacts are minimal in most superficial brain regions such as the hippocampus and the cortex. However, it is frequently detected during optical imaging in the amygdala. To overcome this problem, this protocol suggests implanting the GRIN lens 50 µm away from viral injection site to the lateral side (Figure 1C), which greatly improves the image acquisition process by reducing the potential motion artifacts originated from the lateral ventricle. Although we set 50 µm relative to the viral injection site, the target coordinate for lens implantation may also be set relative to the lateral ventricle. In this protocol, we reasoned that it is more critical to precisely target the viral expressing site for successful performance of imaging. Thus, we used a viral injection coordinate as a reference to set the target coordinate of lens implantation. Through repeated trials, we established an optimal condition that allowed the GRIN lens for efficiently targeting viral expressing site while avoiding motion artifacts caused by lateral ventricle movement. Eventually, the method that can efficiently and accurately correct any motion artifacts would be of great help for accessing optical imaging to deeper brain regions in the future.
Although the in vivo optical calcium imaging with head-mount miniature microscope is a powerful tool and has been optimized, there is still room for improvement in many aspects. This protocol will facilitate studies that aim to investigate real-time neural activity in the amygdala of freely behaving animals.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by grants from Samsung Science and Technology Foundation (Project Number SSTF-BA1801-10).
Materials
| 26G needle | BD | 302002 | Surgery |
| AAV1-Syn-GCaMP7b-WPRE | Addgene | 104493-AAV1 | Surgery |
| AAV2/1-CaMKiiα-GFP | custom made | Surgery | |
| Acrylic-Dental cement (Ortho-jet Acrylic Pink) | Lang | 1334-pink | Surgery & Baseplate Attachment |
| Air flow manipulator | Neurotar | NTR000253-04 | Baseplate Attachment |
| Amoxicillin | SIGMA | A8523-5G | Surgery |
| Baseplate | INSCOPIX | 1050-002192 | Baseplate Attachment |
| Baseplate cover | INSCOPIX | 1050-002193 | Baseplate Attachment |
| Behavioral apparatus (chamber) | Coulbourn Instrument | Testcage | Behavior test |
| Behavioral apparatus (software) | Coulbourn Instrument | Freeze Frame | Behavior test |
| Carbon cage | Neurotar | 180mm x 70mm | Baseplate Attachment |
| Carprofen | SIGMA | PHR1452-1G | Surgery |
| Data processing software | INSCOPIX | INSCOPIX Data Processing Software | Baseplate Attachment & Behavior test |
| Dexamethasone | SIGMA | D1756-500MG | Surgery |
| Drill | Seyang | marathon-4 | Surgery |
| Drill bur | ELA | US1/2, Shank104 | Surgery |
| Glass needle | WPI | PG10165-4 | Surgery |
| GRIN lens (INSCOPIX Proview Lens Probe) | INSCOPIX | 1050-002208 | Surgery |
| Hamilton Syringe | Hamilton | 84875 | Surgery |
| Head plate | Neurotar | Model 5 | Surgery |
| Hex-key | INSCOPIX | 1050-004195 | Baseplate Attachment |
| Laptop computer | Samsung | NT950XBV | Surgery & Baseplate Attachment |
| Lens holder, Stereotaxic rod (INSCOPIX proview implant kit) | INSCOPIX | 1050-004223 | Surgery |
| Microscope gripper | INSCOPIX | 1050-002199 | Baseplate Attachment |
| Microscope, DAQ software, hardware | INSCOPIX | nVista 3.0 | Baseplate Attachment & Behavior test |
| Mobile homecage | Neurotar | MHC V5 | Baseplate Attachment |
| Moterized arm | Neurostar | Customized | Surgery |
| Moterized arm software | Neurostar | Customized | Surgery |
| NI board | National instrument | Behavior test | |
| Removable epoxy bond | WPI | Kwik-Cast | Surgery |
| Resin cement (Super-bond) | Sun medical | Super bond C&B | Surgery |
| Skull screw | Stoelting | 51457 | Surgery |
| Stereotaxic electrode holder | ASI | EH-600 | Surgery |
| Stereotaxic frame | Stoelting | 51600 | Surgery |
| Stereotaxic manipulator | Stoelting | 51600 | Baseplate Attachment |
References
- Gonzalez, W. G., Zhang, H., Harutyunyan, A., Lois, C. Persistence of neuronal representations through time and damage in the hippocampus. Science. 365 (6455), 821-825 (2019).
- Ghandour, K., et al. Orchestrated ensemble activities constitute a hippocampal memory engram. Nature Communications. 10 (1), 2637 (2019).
- Grundemann, J., et al. Amygdala ensembles encode behavioral states. Science. 364 (6437), (2019).
- Krabbe, S., et al. Adaptive disinhibitory gating by VIP interneurons permits associative learning. Nature Neuroscience. 22 (11), 1834-1843 (2019).
- Betley, J. N., et al. Neurons for hunger and thirst transmit a negative-valence teaching signal. Nature. 521 (7551), 180-185 (2015).
- Jennings, J. H., et al. Visualizing hypothalamic network dynamics for appetitive and consummatory behaviors. Cell. 160 (3), 516-527 (2015).
- LeDoux, J. E. Emotion circuits in the brain. Annual Review of Neuroscience. 23, 155-184 (2000).
- Burgoyne, R. D. Neuronal calcium sensor proteins: generating diversity in neuronal Ca2+ signalling. Nature Reviews Neuroscience. 8 (3), 182-193 (2007).
- Miyakawa, H., et al. Synaptically activated increases in Ca2+ concentration in hippocampal CA1 pyramidal cells are primarily due to voltage-gated Ca2+ channels. Neuron. 9 (6), 1163-1173 (1992).
- Denk, W., Yuste, R., Svoboda, K., Tank, D. W. Imaging calcium dynamics in dendritic spines. Current Opinion in Neurobiology. 6 (3), 372-378 (1996).
- Mank, M., et al. A genetically encoded calcium indicator for chronic in vivo two-photon imaging. Nature Methods. 5 (9), 805-811 (2008).
- Akerboom, J., et al. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Frontiers in Molecular Neuroscience. 6, 2 (2013).
- Nakai, J., Ohkura, M., Imoto, K. A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nature Biotechnology. 19 (2), 137-141 (2001).
- Akerboom, J., et al. Optimization of a GCaMP calcium indicator for neural activity imaging. Journal of Neuroscience. 32 (40), 13819-13840 (2012).
- Chen, T. W., et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 499 (7458), 295-300 (2013).
- Dana, H., et al. High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nature Methods. 16 (7), 649-657 (2019).
- Zariwala, H. A., et al. A Cre-dependent GCaMP3 reporter mouse for neuronal imaging in vivo. Journal of Neuroscience. 32 (9), 3131-3141 (2012).
- Mittmann, W., et al. Two-photon calcium imaging of evoked activity from L5 somatosensory neurons in vivo. Nature Neuroscience. 14 (8), 1089-1093 (2011).
- Huber, D., et al. Multiple dynamic representations in the motor cortex during sensorimotor learning. Nature. 484 (7395), 473-478 (2012).
- Grewe, B. F., et al. Neural ensemble dynamics underlying a long-term associative memory. Nature. 543 (7647), 670-675 (2017).
- Cai, D. J., et al. A shared neural ensemble links distinct contextual memories encoded close in time. Nature. 534 (7605), 115-118 (2016).
- Lin, M. Z., Schnitzer, M. J. Genetically encoded indicators of neuronal activity. Nature Neuroscience. 19 (9), 1142-1153 (2016).
- Zhang, L., et al. Miniscope GRIN Lens System for Calcium Imaging of Neuronal Activity from Deep Brain Structures in Behaving Animals. Current Protocols in Neuroscience. 86 (1), 56 (2019).
- Jacob, A. D., et al. A Compact Head-Mounted Endoscope for In vivo Calcium Imaging in Freely Behaving Mice. Current Protocols in Neuroscience. 84 (1), 51 (2018).
- Ghosh, K. K., et al. Miniaturized integration of a fluorescence microscope. Nature Methods. 8 (10), 871-878 (2011).
- Xiong, B., et al. Precise Cerebral Vascular Atlas in Stereotaxic Coordinates of Whole Mouse Brain. Frontiers in Neuroanatomy. 11, 128 (2017).
- Mukamel, E. A., Nimmerjahn, A., Schnitzer, M. J. Automated analysis of cellular signals from large-scale calcium imaging data. Neuron. 63 (6), 747-760 (2009).
- Millhouse, O. E., DeOlmos, J. Neuronal configurations in lateral and basolateral amygdala. 神经科学. 10 (4), 1269-1300 (1983).
- McDonald, A. J. Neuronal organization of the lateral and basolateral amygdaloid nuclei in the rat. Journal of Comparative Neurology. 222 (4), 589-606 (1984).
- McDonald, A. J. Neurons of the lateral and basolateral amygdaloid nuclei: a Golgi study in the rat. Journal of Comparative Neurology. 212 (3), 293-312 (1982).

