Assessment of Submitochondrial Protein Localization in Budding Yeast Saccharomyces cerevisiae
Summary
Despite recent advances, many yeast mitochondrial proteins still remain with their functions completely unknown. This protocol provides a simple and reliable method to determine the submitochondrial localization of proteins, which has been fundamental for the elucidation of their molecular functions.
Abstract
Despite recent advances in the characterization of yeast mitochondrial proteome, the submitochondrial localization of a significant number of proteins remains elusive. Here, we describe a robust and effective method for determining the suborganellar localization of yeast mitochondrial proteins, which is considered a fundamental step during mitochondrial protein function elucidation. This method involves an initial step that consists of obtaining highly pure intact mitochondria. These mitochondrial preparations are then subjected to a subfractionation protocol consisting of hypotonic shock (swelling) and incubation with proteinase K (protease). During swelling, the outer mitochondrial membrane is selectively disrupted, allowing the proteinase K to digest proteins of the intermembrane space compartment. In parallel, to obtain information about the topology of membrane proteins, the mitochondrial preparations are initially sonicated, and then subjected to alkaline extraction with sodium carbonate. Finally, after centrifugation, the pellet and supernatant fractions from these different treatments are analyzed by SDS-PAGE and western blot. The submitochondrial localization as well as the membrane topology of the protein of interest is obtained by comparing its western blot profile with known standards.
Introduction
Mitochondria are essential organelles of eukaryotic cells that play crucial roles in bioenergetics, cellular metabolism, and signaling pathways1. To properly execute these tasks, mitochondria rely on a unique set of proteins and lipids responsible for their structure and function. The budding yeast Saccharomyces cerevisiae has been widely used as a model system for investigations on mitochondrial processes, as well as for other organelles2. The mitochondrial genome codes for only eight proteins in yeast; the vast majority of mitochondrial proteins (~99%) are encoded by nuclear genes, which are translated on cytosolic ribosomes, and subsequently imported into their correct submitochondrial compartments by sophisticated protein import machineries3,4,5. Thus, mitochondrial biogenesis depends on the coordinated expression of both the nuclear and mitochondrial genomes6,7. Genetic mutations causing defects in mitochondrial biogenesis are associated with human diseases8,9,10.
In the past two decades, high-throughput proteomic studies targeting highly-purified mitochondria resulted in a comprehensive characterization of yeast mitochondrial proteome, which has been estimated to be composed of at least 900 proteins11,12,13,14. Although these studies provided valuable information, the suborganellar localization of each protein in the four mitochondrial subcompartments, namely, the outer membrane (OM), intermembrane space (IMS), inner membrane (IM), and matrix, is still required. This question was partially addressed with proteomic-wide studies of the two smaller mitochondrial subcompartments (OM and IMS)15,16. More recently, Vögtle and collaborators made a major step forward by generating a high-quality global map of submitochondrial protein distribution in yeast. Using an integrated approach combining SILAC-based quantitative mass spectrometry, different submitochondrial fractionation protocols, and the data set from the OM and IMS proteomes, the authors assigned 818 proteins into the four mitochondrial subcompartments13.
Despite the advances achieved by these high-throughput proteomic studies, our knowledge about the submitochondrial proteome composition is far from being complete. Indeed, among 986 proteins reported by Vögtle and collaborators as being localized into yeast mitochondria, 168 could not be assigned in any of the four submitochondrial compartments13. Moreover, the authors did not provide information about the membrane topology of proteins that were predicted to be peripherally attached to the periphery of mitochondrial membranes. For example, it is not possible to know if a protein that was assigned as peripherally attached to the inner membrane is facing the matrix or the intermembrane space. Apart from these missing data from the proteome-wide studies, there are conflicting information about the suborganellar localization of a significant number of mitochondrial proteins. One example is the protease Prd1, which has been assigned as an intermembrane space protein in the common databases such as Saccharomyces Genome Database (SGD) and Uniprot. Surprisingly, using a subfractionation protocol similar to that described here, Vögtle and collaborators clearly showed that Prd1 is a genuine matrix protein13. As mentioned above, the submitochondrial localization of many mitochondrial proteins needs to be elucidated or reevaluated. Here, we provide a simple and reliable protocol to determine the suborganellar localization of yeast mitochondrial proteins. This protocol was developed and optimized by various research groups and has been routinely used to determine the submitochondrial localization, as well as the membrane topology of many mitochondrial proteins.
Protocol
1. Growth of yeast cells
- Isolate single colonies of the strain of interest by streaking a small portion of the cells from a -80 °C glycerol stock onto a YPD (1% yeast extract, 2% peptone, 2% glucose) agar plate. Incubate the plate at 30 °C for 2-3 days.
NOTE: The S. cerevisiae strain used in this protocol is derived from BY4741 (MATα; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0). With the exception of the auxotrophic marker genes, this strain does not contain any deleted gene and does not carry any plasmid. Thus, it can be successfully cultivated in a rich medium, stimulating vigorous cell growth. When working with strains transformed with plasmids, use the appropriate minimal medium for plasmid selection. - Prepare a starter culture by inoculating 2-3 individual colonies from YPD agar plate in 10-20 mL of YPGal medium (1% yeast extract, 2% peptone, 2% galactose) in a 100 mL Erlenmeyer flask. Incubate at 30 °C for 24 h with vigorous shaking.
NOTE: The choice of the growth medium depends on the yeast strain used in the protocol. Both YPD and YPGal contain fermentable carbon sources, which allow the growth of strains that do not perform mitochondrial respiration. However, since glucose represses the expression of many mitochondrial genes, it is not recommended to use this carbon source since it will produce lower amounts of mitochondria. When working with respiratory competent strains that can respire, it is also possible to use carbon sources such as glycerol and ethanol in an attempt to obtain a higher yield of mitochondria. - Dilute the starter culture into 1 L of fresh YPGal medium to an OD600 less than 0.1. Cultivate the cells at 30 °C with vigorous shaking until OD600 reaches 1-1.5.
NOTE: Determine the growth rate (doubling time) for each yeast strain before performing the experiment. This will provide an accurate estimate of the time of incubation required for the culture to reach an OD600 of 1-1.5.
2. Isolation of highly purified mitochondria
NOTE: This protocol is adapted from17, with minor modifications.
- Harvest the cells by centrifugation at 3,000 x g for 5 min at room temperature.
- Wash the cells with distilled water and collect them by centrifugation at 3,000 x g for 5 min at room temperature.
- Determine the wet weight of the cells.
NOTE: The easiest way to measure the weight of the cell pellet from step 2.2 is to determine the weight of the empty centrifuge tube just before the collection of the cells. After centrifugation, discard the supernatant and measure the weight of the same tube with the cells. The weight of the cells is the difference between the two measurements. - Resuspend the cells in DTT buffer (2 mL per 1 g of cells) using a Pasteur pipette or P5000 tip. See Table 1 for DDT buffer composition.
- Incubate the cells at 30 °C for 20 min with gentle shaking (~70 rpm).
- Centrifuge at 3,000 x g for 5 min at room temperature to pellet the cells.
- Wash the cells with Zymolyase buffer without the enzyme (about 7 mL per 1 g of cells).
See Table 1 for Zymolyase buffer composition. - Centrifuge at 3,000 x g for 5 min at room temperature to pellet the cells.
- Resuspend the cells in the buffer without Zymolyase (7 mL per 1 g of cells).
- Transfer the cell suspension to a 250 mL Erlenmeyer flask and add Zymolyase-20T (3 mg per g wet weight).
- Incubate the cells at 30 °C for 30-40 min with gentle shaking (~70 rpm). Check the efficiency of this process by comparing the turbidity of the cell suspension before and after Zymolyase treatment.
NOTE: In this step, the cells will be converted into spheroplasts due to cell-wall digestion by Zymolyase.- For this, add 50 µL of each cell suspension to separate glass tubes containing 2 mL of water. After mixing vigorously, the turbidity of the cell suspension treated with Zymolyase should rapidly decrease due to the osmotic disruption of spheroplasts. On the other hand, the turbidity of the non-treated cell suspension should remain unchanged.
NOTE: The effects of turbidity can also be monitored by simple visual inspection or by measuring the OD600 of both samples. In the second case, the OD600 of the Zymolyase treated sample should be 10%-20% of the non-treated sample. An alternate method involves counting the cells in both samples by using light microscopy. - If the yield of spheroplasts formation is low, add more Zymolyase and incubate for a further 15 min interval.
- For this, add 50 µL of each cell suspension to separate glass tubes containing 2 mL of water. After mixing vigorously, the turbidity of the cell suspension treated with Zymolyase should rapidly decrease due to the osmotic disruption of spheroplasts. On the other hand, the turbidity of the non-treated cell suspension should remain unchanged.
- Harvest the spheroplasts by centrifugation at 2500 x g for 5 min at 4 °C.
NOTE: All the further steps should be carried out fast and on ice or at 4 °C to avoid protein degradation by hydrolytic enzymes. - Wash the spheroplasts twice with ice-cold homogenization buffer (about 6.5 mL per 1 g of cells) and pellet by centrifuging at 2,500 x g for 5 min at 4 °C. See Table 1 for homogenization buffer composition.
- Resuspend the spheroplasts in ice-cold homogenization buffer (6.5 mL per 1 g of cells) and transfer it to a pre-chilled glass Dounce homogenizer. Use a large glass homogenizer of approximately 30 mL.
- Homogenize the spheroplasts with 15 strokes using a pestle.
NOTE: The number of strokes should be adjusted depending on the pestle fitting. For tight pestle, 15 strokes are sufficient. On the other hand, if using a loose pestle, it is recommended to perform up to 25 strokes. - Transfer the homogenate to a 50 mL centrifuge tube and add 1 volume of ice-cold homogenization buffer.
- Centrifuge the homogenate at low speed, 1,500 x g for 5 min at 4 °C to pellet nuclei, cell debris, and unbroken cells.
- Transfer the supernatant to a new 50 mL centrifuge tube using a Pasteur pipette or P5000 tip taking care to avoid disrupting the pellet.
- Centrifuge at 4,000 x g for 5 min at 4 °C.
- Transfer the supernatant to a high-speed centrifuge tube, and centrifuge at 12,000 x g for 15 min at 4 °C to pellet the crude mitochondria fraction.
- Discard the supernatant and gently wash the crude mitochondrial pellet in 20-30 mL ice-cold homogenization buffer by gentle pipetting using a P5000 tip.
- Transfer the suspension to a 50 mL centrifuge tube and centrifuge at 4,000 x g for 5 min at 4 °C to pellet the remaining cell debris.
- Transfer the supernatant to a high-speed centrifuge tube, and centrifuge at 12,000 x g for 15 min at 4 °C to pellet the crude mitochondria fraction.
- Discard the supernatant and gently resuspend the crude mitochondrial pellet in a small volume (typically 1000 µL) of ice-cold SEM buffer by gentle pipetting using a P1000 tip. See Table 1 for SEM buffer composition.
NOTE: Although this crude mitochondrial fraction can be used directly in some applications such as in organello protein import assays, it contains substantial amounts of other cellular components. These contaminations might lead to misinterpretations of the results when determining the submitochondrial localization of a protein. Therefore, further purification steps are required to get highly purified mitochondrial preparation, as described below. - Prepare sucrose solutions in the EM buffer at concentrations of 60%, 32%, 23%, and 15% (w/v). These solutions are stable for up to 1 month at 4 °C. See Table 1 for EM buffer composition.
- Prepare a 4-step sucrose gradient in an ultracentrifuge tube as follows: Place 1.5 mL of 60% (w/v) sucrose onto the bottom of the centrifuge tube. Next, pipette carefully stepwise: 4 mL of 32%, 1.5 mL of 23%, and 1.5 mL of 15% sucrose (w/v). Take care to avoid disrupting the phases.
- Carefully load the crude mitochondrial fraction on top of the sucrose gradient.
- Centrifuge for 1 h at 134,000 x g at 4 °C in a swinging bucket rotor.
- Carefully keep removing the sucrose solution until the highly purified mitochondria fraction is reached which is represented by a brown band at the 60%/32% sucrose interface.
- Recover the purified mitochondria using a P1000 cut tip and place it into a pre-chilled high-speed centrifuge tube.
- Dilute the recovered mitochondria with 5-10 volumes of ice-cold SEM buffer.
- Centrifuge for 30 min at 12,000 x g at 4 °C.
- Resuspend the pure mitochondria in 500 µL of ice-cold SEM buffer by gentle pipetting using a P1000 cut tip.
- Determine the protein concentration of the highly purified mitochondrial preparation using the Bradford procedure following the manufacturer's instructions. Adjust the protein concentration to 10 mg of protein/mL with ice-cold SEM buffer.
NOTE: For the submitochondrial fractionation protocol described below, it is recommended to use freshly prepared mitochondria. However, Vögtle and collaborators performed a detailed quality control analysis of the mitochondrial intactness and showed that frozen organelles can also be used in this protocol13. For this, make aliquots of 40 µL and freeze them in liquid nitrogen. Store at -80 °C.
3. Submitochondrial fractionation protocol
NOTE: This protocol is adapted from reference18 and is composed of two steps: (1) hypotonic swelling in the presence or absence of proteinase K, and (2) sonication followed by carbonate extraction. Perform all the steps of both the protocols on ice or at 4 °C to avoid protein degradation.
- Hypotonic swelling in the presence of proteinase K
- Transfer 40 µL of highly purified mitochondria at 10 mg/mL (400 µg) into four 1.5 mL pre-chilled labeled microcentrifuge tubes.
- Add 360 µL of SEM buffer in tubes 1 and 2.
- Add 360 µL of EM buffer in tubes 3 and 4.
- Add 4 µL of proteinase K (10 mg/mL) in tubes 2 and 4. Use the pipetting scheme listed in Table 2 to avoid mistakes.
NOTE: Prepare a 10 mg/mL solution of proteinase K in water immediately before use. The final concentration of proteinase K in the experiment should be around 50-100 µg/mL. Please see the Discussion section for further details. - Mix all the tubes gently and incubate on ice for 30 min with occasional mixing.
- Add 4 µL of 200 mM PMSF to all the four tubes to stop proteinase K activity.
CAUTION: PMSF is highly toxic. Wear gloves when working with solutions containing PMSF. - Centrifuge at 20,000 x g for 30 min at 4 °C.
- Collect the supernatant and transfer it to a new 1.5 mL pre-chilled labeled microcentrifuge tube. Take care to avoid disrupting the pellet.
NOTE: The pellet can be directly resuspended in the sample buffer for further SDS-PAGE and western blot analysis. However, traces of proteinase K can remain active even after PMSF treatment and eventually can digest some proteins after the pellet has been dissolved in the SDS-containing sample buffer. To avoid this problem, proteinase K could be completely inactivated by treatment of the samples with trichloroacetic acid (TCA) as described below.
CAUTION: TCA is highly toxic. Wear gloves when working with solutions containing TCA. - Resuspend the pellet from step 3.1.8 in 400 µL of ice-cold SEM buffer.
- Precipitate the supernatant (from step 3.1.8) and the resuspended pellet (from step 3.1.9) with TCA to a final concentration of 10% (w/v).
- Incubate all the tubes on ice for 10 min.
- Centrifuge the TCA-treated samples for 10 min at 12,000 x g at 4 °C.
- Remove the supernatant and resuspend the pellet in 200 µL of sample buffer.
NOTE: It is possible that the bromophenol blue pH indicator turns yellow because of the acid treatment. If this happens, add small aliquots (1-5 µL) of 1 M Tris base until it turns blue. - Add 4 µL of 200 mM PMSF to all the tubes.
- Store all the samples at -80 °C until further analysis by SDS-PAGE and western blot.
- Sonication and carbonate extraction
NOTE: In this protocol, it is not necessary to use freshly prepared mitochondria once the sonication causes the rupture of the mitochondrial membranes.- Transfer 200 µL of highly purified mitochondria at 10 mg/mL (2 mg protein) into a 1.5 mL pre-chilled microcentrifuge tube.
- Dilute mitochondria one-fold with ice-cold SEM buffer.
- Sonicate mitochondria for 3 x 30 s on ice. Use a sonicator compatible for small volumes.
- Centrifuge the sample for 30 min at 100,000 x g at 4 °C.
- Collect the supernatant and transfer it to a new 1.5 mL pre-chilled microcentrifuge tube. Keep it on ice. This sample will be named soluble protein fraction (S).
- Resuspend the pellet from step 3.2.4 in 400 µL of ice-cold SEM buffer.
- Take 100 µL of the resuspended pellet from step 3.2.6 and transfer it to a new 1.5 mL pre-chilled microcentrifuge tube. Keep it on ice. This sample will be named submitochondrial particles fraction (SMP).
- Dilute the remaining 300 µL from step 3.2.6 one-fold with freshly prepared 200 mM sodium carbonate.
- Incubate the sample from step 3.2.8 on ice for 30 min.
- Centrifuge the sample for 30 min at 100,000 x g at 4 °C.
- Collect the supernatant and transfer it to a new 1.5 mL pre-chilled microcentrifuge tube. Keep it on ice. This sample will be named carbonate supernatant fraction (CS).
- Resuspend the pellet from step 3.2.10 in 400 µL of ice-cold SEM buffer. This sample will be named carbonate precipitated fraction (CP).
- Precipitate all the samples (S, SMP, CS, and CP) with TCA to a final concentration of 10% (w/v).
- Incubate all the tubes on ice for 10 min.
- Centrifuge the TCA-treated samples for 10 min at 12,000 x g at 4 °C.
- Remove the supernatant and resuspend each pellet in the sample buffer. If the sample buffer becomes yellow, add small aliquots (1-5 µL) of 1 M Tris base until it turns blue.
- Add 1 µL of 200 mM PMSF to all the tubes.
- Store all the samples at -80 °C until further analysis by SDS-PAGE and western blot.
Representative Results
The success of submitochondrial fractionation protocol depends on obtaining highly purified intact mitochondria. For this, it is essential that during the yeast cell lysis, the intactness of the organelles remains almost totally preserved. This is achieved by using a cell lysis protocol that combines the enzymatic digestion of the cell wall followed by physical disruption of the plasma membrane by using a Dounce homogenizer. The mitochondrial contents are then collected by differential centrifugation. This subcellular fractionation yields an enriched mitochondrial fraction, as confirmed by the presence of high levels of porin (Por1), a mitochondrial marker protein (Figure 1, lane 2). However, this is a crude mitochondria fraction, which contains substantial amounts of other cellular compartments, including endoplasmic reticulum, vacuole, cytosol, and endosome (Figure 1, lane 2). These contaminations may introduce artifacts in some applications, such as submitochondrial protein localization experiments. To decrease the amount of these contaminations, the crude mitochondrial fraction is further purified on sucrose density gradient centrifugation. This additional purification step generates a highly pure mitochondrial fraction, as evidenced by a significant reduction in the contents of the protein markers for other cellular compartments (Figure 1, lane 3).
In order to determine the submitochondrial localization of proteins, the highly purified mitochondria are further fractionated into their subcompartments (Figure 2A). This protocol involves the conversion of mitochondria into mitoplasts by hypotonic osmotic shock. In this process, intact mitochondria are incubated in a hypoosmotic buffer resulting in swelling of the organelle. During swelling, the outer mitochondrial membrane is selectively ruptured by osmotic unbalance and the intermembrane space protein content is released into the supernatant. All this procedure is performed in the presence or absence of proteinase K. As a consequence of the outer membrane disruption, the protease gains access to intermembrane space protein content and promotes the degradation of the corresponding proteins. In contrast, the protein content of the mitochondrial matrix remains protected from attack of the protease due to the integrity of the inner mitochondrial membrane. After these treatments, the protein content of the different samples is evaluated by SDS-PAGE and western blot analyzes.
The efficient conversion of mitochondria to mitoplasts by osmotic shock (swelling) can be monitored in two ways: (1) disappearance of the soluble intermembrane space marker protein (e.g., cytochrome Cyt. b2) in the pellet fraction from mitoplasts with its concomitant appearance in the supernatant fraction (Figure 2B, compare lane 3 with lane 7); (2) selective degradation of the inner-membrane marker protein facing the intermembrane space (e.g., ScoI) by proteinase K only in mitoplasts (Figure 2B, lane 4). In addition, the protection of the markers Cyt. b2 and ScoI against proteinase K degradation in the pellet fraction from mitochondria are used to confirm the integrity of the outer mitochondrial membrane (Figure 2B, lane 2). On the other hand, the integrity of the inner mitochondrial membrane is confirmed by the protection of the matrix soluble protein marker α-KGD against proteinase K degradation (Figure 2B, lane 4). To determine the submitochondrial localization of a protein of interest, simply compare its western blot profile with the profiles of these standards with known localization.
In the case of the protein depicted in Figure 2B (Prx1), its western blot profile is indicative of a protein with dual mitochondrial localization: intermembrane space and matrix. At first glance, its fractionation profile is similar to α-KGD, indicating a matrix localization. However, its presence in the supernatant of mitoplasts also indicates an intermembrane space localization. The fractionation profiles of protein markers described above eliminate the possible artifacts associated with the integrity of the mitochondrial preparation and corroborate the dual localization of Prx119.
To investigate the topology of proteins on mitochondrial membranes, mitochondria are submitted to two additional treatments: sonication and carbonate extraction (Figure 3A). While sonication releases only soluble proteins into the supernatant fraction13, alkaline extraction with sodium carbonate additionally solubilizes peripherally membrane-associated proteins13,20. In both the treatments, integral membrane proteins remain in the pellet fraction13. These assumptions are confirmed by western blot analysis of the pellet and supernatant fractions from both treatments (Figure 3B). An integral mitochondrial membrane protein (e.g., porin Por1) is expected to be found totally in the pellet fraction, even after the alkali treatment with sodium carbonate (Figure 3B, lanes 3 and 5). On the other hand, a soluble matrix protein (e.g., α-KGD) is expected to be completely solubilized in both the treatments (Figure 3B, lanes 2 and 4). The significant retention of α-KGD in the pellet from sonication (Figure 3B, lane 3, SMP fraction) might be due to slight variations in the sonication parameters that can affect the formation of the so-called submitochondrial particles, which are efficiently sedimented by ultracentrifugation. The behavior of these proteins with known solubility profiles are then used to determine the mitochondrial solubility of a protein of interest. In the case of Prx1, its western blot profile is suggestive of a protein associated with the membrane periphery and alkaline treatment induces its solubilization (Figure 3B, lane 4).
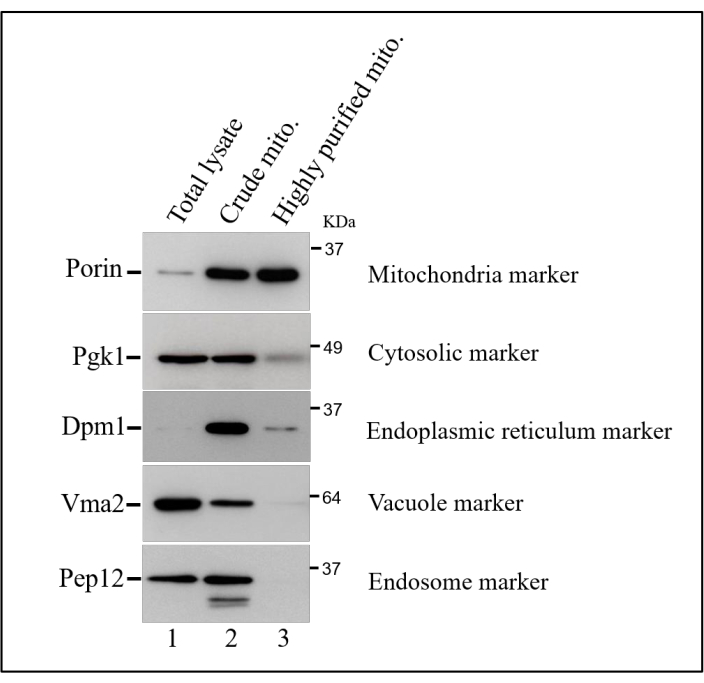
Figure 1: Isolation of highly purified mitochondria. Western blot analysis of total lysate fraction (lane 1), a crude mitochondrial fraction (lane 2), and highly purified mitochondrial fraction (lane 3). The fractions were separated by SDS-PAGE on a 12% polyacrylamide gel, transferred to nitrocellulose and probed with antibodies raised against markers for distinct cellular compartments as described on the right side of the gel. This figure has been modified from reference19. Please click here to view a larger version of this figure.
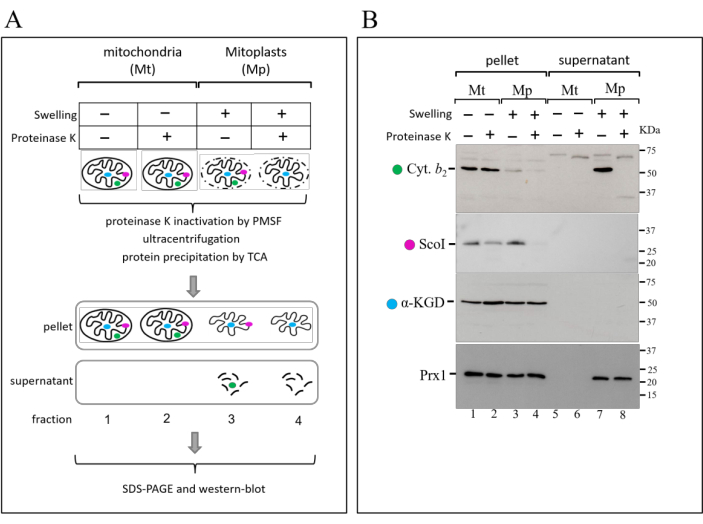
Figure 2: Submitochondrial fractionation protocol by hypotonic swelling in the presence of proteinase K. (A) Schematic representation of submitochondrial fractionation protocol. Highly purified mitochondria are separately subjected to isotonic or hypotonic treatments (swelling) in the presence (+) or absence (-) of proteinase K. After treatment, the activity of proteinase K is inhibited by the addition of PMSF, and mitochondria and mitoplasts are recovered by centrifugation. The protein content from the resulting pellet and supernatant fractions is precipitated by TCA, and then analyzed by SDS-PAGE and western blot. The color spheres represent submitochondrial protein markers for: a soluble intermembrane space protein (green), an inner membrane protein that faces the intermembrane space (pink), and a soluble matrix protein (light blue). (B) Western blot analysis of pellet and supernatant fractions from submitochondrial fractionation protocol. The fractions were separated by SDS-PAGE on a 12% polyacrylamide gel, transferred to nitrocellulose and probed with antibodies raised against markers for distinct submitochondrial compartments as depicted in A. See text for more details. This figure has been modified from19. Please click here to view a larger version of this figure.
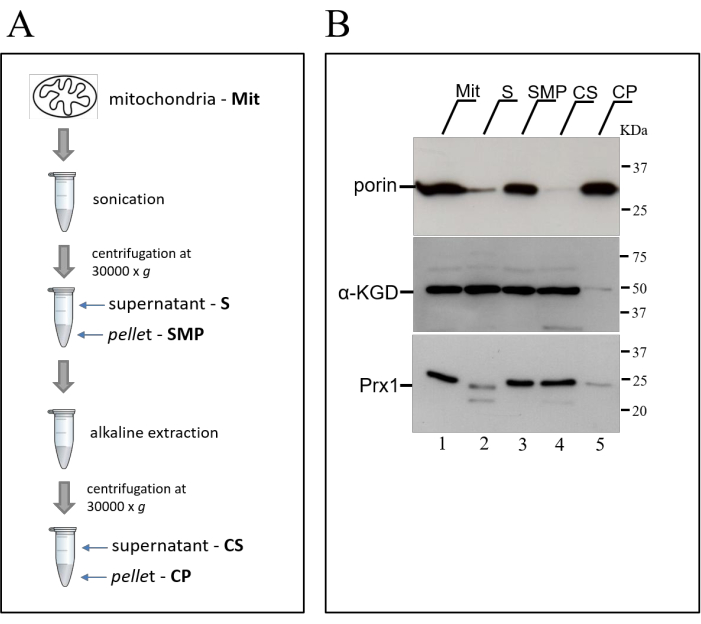
Figure 3: Submitochondrial fractionation protocol by sonication and carbonate extraction. (A) Schematic representation of the protocol used to determine the solubility and membrane topology of mitochondrial proteins. Mitochondria are initially sonicated and centrifuged, resulting in a soluble protein fraction (S), and the compartmentalized membranous product, called submitochondrial particle (SMP). The pellet from the sonication step is subsequently submitted to an alkaline treatment with sodium carbonate (Na2CO3) and centrifuged, resulting in carbonate supernatant (CS) and carbonate-precipitated fractions (CP). (B) Western blot analysis of pellet and supernatant fractions from sonication and carbonate extraction protocol. The fractions were separated by SDS-PAGE on a 12% polyacrylamide gel, transferred to nitrocellulose and probed with antibodies raised against protein markers showing distinct levels of solubility. See text for more details. This figure has been modified from19. Please click here to view a larger version of this figure.
| Solution | Components | Comments | |
| YPD medium | 1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) glucose | Dissolve 10 g Bacto Yeast extract, 20 g Bacto Peptone and 20 g glucose in 900 m of distilled water added. Fill up to 1000 ml and sterilize by autoclaving. | |
| YPGal medium | 1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) galactose | Dissolve 10 g Bacto Yeast extract, 20 g Bacto Peptone and 20 g galactose in 900 m of distilled water added. Fill up to 1000 ml and sterilize by autoclaving. | |
| DTT buffer | 100 mM Tris-H2SO4 (pH 9.4), 10 mM dithiothreitol (DTT) | To make 15 ml: mix 1.5 ml of 1 M Tris-H2SO4, pH 9.4 with 150 µL of 1M DTT prewarmed at 30 °C. Volume to 15 ml with ddH2O Prepare freshly prior to use |
|
| Zymolyase buffer | 20 mM potassium phosphate buffer (pH 7.4), 1.2 M sorbitol | To make 100 ml: mix 2 ml of 1 M potassium phosphate buffer (pH 7.4) with 60 ml of 2 M sorbitol Volume to 15 ml with ddH2O Dissolve the powder (3 mg per gram wet weight) of Zymolyase-20T from Arthrobacter luteus (MP Biomedicals, Irvine, CA) in the buffer just before use |
|
| Homogenization buffer | 10 mM Tris-HCl (pH 7.4), 0.6 M sorbitol, 1 mM EDTA, 0.2% (w/v) bovine serum albumin (BSA), 1 mM phenylmethylsulfonyl fluoride (PMSF) | To make 250 ml: mix 2.5 ml of 1 M Tris-HCl buffer (pH 7.4) with 75 ml of 2 M sorbitol, 500 μL of 500 mM EDTA and 0.5 g of BSA (essentially fatty acid-free) Volume to 250 ml with ddH2O Pre-cool at 4°C Add PMSF and BSA in the buffer just before use |
|
| SEM buffer | 10 mM MOPS-KOH (pH 7.2), 250 mM sucrose, 1 mM EDTA | To make 250 ml: mix 2.5 ml of 1 M MOPS-KOH buffer (pH 7.2) with 31.25 ml of 2 M sucrose and 500 μL of 500 mM EDTA Volume to 250 ml with ddH2O Pre-cool at 4°C just before use. |
|
| EM buffer | 10 mM MOPS-KOH (pH 7.2), 1 mM EDTA | To make 250 ml: mix 2.5 ml of 1 M MOPS-KOH buffer (pH 7.2) with 500 μL of 500 mM EDTA Volume to 250 ml with ddH2O Pre-cool at 4°C just before use |
|
| sample buffer | 2% (w/v) sodium dodecylsulfate (SDS), 50 mM DTT, 10% (v/v) glycerol, 0.02% bromophenol blue, 60 mM Tris-HCl (pH 6.8), |
||
Table 1: Media, solutions, and buffers.
| Reagent | 1 | 2 | 3 | 4 | |
| Mitochondria (10 mg/mL) | 40 µL | 40 µL | 40 µL | 40 µL | |
| SEM buffer | 360 µL | 360 µL | – | – | |
| EM buffer | – | – | 360 µL | 360 µL | |
| Proteinase K (10 mg/mL) | – | 4 µL | – | 4 µL | |
Table 2: Pipetting scheme to perform hypotonic swelling.
Discussion
The protocol presented here has been successfully used and continuously optimized for a long-time to determine the protein localization in the submitochondrial compartments13,14,18,21,22,23. The reliability and reproducibility of this protocol are strongly dependent on the purity and integrity of mitochondrial preparations18. Both of these requirements are achieved by adding an additional purification step (sucrose density gradient centrifugation) to crude mitochondrial preparations13,24,25(Figure 1). Besides eliminating unwanted nonmitochondrial contaminants, this additional purification step also eliminates broken mitochondria and mitoplasts that can arise from the numerous physical manipulations of the sample during the protocol18. Thus, despite increasing the time of the procedure, this sucrose gradient purification step generates highly purified intact mitochondria, which is considered to be essential for the success of the submitochondrial fractionation protocol18,22.
Vögtle and collaborators reported that frozen organelles (at -80 °C) could also be successfully used for submitochondrial fractionation13; we, however, recommend using fresh mitochondrial preparation. Independent of choice, the intactness of purified mitochondria can be confirmed by checking the sensitivity of intermembrane space proteins against externally added proteinase K. If the organelles are intact, proteins of this compartment such as Cyt. b2 and Sco1 should remain protected from proteolytic degradation due to the barrier provided by the outer membrane (Figure 2B, lane 2). In contrast, when the organelles are incubated in a hypoosmotic buffer, the rupture of the outer membrane due to osmotic shock (swelling) makes these proteins prone to protease degradation (Figure 2B, lane 4). On the other hand, the proteins present in the matrix compartment, such as α-KGD, should remain protected against degradation in both mitochondria and mitoplasts because of the integrity of the inner membrane (Figure 2B, lanes 2 and 4). Thus, the profiles of these well-studied mitochondrial marker proteins can be used either to assess the integrity of mitochondrial preparations or the success of the fractionation protocol. Furthermore, the submitochondrial localization of a protein of interest is obtained simply by comparison of its fractionation profile with those from the marker proteins18,22(Figure 2B). Importantly, if the mitochondrial marker proteins show a behavior distinct from that described above, the integrity of the organelles is likely compromised and, thus, the mitochondrial preparation should not be used for the purpose of this protocol.
Although hypotonic shock has proven to be a reliable method to distinguish between intermembrane space proteins versus matrix, this method does not provide information whether a matrix protein is soluble or attached into the mitochondrial inner membrane. This information can be achieved by submitting mitochondria to sonication followed by alkaline extraction with sodium carbonate (Figure 3). While soluble matrix proteins are expected to be released into the supernatant fraction upon sonication, proteins associated with membranes tend to be in the precipitate13. On the other hand, alkaline extraction with sodium carbonate efficiently solubilizes proteins peripherally attached to membranes, but not integral membrane proteins13,20. Thus, if a protein is resistant against protease degradation after swelling but is released into the supernatant by alkali treatment, it is probably a peripherally attached inner-membrane protein facing the matrix compartment. It is important to keep in mind that the success of protease-sensitivity assay is strikingly dependent on the sensitivity of the protein of interest against the protease used for digestion. Some mitochondrial proteins seem to be resistant against proteolytic degradation at the standard proteinase K concentration (0.1 mg/mL) usually employed in subfractionation protocols (Figure 2B, lane 8)19. Doubling proteinase K concentration (0.2 mg/mL) seems to be sufficient to enable a complete proteolytic degradation. However, keep in mind that higher concentrations of protease might destabilize the outer mitochondrial membrane and eventually compromise the effectiveness of the protocol.
There is no doubt that to elucidate the function of mitochondrial proteins, it is essential to determine their submitochondrial localization. In this regard, this protocol represents a powerful tool for researchers that are beginning to study mitochondria. Despite being directed to yeast, many of its principles could be easily applicable to other organisms. Indeed, with the exception of mitochondrial purification steps, the rest of the protocol is very similar to those reported for other organisms26,27,28. Another important contribution of this protocol is its applicability in the study of mutants of S. cerevisiae that show altered mitochondrial biogenesis. It has been widely reported that yeast mutants for mitochondrial protein import machinery show an altered distribution of mitochondrial proteins5,29. Thus, this protocol can be routinely used to investigate the consequences on mitochondrial protein distribution caused by mutations that can alter mitochondrial biogenesis. Finally, the protocol can be particularly useful for investigating proteins that show dual mitochondrial localization19,30.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Dr. A. Tzagoloff (Columbia University) for providing antibodies raised against submitochondrial marker proteins Cyt. b2, αKGD, and Sco1. We also thank Dr. Mario Henrique de Barros (Universidade de São Paulo) for helpful discussion and comments during the establishment of this protocol.
This work was supported by research grants from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (grant 2013/07937-8).
Fernando Gomes and Helena Turano are also supported by FAPESP, grants 2017/09443-3 and 2017/23839-7, respectively. Angélica Ramos is also supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Materials
| Bacto Peptone | BD | 211677 | |
| Bacto Yeast extract | BD | 212750 | |
| Beckman Ultra-Clear Centrifuge Tubes, 14 x 89 mm | Beckman Coulter | 344059 | |
| Bovine serum albumin (BSA fatty acid free) | Sigma-Aldrich | A7030 | Component of Homogenization buffer |
| DL-Dithiothreitol | Sigma-Aldrich | 43815 | Component of DDT buffer |
| D-Sorbitol | Sigma-Aldrich | S1876 | |
| Ethylenediaminetetraacetic acid (EDTA) | Sigma-Aldrich | E9884 | |
| Galactose | Sigma-Aldrich | G0625 | |
| Glucose | Sigma-Aldrich | G7021 | |
| MOPS | Sigma-Aldrich | M1254 | |
| Phenylmethylsulfonyl fluoride (PMSF) | Sigma-Aldrich | P7626 | Used to inactivate proteinase K |
| Potassium phosphate dibasic | Sigma-Aldrich | P3786 | |
| Potassium phosphate monobasic | Sigma-Aldrich | P0662 | |
| Proteinase K | Sigma-Aldrich | ||
| Sucrose | Sigma-Aldrich | S8501 | |
| Trichloroacetic acid (TCA) | Sigma-Aldrich | T6399 | |
| Trizma Base | Sigma-Aldrich | T1503 | |
| Zymolyase-20T from Arthrobacter luteus | MP Biomedicals, Irvine, CA | 320921 | Used to lyse living yeast cell walls to produce spheroplast |
References
- Pfanner, N., Warscheid, B., Wiedemann, N. Mitochondrial proteins: from biogenesis to functional networks. Nature Reviews Molecular Cell Biology. 20 (5), 267-284 (2019).
- Malina, C., Larsson, C., Nielsen, J. Yeast mitochondria: An overview of mitochondrial biology and the potential of mitochondrial systems biology. FEMS Yeast Research. 18 (5), 1-17 (2018).
- Wiedemann, N., Pfanner, N. Mitochondrial machineries for protein import and assembly. Annual Review of Biochemistry. 86, 685-714 (2017).
- Chacinska, A., Koehler, C. M., Milenkovic, D., Lithgow, T., Pfanner, N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 138 (4), 628-644 (2009).
- Schmidt, O., Pfanner, N., Meisinger, C. Mitochondrial protein import: from proteomics to functional mechanisms. Nature Reviews. Molecular Cell Biology. 11 (9), 655-667 (2010).
- Couvillion, M. T., Soto, I. C., Shipkovenska, G., Churchman, L. S. Synchronized mitochondrial and cytosolic translation programs. Nature. 533 (7604), 499-503 (2016).
- Richter-Dennerlein, R., et al. Mitochondrial protein synthesis adapts to influx of nuclear-encoded protein. Cell. 167 (2), 471-483 (2016).
- Suomalainen, A., Battersby, B. J. Mitochondrial diseases: The contribution of organelle stress responses to pathology. Nature Reviews Molecular Cell Biology. 19 (2), 77-92 (2018).
- Nicolas, E., Tricarico, R., Savage, M., Golemis, E. A., Hall, M. J. Disease-associated genetic variation in human mitochondrial protein import. American Journal of Human Genetics. 104 (5), 784-801 (2019).
- Calvo, S. E., Mootha, V. K. The mitochondrial proteome and human disease. Annual Review of Genomics and Human Genetics. 11, 25-44 (2010).
- Reinders, J., Zahedi, R. P., Pfanner, N., Meisinger, C., Sickmann, A. Toward the complete yeast mitochondrial proteome: Multidimensional separation techniques for mitochondrial proteomics. Journal of Proteome Research. 5 (7), 1543-1554 (2006).
- Sickmann, A., et al. The proteome of Saccharomyces cerevisiae mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 100 (23), 13207-13212 (2003).
- Vögtle, F. N., et al. Landscape of submitochondrial protein distribution. Nature Communications. 8 (1), 00359 (2017).
- Morgenstern, M., et al. Definition of a high-confidence mitochondrial proteome at quantitative scale. Cell Reports. 19 (13), 2836-2852 (2017).
- Zahedi, R. P., et al. Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Molecular Biology of the Cell. 17 (3), 1436-1450 (2006).
- Vögtle, F. -. N., et al. Intermembrane space proteome of yeast mitochondria. Molecular & Cellular Proteomics. 11 (12), 1840-1852 (2012).
- Gregg, C., Kyryakov, P., Titorenko, V. I. Purification of mitochondria from yeast cells. Journal of Visualized Experiments: JoVE. (30), e1417 (2009).
- Boldogh, I. R., Pon, L. A. Purification and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods in Cell Biology. 80 (06), 45-64 (2007).
- Gomes, F., et al. Proteolytic cleavage by the inner membrane peptidase (IMP) complex or Oct1 peptidase controls the localization of the yeast peroxiredoxin Prx1 to distinct mitochondrial compartments. Journal of Biological Chemistry. 292 (41), 17011-17024 (2017).
- Fujiki, Y., Hubbard, L., Fowler, S., Lazarow, P. B. Isolation of intracellular membranes by means of sodium carbonate treatment: Application to endoplasmic reticulum. Journal of Cell Biology. 93 (1), 97-102 (1982).
- Glick, B. S., et al. Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell. 69 (5), 809-822 (1992).
- Diekert, K., de Kroon, A. I., Kispal, G., Lill, R. Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods in Cell Biology. 65, 37-51 (2001).
- Glick, B. S. Pathways and energetics of mitochondrial protein import in Saccharomyces cerevisiae. Methods in Enzymology. 260 (1992), 224-231 (1995).
- Meisinger, C., Sommer, T., Pfanner, N. Purification of Saccharomcyes cerevisiae mitochondria devoid of microsomal and cytosolic contaminations. Analytical Biochemistry. 287 (2), 339-342 (2000).
- Meisinger, C., Pfanner, N., Truscott, K. N. Isolation of yeast mitochondria. Methods in molecular biology. 313 (1), 33-39 (2006).
- Kang, Y., et al. Tim29 is a novel subunit of the human TIM22 translocase and is involved in complex assembly and stability. eLife. 5, 17463 (2016).
- Wrobel, L., Sokol, A. M., Chojnacka, M., Chacinska, A. The presence of disulfide bonds reveals an evolutionarily conserved mechanism involved in mitochondrial protein translocase assembly. Scientific Reports. 6, 27484 (2016).
- Callegari, S., et al. TIM29 is a subunit of the human carrier translocase required for protein transport. FEBS letters. 590 (23), 4147-4158 (2016).
- Neupert, W., Herrmann, J. M. Translocation of proteins into mitochondria. Annual Review of Biochemistry. 76, 723-749 (2007).
- Meineke, B., et al. The outer membrane form of the mitochondrial protein Mcr1 follows a TOM-independent membrane insertion pathway. FEBS Letters. 582 (6), 855-860 (2008).

