Capillary Electrophoresis Mass Spectrometry Approaches for Characterization of the Protein and Metabolite Corona Acquired by Nanomaterials
Summary
Here we present a protocol to characterize the complete biomolecular corona, proteins, and metabolites, acquired by nanomaterials from biofluids using a capillary electrophoresis – mass spectrometry approach.
Abstract
The adsorption of biomolecules from surrounding biological matrices to the surface of nanomaterials (NMs) to form the corona has been of interest for the past decade. Interest in the bio-nano interface arises from the fact that the biomolecular corona confers a biological identity to NMs and thus causes the body to identify them as “self”. For example, previous studies have demonstrated that the proteins in the corona are capable of interacting with membrane receptors to influence cellular uptake and established that the corona is responsible for cellular trafficking of NMs and their eventual toxicity. To date, most research has focused upon the protein corona and overlooked the possible impacts of the metabolites included in the corona or synergistic effects between components in the complete biomolecular corona. As such, this work demonstrates methodologies to characterize both the protein and metabolite components of the biomolecular corona using bottom-up proteomics and metabolomics approaches in parallel. This includes an on-particle digest of the protein corona with a surfactant used to increase protein recovery, and a passive characterization of the metabolite corona by analyzing metabolite matrices before and after NM exposures. This work introduces capillary electrophoresis – mass spectrometry (CESI-MS) as a new technique for NM corona characterization. The protocols outlined here demonstrate how CESI-MS can be used for the reliable characterization of both the protein and metabolite corona acquired by NMs. The move to CESI-MS greatly decreases the volume of sample required (compared to traditional liquid chromatography – mass spectrometry (LC-MS) approaches) with multiple injections possible from as little as 5 µL of sample, making it ideal for volume limited samples. Furthermore, the environmental consequences of analysis are reduced with respect to LC-MS due to the low flow rates (<20 nL/min) in CESI-MS, and the use of aqueous electrolytes which eliminates the need for organic solvents.
Introduction
Bio-nano interactions, at the interface between nanomaterial (NM) surfaces and biological matrices from which biomolecules adsorb to the NM, is an intense area of research underpinning nanosafety and nanomedicine1. This layer of adsorbed biomolecules confers a biological identity to NMs, thus cells identify them as “self”, subsequently influencing cellular uptake, distribution and biological endpoints2,3,4. The vast majority of bio-nano studies have focused upon the proteins within the corona, and have demonstrated that proteins vary quantitatively and qualitatively between NMs of different compositions5. This variation is dependent upon both the properties of the NM such as size, functionalization, and material in addition to the properties of the biological matrix including composition and salt content5. A rising area of interest in the bio-nano interface are the metabolites that adsorb to NMs6. Several studies have demonstrated that, as with the proteins, NM properties are a key factor in determining the composition of the metabolites in the corona and that they can influence biological outcomes of NM exposure6,7,8.
In studies of the biomolecular corona there are four distinct phases to its characterization, corona formation, the isolation of the biomolecules within the corona, their detection via qualitative or quantitative mass spectrometry and identification9. To date, a range of techniques have been adopted to isolate the protein corona including boiling in SDS-PAGE running buffer, solvent and salt extractions or direct digestion of proteins in situ on the surface of NMs. In terms of the metabolites in the corona, the isolation is more complex with various methods using solvents or even the dissolution of the NMs proposed as solutions8,10,11. However, unlike the protein corona a “one size fits all” approach is unlikely to apply to the isolation of metabolites from NMs, due to the wide chemical space occupied by the metabolome and the diversity of possible NMs. An alternative approach is to characterize the biological matrix before and after NM exposure with the difference in metabolite concentrations attributed to the adsorption of metabolites to NMs.12
This alternative approach would be applicable to all biofluids and NMs and although it does require twice as much analysis, it consequently offers a much more precise measure of the metabolite corona and has no risk of low metabolite recoveries from the NM surface being mistaken for low binding of the specific metabolite.
The second step to the biomolecular corona analysis is the detection of the biomolecules. Traditionally for the proteins in the corona, this has been the remit of nano-LC-MS, the workhorse of proteomics; other approaches such as NMR13, 1D and 2D SDS-PAGE have also been applied. In terms of the metabolite corona LC-MS7, GC-MS8 and direct infusion mass spectrometry have been utilized14. However, a new approach has recently started to gain traction, namely capillary electrophoresis-mass spectrometry (CE-MS)11,15 and has been present in NM labs as a standalone technique to characterize various physical and chemical properties of NMs16. CE-MS is an orthogonal separation technique to nanoLC-MS and GC-MS and is capable of enabling the detection of highly polar and charged metabolites17,18. Furthermore, CE-MS is well suited for the analysis of proteins and their posttranslational modifications such as phosphorylation and glycosylation19,20,21,22. The final step, identification and data analysis can be performed in several ways depending upon if a quantitative / qualitative or targeted / untargeted approach is being used, this, however, falls outside the remit of this protocol.
CESI-MS, a combination of CE and a nanoESI interface, is a recent advance in CE technology utilizing a sheathless interface. This enables direct connection of ultra-low flow CE (<20 nL/min) to a high resolution mass spectrometer with no dilution, resulting in significantly improved detection sensitivity23,24,25. CESI-MS enables volume limited samples (< 10 µL) to be analyzed while maintaining a highly sensitive analysis26. CESI-MS also compares favorably to traditional low flow rate nanoLC-MS approaches used in proteomics and metabolomics in terms of reproducibility27,28, throughput, and carry over making it an exciting prospect for the complete biomolecular corona characterization.
To highlight the potential of CESI-MS for the analyses of bio-nano interactions this work describes the sample preparation required to isolate the NM biomolecular corona from a single human plasma sample in such a way as to maximize data from both the protein and metabolite aspects of the complete biomolecular NM corona. While we report on the use of human plasma, this protocol would be equally appropriate for other biofluids such as blood serum, complete cell culture medium, cell lysate, cerebrospinal fluid or urine. The analyses of these two components (proteins and metabolites) will then be described using neutral coated CESI capillaries for the protein corona and bare fused silica capillaries for both cationic and anionic metabolites.
Protocol
Use of human biofluid was approved as per the IRB protocol guidelines from University of Leiden and Innsbruck Medical University. When human or animal biofluids are being investigated, ethical approval from the research institution is required, and has been obtained in the case of the results shown in this protocol. Furthermore adequate reporting should also be performed to ensure transparency and reusability of data in future work9,29,30.
1. Preparation of background electrolytes (BGE)
NOTE: All solvents should be prepared in a suitable fume hood and adequate personal protective equipment must be used for all steps (labcoat, gloves and goggles). In each step, low binding plastic vials are required to minimize analyte loss and contamination of sample with salts leached from glassware.
- Prepare 10% acetic acid BGE (v/v), pH 2.2 on the daily basis for metabolomics.
- Into a 10 mL volumetric flask add 1 mL of acetic acid, followed by the addition of deionized (DI) water to the marked line and thorough mixing.
- Prepare 100 mM acetic acid, pH 2.9 on the daily basis for proteomics.
- Into a 10 mL volumetric flask add 57 µL of acetic acid, followed by the addition of DI water to the marked line and thorough mixing.
2. NM preparation
- Vigorously disperse NMs in water using published guidelines31,32,33.
NOTE: In the development of this protocol, 7 NMs were used as follows: 100 and 1,000 nm carboxylated polystyrene were used for protein and metabolite corona, respectively. 100 nm unmodified polystyrene NMs, 13 nm anatase TiO2 NMs which were unmodified or coated with either polyacrylate or PVP polymers and 22 nm unmodified SiO2 NMs were used for both protein and metabolite coronas. While the method was developed and demonstrated using these NMs, it should be noted that this approach would be applicable for a wide array of NM compositions, sizes, shapes, and morphologies. - Characterize particle size using either dynamic light scattering34,35, single particle inductively coupled plasma mass spectrometry36, nanoparticle tracking analysis37, or transmission electron microscopy and surface charge as zeta potential using a zeta sizer34.
3. Biomolecular corona formation
- Split 2 mL of human plasma (or biofluid of choice) into 1 mL aliquots, one for the metabolite and protein corona and the second for plasma metabolome characterization.
- Incubate 1 mg/mL of NMs in the human plasma for 1 h at 37 ˚C while gently mixing at 500 rpm in a thermomixer. Following incubation centrifuge the sample at 4,000 x g for 15 min at 4 ˚C to pellet the NMs.
- Collect the plasma supernatant for metabolite corona analysis. Retain the NM-protein corona complex for protein corona characterization.
4. Protein corona isolation
- Resuspend the NM-protein complex (the pellet) in 1 mL of 10x PBS Buffer and vortex vigorously for 2 min to remove unbound proteins. Centrifuge the PBS solution at 4,000 x g for 15 min at 4 ˚C to pellet the NMs, and carefully remove the supernatant without disturbing the pellet.
- Resuspend the pellet in 1 mL of ammonium bicarbonate buffer (ABC) (100 mM, pH 8.0) and vortex vigorously for 2 min to remove unbound proteins and undesired salts. Centrifuge at 4,000 x g for 15 min at 4 ˚C to pellet the NMs; remove and discard the supernatant. Repeat this step.
- Reduce protein disulfide bonds by dissolving the NM-protein pellet in 20 µL of ABC buffer (100 mM pH 8) containing 10 mM dithiotheritol. Incubate the reduction solution for 30 min at 56 ˚C.
- To digest proteins, add 2 µg of sequencing grade Trypsin to 20 µL of ABC containing 0.1% surfactant. Incubate the digest solution for 16 h at 37 ˚C.
- Alkylate the sample by the addition of 20 µL of iodoacetamide at 55 mM in 100 mM ABC and incubate at room temperature for 20 min.
- Add 20 µL of 0.1 M HCl to cleave the surfactant and leave for 10 min at room temperature.
- Enrich and desalt peptides using a C18 packed desalting pipette tip.
- Wet the tip with 80 µL 0.1% formic acid 50% acetonitrile 5 times, clean with 80 µL 0.1% formic acid 5 times, draw up and elute sample 10 times, desalt sample by flushing 80 µL 0.1 % formic acid 5 times and elute the sample 2 times with 80 µL 0.1% formic acid 70% acetonitrile into a clean low binding PCR tube.
- Lyophilize the samples and store at -20 ˚C until analysis.
5. Metabolite corona isolation
- Take the control plasma and the supernatant from the NM plasma incubation from Step 3.3 and keep on ice during sample preparation.
- Take 50 µL from each into separate vials and dilute 1 in 10 with DI water. Take 50 µL of each diluted sample and move to new vials for step 5.3.
- Add 200 µL chloroform, 250 µL methanol and 350 µL DI water and vigorously vortex mixture for 2 min. Centrifuge for 10 min at 20,800 x g at 4 ˚C.
- Take 500 µL of supernatant and filter through a 3 kDa centrifugal filter for 2 h at 10,000 x g at 4 ˚C. Take 430 µL of ultrafiltrate and dry in a speed vac. Samples can now be frozen prior to analysis.
6. Preparation of CESI-MS system38
NOTE: Prior to the installation of the capillaries, check that the inlet and outlet end of the capillaries are intact and that there are no visible breakages in the capillary.
- Installation of a neutral coated capillary for protein corona analysis39.
- Place a new neutral coated capillary (90 cm length with 30 µm internal diameter) into the CESI instrument following the manufacturer guidelines.
- Preparation of CESI capillaries for proteomic analysis.
- Flush the separation capillary in the forward direction using 100 mM of HCl for 5 min at 100 psi and check for droplet formation at the outlet of the capillary. Flush separation capillary with BGE at 100 psi for 10 min and then DI water at 100 psi for 30 min (this is only required for new capillaries).
- Flush both the separation capillary and the conductive capillary with BGE at 90 psi for 5 min and ensure droplets form at the outlet end of both capillaries.
- Flush the separation capillary with DI water at 90 psi for 10 min, then 50 psi for 10 min with 0.1 M HCl, followed again by 90 psi for 10 min with DI water and finally BGE at 90 psi for 10 min. Couple CESI to the MS using a nanospray source and adapter as previously described38.
- Installation of bare fused silica capillary for metabolite corona characterization38.
- Place a new bare fused silica capillary (90 cm length with a 30 µm internal diameter) into the CESI instrument following manufacturer guidelines.
- Preparation of CESI capillaries for metabolomics analysis.
- Flush the separation capillary in the forward direction using 100% MeOH at 50 psi for 15 min and check for droplet formation at the outlet of the capillary. Flush both the separation capillary and the conductive capillary with BGE at 90 psi for 5 min and ensure droplets form at the outlet of both capillaries.
- Flush the separation capillary with DI water at 90 psi for 10 min, then 50 psi for 10 min with 0.1 M NaOH, followed again by 90 psi for 10 min with DI water and finally BGE at 90 psi for 10 min. Couple CESI to the ESI-MS using a nanospray source and adapter as previously described.38
7. CE-MS for protein corona analysis
- Resuspend samples from step 4.8 in 20 µL of 50 mM ammonium acetate pH 4.0.
- Perform 5 psi 10 s hydrodynamic injection of sample. Separate using a 30 kV separation voltage with the following pressure gradient: 0-10 min at 1 psi, 10-35 min at 1.5 psi and 35-45 min at 5 psi using 100 mM, pH 2.9 acetic acid as BGE.
- Collect mass spectra between 250-2000 m/z with top-10 data dependent fragmentation acquisition.
- Between samples, rinse capillary with 0.1 M HCl for 3 min at 100 psi and 10 min 100 psi of BGE for separation capillary and for 3 min at 100 psi for the conductive liquid capillary.
8. CESI-MS for metabolite corona analysis
NOTE: All samples must be analyzed twice, once in positive mode for the detection of cations and once in negative mode for the detection of anions. The control plasma samples need to be analyzed at least 5 times.
- Resuspend NM exposed and unexposed plasma samples from step 5.4 in 430 µL of DI water and vigorously vortex for 2 min. Filter through a 0.1 µm membrane filter.
- Take 95 µL of filtrate and add 5 µL of internal standards at 200 µM for cations (L-methionine sulfone) and 400 µM for anions (2,2,4,4-D4-citric acid) and vortex vigorously. Centrifuge at 4,000 x g at 4 ˚C for 10 min prior to CESI-MS analysis.
- Inject sample hydrodynamically for 30 s (cations) and 40 s (anions) at 2 psi.
- Separate metabolites in 10% acetic acid with a separation voltage of 30 kV in forward (cations) and reverse (anions) mode. Reverse separation mode is assisted with 0.5 psi forward pressure on the separation capillary.
- Set mass spectrometer to collect data in positive (cations) or negative (anions) mode over the mass range 65-1000 m/z. Between samples, rinse capillary with 0.1 M HCl, 0.1 M NaOH, DI water and BGE at 50 psi for 2 min.
Representative Results
The described method is the first to characterize both the proteins and metabolites in the NM biomolecular corona using the same human plasma sample. This method is capable of detecting >200 proteins and >150 metabolites, thus enabling the most comprehensive overview of the complete biomolecular corona to be determined, enabling enhanced understanding of NMs cellular attachment, uptake and impacts.
The use of CESI-MS for both proteomics (Figure 1) and metabolomics (Figure 2) separation and detection shows good separation windows on both capillaries for each approach.
This method is capable of distinguishing between the proteins in the corona across a wide range of NMs compositions, thus demonstrating the methods’ applicability to NMs across a broad range of physical and chemical characteristics (Table 1).
The unique fingerprints of the metabolite corona can also be distinguished using the metabolomic branch of this methodology (Figure 3). Although this approach utilizes a passive approach to characterize the NM metabolite corona it is still capable of uncovering interesting insights into the role of metabolites in the biomolecular corona such as the differential adsorption of isomers (Figure 4).
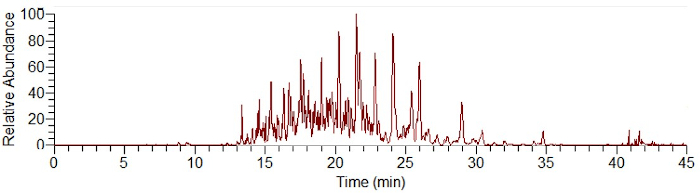
Figure 1: Exemplar electropherogram of a SiO2 protein corona separation using CESI-MS following an on-particle digest. Please click here to view a larger version of this figure.
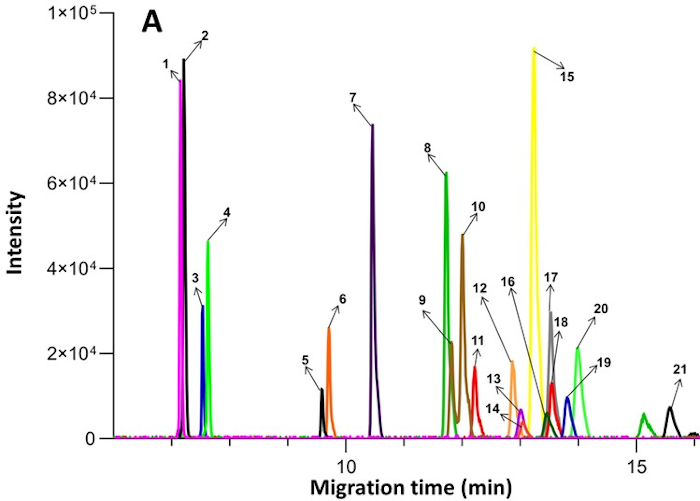
Figure 2: Example of extracted ion electropherograms obtained from endogenous compounds in plasma by CE-MS. The numbered compounds are as follows: 1. Ornithine; 2. Lysine; 3. Arginine; 4. Histidine; 5. Creatine; 6. Glycine; 7. Alanine; 8. Valine; 9. Isoleucine; 10. Leucine; 11. Serine; 12. Threonine; 13. Asparagine; 14. Methionine; 15. Glutamine; 16. Glutamic acid; 17. Phenyl-D5-alanine; 18. Phenylalanine; 19. Tyrosine; 20. Proline; 21. Methionine sulfone (internal standard). Published under an open access Creative Commons CC BY license and reproduced from Zhang et al17. Please click here to view a larger version of this figure.
Table 1: Analysis of protein corona across an array of 6 NMs. Heat map of protein classes identified on silica (SiO2), titania (TiO2), PVP capped titania (TiO2-PVP), dispex capped titania (TiO2-Dispex), polystyrene and carboxylated polystyrene NMs. Percentages refer to the proteins’ abundance relative to total protein content based upon top 3 peptides abundance for each protein. Given the differences observed in the relative abundances of proteins in the different NM coronas, it is clear that this proposed protocol is sufficiently sensitive to differentiate between the protein corona of different NMs. Figure reproduced from a CESI-MS analysis of the protein corona, published under an open access Creative Commons CC BY license11. Please click here to download this table.
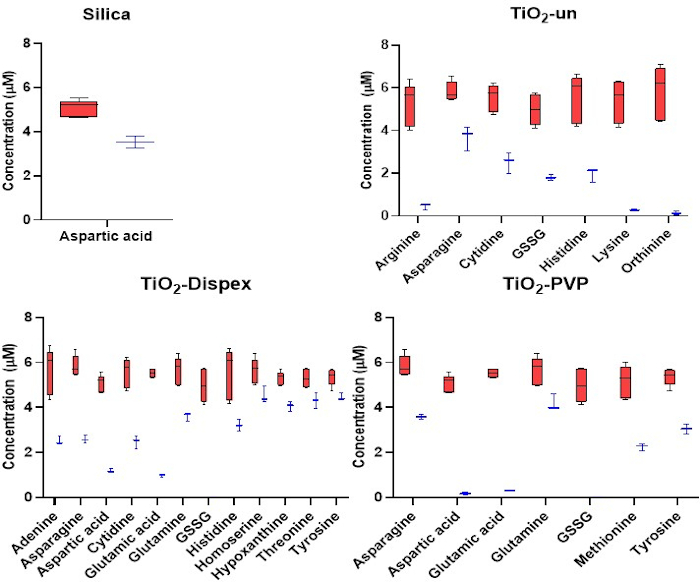
Figure 3: Targeted analysis of the cationic metabolites in the NM corona. Adsorption of cations to SiO2 and TiO2 NMs. Red refers to the initial concentration of metabolites whereas blue refers to the concentration of metabolites following a 1 h incubation with the NMs at 37 ˚C. The reduction in metabolite concentration in the solution is a result of metabolite adsorption to the NM surface. No significant adsorption of metabolites was observed for the polystyrene NMs. Box plots represent the minimum and maximum values, median and interquartile ranges. Figure reproduced from a CESI-MS targeted analysis of the metabolite corona published under an open access Creative Commons CC BY license15. Please click here to view a larger version of this figure.
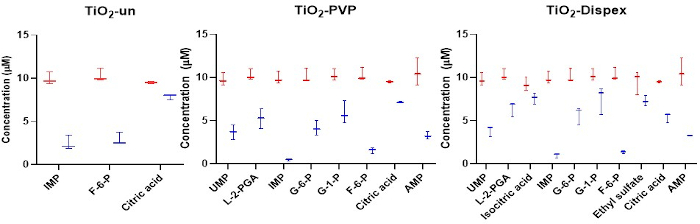
Figure 4: Targeted analysis of the anionic metabolites in the NM corona with a focus on isomer adsorption. Adsorption of anions to a range of titania NMs shows clear differences between various isomers such as the sugar phosphates. Red refers to the initial concentration of metabolites whereas blue refers to the concentration of metabolites following a 1 h incubation with the NM at 37 ˚C. The reduction in metabolite concentration in solution is a result of metabolite adsorption to the NM surface. No significant adsorption of metabolites was observed for the SiO2 and the polystyrene NMs. Box plots represent the minimum and maximum values, median and interquartile ranges. Figure reproduced from a CESI-MS targeted analysis of the metabolite corona published under an open access Creative Common CC BY license15. Please click here to view a larger version of this figure.
| Analyte | Mean RSD peak area | Mean RSD migration time |
| 1928 peptides intraday (n=3) | 15% | 0.30% |
| 27 cations intra-day (n=16) | 5.80% | 2.20% |
| 27 cation inter-day (n=36) | 8.60% | 1.80% |
Table 2: Reproducibility of CESI-MS technical replicates for peptides and cationic metabolites. RSD: relative standard deviation, as a measure of reproducibility. The reproducibility of CESI-MS in-terms of peak areas and migration times is very good, with all analytes having a mean RSD <15% and migration times <2.2%. These results demonstrate the high degree of confidence that can be attributed to the data collected using this technique and confirm that variations detected are due to genuine differences in sample composition (enrichment on NMs) rather than analytical variation. Table 2 is generated from results presented previously11,15.
Discussion
This is the first method proposed to characterize the complete biomolecular corona incorporating proteins and metabolites, from the same sample, using CESI-MS. The ability to characterize both the proteins and metabolites from the same sample will significantly increase the amount of data collected from a single experimental exposure enabling new insights into the process of corona formation and its impacts for NMs toxicity and efficacy as nanomedicines. Furthermore, CESI-MS is an ideal platform to characterize both classes of biomolecules with just a change of capillary required. Moving forward, new methods developed for non-aqueous CE will enable lipidomics to be performed using CE-MS40 thus it is now possible to analyze proteins, metabolites and lipids using a single platform. The current conventional approaches would require two dedicated LC-MS platforms, one for the protein corona and one for the metabolite corona. Thus, this approach can collect twice as much data using a single analytical platform.
There are some caveats to this approach particularly with the metabolite corona as this protocol proposes an indirect measurement of the corona. As such, problem may arise when the metabolite levels are present at high concentrations relative to the NMs and the subsequent drop in metabolite concentration in the matrix is small. However, unlike with the protein corona, it is unlikely that a “one size fits all” approach to isolating the metabolite corona will be developed due to each NM having unique surface chemistries. Going forward it is more likely that NM specific approaches to isolate the metabolite corona will be developed and validated however; this will be applicable to only that specific NM. Despite this, the CESI-MS will still be a highly suitable platform for the characterization of the polar charged metabolites regardless of how they are isolated. Also noteworthy is that if a pellet does not form during the centrifugation steps designed to pellet the NMs, a higher centrifugal force may be required; this is most likely to occur with less dense NMs. It is also critical that all filtering steps are adhered to within the protocol due to the narrow bore of the capillaries these can easily become blocked if any particulate matter has not been adequately eliminated from the sample.
While the protein corona has been an intense area of research for a number of years the ability to combine that with the metabolite corona is a new field of interest for scientists investigating the bio-nano interface6,14. To date, the majority of these studies have focused upon the non-polar, lipid fraction of the metabolite corona9,28. This current method enables the characterization of the highly polar and charged metabolites in the corona which includes important metabolites involved in energy metabolism, glycolysis, and DNA synthesis. As a result, when combined with the proteomics workflow, a more holistic characterization of the biomolecular corona is possible. This enables a more in-depth analysis of bio-nano interactions moving forward. This method enables enhanced mechanistic insights into receptor-mediated endocytosis via protein and metabolite interactions with membrane receptors. Furthermore, inter and intra corona interactions between proteins and small molecules may be explored; for example, it has been recently shown that the proteins in the corona play a key role in the recruitment of metabolites15. It is worth noting that the representative results in Figure 3 and Figure 4 are absolute quantification of metabolites, however, an untargeted approach using multivariate data analysis to compare all CE-MS features in controls compared to exposed plasma would also elucidate metabolite binding. Thus, there is a significant scope to investigate how, and which, metabolites and proteins influence their respective recruitment into NM coronas. These additional studies may help design coronas to optimize delivery of nanomedicines or uncover further hurdles to their development such as the suppression of pharmaceutical action because of the biomolecular corona blocking or masking targeting functionalities.
CESI-MS with its nano scale flow rates of around 20 nL/min and minimal use of organic solvents offers an improvement in green and economic credentials over standard LC-MS and nanoLC-MS in terms of solvent use, the main challenges for metabolomics and proteomics studies, respectively. CESI-MS offers highly reproducible results for both migration time and peak area which compare favorably with LC-MS based methods11,15. Furthermore, compared to nanoLC-MS, carry-over is not an issue in CESI-MS. Therefore, extensive system clean-up between sample analyses is not required, which greatly improves sample throughput11.
In summary, the proposed workflow is the first to detail an approach to characterize both the protein and metabolite components of the complete biomolecular corona of NMs. This unlocks a new aspect of bio-nano interactions, the metabolite corona, thus enabling the collection of twice as much data from the same sample, enabling a more complete understanding of interactions at the bio-nano interface.
Disclosures
The authors have nothing to disclose.
Acknowledgements
A.J.C, J.A.T and I.L acknowledge funding from the European Commission via Horizon 2020 project ACEnano (Grant no. H2020-NMBP-2016-720952). W.Z acknowledges PhD funding from the China Scholarship Council (CSC, No. 201507060011). R.R acknowledges the financial support of the Vidi grant scheme of the Netherlands Organization of Scientific Research (NWO Vidi 723.0160330).
Materials
| 1,4-Dithiothreitol | Sigma | 10197777001 | |
| 2,2,4,4-D4-citric | Simga | 485438-1G | Internal standard anion |
| Ammonium Acetate >98% | Sigma | A1542 | |
| Ammonium Bicarbonate >99.5% | Sigma | 09830 | |
| Capillary cartridge coolant | Sciex | 359976 | |
| CESI 8000 instrument | Sciex | A98089 | |
| CESI Cap | Sciex | B24699 | |
| CESI Vials | Sciex | B11648 | |
| Chloroform | Sigma | 650498 | Toxic, use in a fume hood |
| Glacial Acetic acid | Sigma | A6283 | |
| Hydrochloric acid 1mol/L | Sigma | 43617 | |
| Iosoacetamide | Sigma | I1149 | |
| L-methionine sulfone | Sigma | M0876-1G | Internal standard cation |
| Methonal (LC-MS Ultra Chromasolve) | Sigma | 14262 | Use in a fume hood |
| Microvials | Sciex | 144709 | |
| nanoVials | Sciex | 5043467 | |
| Neutral Surface Cartridge 30 µM ID x 90 cm total length | Sciex | B07368 | |
| OptiMS Adapter for Sciex Nanospray III source | Sciex | B07363 | B07366 and B83386 for Thermo mass spectrometers, B85397 for Waters mass spectrometers, B86099 for Bruker mass spectrometers |
| OptiMS Fused-Silica Cartridge 30 µm ID x 90 cm total length | Sciex | B07367 | |
| Phospahte buffered saline 10x | Sigma | P7059 | |
| Pierce C18 Tips, 100 µL bed | Thermo Fisher Scientific | 87784 | |
| Polystyrene 100 nm | Polysciences Inc | 876 | |
| Polystyrene carboxylated 100 nm | Polysciences Inc | 16688 | |
| Polystyrene carboxylated 1000 nm | Polysciences Inc | 08226 | |
| Rapigest SF (surfactant) | Waters | 186001860 | |
| Sequencing Grade modified Trypsin | Promega | V5111 | |
| SiO2 Ludox TM-40 | Sigma | 420786 | |
| Sodium Hydroxide solution | Simga | 72079 | |
| TiO2 Dispex coated | Promethion particles | Bespoke particles | |
| TiO2 PVP coated | Promethion particles | Bespoke particles | |
| TiO2 uncoated | Promethion particles | Bespoke particles |
References
- Ke, P. C., Lin, S., Parak, W. J., Davis, T. P., Caruso, F. A Decade of the Protein Corona. ACS Nano. 11 (12), 11773-11776 (2017).
- Sasidharan, A., Riviere, J. E., Monteiro-Riviere, N. A. Gold and silver nanoparticle interactions with human proteins: impact and implications in biocorona formation. Journal of Materials Chemistry B. 3 (10), 2075-2082 (2015).
- Albanese, A., et al. Secreted biomolecules alter the biological identity and cellular interactions of nanoparticles. ACS Nano. 8 (6), 5515-5526 (2014).
- Lynch, I., Dawson, K. A. Protein-nanoparticle interactions. Nano Today. 3 (1-2), 40-47 (2008).
- Tenzer, S., et al. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nature Nanotechnology. 8 (10), 772-781 (2013).
- Chetwynd, A. J., Lynch, I. The rise of the nanomaterial metabolite corona, and emergence of the complete corona. Environmental Science: Nano. 7 (4), 1041-1060 (2020).
- Lee, J. Y., et al. Analysis of lipid adsorption on nanoparticles by nanoflow liquid chromatography-tandem mass spectrometry. Analytical and Bioanalytical Chemistry. , 1-10 (2018).
- Pink, M., Verma, N., Kersch, C., Schmitz-Spanke, S. Identification and characterization of small organic compounds within the corona formed around engineered nanoparticles. Environmental Science: Nano. 5 (6), 1420-1427 (2018).
- Chetwynd, A. J., Wheeler, K. E., Lynch, I. Best practice in reporting corona studies: Minimum information about Nanomaterial Biocorona Experiments (MINBE). Nano Today. 28, 100758 (2019).
- Zhang, H., et al. Quantitative proteomics analysis of adsorbed plasma proteins classifies nanoparticles with different surface properties and size. Proteomics. 11 (23), 4569-4577 (2011).
- Faserl, K., Chetwynd, A. J., Lynch, I., Thorn, J. A., Lindner, H. H. Corona isolation method matters: Capillary electrophoresis mass spectrometry based comparison of protein corona compositions following on-particle versus in-solution or in-gel digestion. Nanomaterials. 9 (6), 898 (2019).
- Nasser, F., Lynch, I. Secreted protein eco-corona mediates uptake and impacts of polystyrene nanoparticles on Daphnia magna. Journal of Proteomics. 137, 45-51 (2016).
- Hellstrand, E., et al. Complete high-density lipoproteins in nanoparticle corona. FEBS Journal. 276 (12), 3372-3381 (2009).
- Grintzalis, K., Lawson, T. N., Nasser, F., Lynch, I., Viant, M. R. Metabolomic method to detect a metabolite corona on amino functionalised polystyrene nanoparticles. Nanotoxicology. , 1-12 (2019).
- Chetwynd, A. J., Zhang, W., Thorn, J. A., Lynch, I., Ramautar, R. The nanomaterial metabolite corona determined using a quantitative metabolomics approach: A pilot study. Small. , (2020).
- Chetwynd, A. J., Guggenheim, E. J., Briffa, S. M., Thorn, J. A., Lynch, I., Valsami-Jones, E. Current application of capillary electrophoresis in nanomaterial characterisation and its potential to characterise the protein and small molecule corona. Nanomaterials. 8 (2), 99 (2018).
- Zhang, W., et al. Assessing the suitability of capillary electrophoresis-mass spectrometry for biomarker discovery in plasma-based metabolomics. Electrophoresis. , 00126 (2019).
- Ramautar, R., Somsen, G. W., de Jong, G. J. CE-MS for metabolomics: Developments and applications in the period 2016-2018. Electrophoresis. 40 (1), 165-179 (2019).
- Sarg, B., Faserl, K., Lindner, H. H. Identification of novel site-specific alterations in the modification level of myelin basic protein isolated from mouse brain at different ages using capillary electrophoresis-mass spectrometry. Proteomics. 17 (19), 1700269 (2017).
- Faserl, K., Sarg, B., Maurer, V., Lindner, H. H. Exploiting charge differences for the analysis of challenging post-translational modifications by capillary electrophoresis-mass spectrometry. Journal of Chromatography A. 1498, 215-223 (2017).
- Zhang, Z., Qu, Y., Dovichi, N. J. Capillary zone electrophoresis-mass spectrometry for bottom-up proteomics. TrAC – Trends in Analytical Chemistry. 108, 23-37 (2018).
- Shen, X., et al. Capillary zone electrophoresis-mass spectrometry for top-down proteomics. TrAC – Trends in Analytical Chemistry. 120, 115644 (2019).
- Ramautar, R., Busnel, J. M., Deelder, A. M., Mayboroda, O. A. Enhancing the coverage of the urinary metabolome by sheathless capillary electrophoresis-mass spectrometry. Analytical Chemistry. 84 (2), 885-892 (2012).
- Faserl, K., Kremser, L., Müller, M., Teis, D., Lindner, H. H. Quantitative proteomics using ultralow flow capillary electrophoresis-mass spectrometry. Analytical Chemistry. 87 (9), 4633-4640 (2015).
- Faserl, K., Sarg, B., Kremser, L., Lindner, H. Optimization and evaluation of a sheathless capillary electrophoresis-electrospray ionization mass spectrometry platform for peptide analysis: comparison to liquid chromatography-electrospray ionization mass spectrometry. Analytical Chemistry. 83 (19), 7297-7305 (2011).
- Segers, K., et al. CE-MS metabolic profiling of volume-restricted plasma samples from an acute mouse model for epileptic seizures to discover potentially involved metabolomic features. Talanta. 217, 121107 (2020).
- Chetwynd, A. J., David, A. A review of nanoscale LC-ESI for metabolomics and its potential to enhance the metabolome coverage. Talanta. 182, 380-390 (2018).
- Drouin, N., et al. Capillary electrophoresis-Mass spectrometry at trial by metabo-ring: Effective electrophoretic mobility for reproducible and robust compound annotation. Analytical Chemistry. 92 (20), 14103-14112 (2020).
- Faria, M., et al. Minimum information reporting in bio-nano experimental literature. Nature Nanotechnology. 13 (9), 777-785 (2018).
- Leong, H. S., et al. On the issue of transparency and reproducibility in nanomedicine. Nature Nanotechnology. 14 (7), 629-635 (2019).
- Kaur, I., et al. Dispersion of nanomaterials in aqueous media: Towards protocol optimization. Journal of Visualized Experiments. (130), e56074 (2017).
- Bihari, P., et al. Optimized dispersion of nanoparticles for biological in vitro and in vivo studies. Particle and Fibre Toxicology. 5 (1), 1-14 (2008).
- Taurozzi, J. S., Hackley, V. A., Wiesner, M. R. Ultrasonic dispersion of nanoparticles for environmental, health and safety assessment – and recommendations. Nanotoxicology. 5 (4), 711-729 (2011).
- Lu, P. J., et al. Methodology for sample preparation and size measurement of commercial ZnO nanoparticles. Journal of Food and Drug Analysis. 26 (2), 628-636 (2018).
- Langevin, D., et al. Inter-laboratory comparison of nanoparticle size measurements using dynamic light scattering and differential centrifugal sedimentation. NanoImpact. 10, 97-107 (2018).
- Lee, S., Bi, X., Reed, R. B., Ranville, J. F., Herckes, P., Westerhoff, P. Nanoparticle size detection limits by single particle ICP-MS for 40 elements. Environmental Science and Technology. 48 (17), 10291-10300 (2014).
- Hole, P., et al. Interlaboratory comparison of size measurements on nanoparticles using nanoparticle tracking analysis (NTA). Journal of Nanoparticle Research. 15 (12), 2101 (2013).
- Zhang, W., Gulersonmez, M. C., Hankemeier, T., Ramautar, R. Sheathless capillary electrophoresis-mass spectrometry for metabolic profiling of biological samples. Journal of Visualized Experiments. , e54535 (2016).
- Faserl, K., Sarg, B., Lindner, H. H. Application of CE-MS for the analysis of histones and histone modifications. Methods. , (2020).
- Azab, S., Ly, R., Britz-Mckibbin, P. Robust method for high-throughput screening of fatty acids by multisegment injection-nonaqueous capillary electrophoresis-mass spectrometry with Sstringent quality control. Analytical Chemistry. 91 (3), 2329-2336 (2019).
- Zhang, X., Pandiakumar, A. K., Hamers, R. J., Murphy, C. J. Quantification of lipid corona formation on colloidal nanoparticles from lipid vesicles. Analytical Chemistry. 19 (24), 14387-14394 (2018).

