Planar Supported Lipid Bilayer Assay: A Microscopy-Based In Vitro Technique to Investigate Septin Assembly on Biological Membrane Mimics
Abstract
Source: Curtis, B. N. et al. Reconstitution of Septin Assembly at Membranes to Study Biophysical Properties and Functions. J. Vis. Exp. (2022)
In this video, we demonstrate a procedure to investigate septin organization on a glass-supported lipid bilayer in vitro using total internal reflection fluorescence microscopy.
Protocol
1. Planar Lipid Bilayers
- Plasma Cleaning of the Slides
NOTE: Plasma cleaning removes organic contaminants and increases the hydrophilicity of the glass, ensuring efficient lipid adsorption. The following steps may change or need to be optimized depending on the plasma cleaner and glass coverslips used.- Purge the plasma cleaner for 5 min with oxygen to remove air from the lines and chamber. This is to ensure that, when the plasma cleaner is run, there is primarily oxygen in the lines and chamber, providing more consistent bilayer preparations.
- While purging, stream an inert gas, such as N2 or Ar, over the micro cover glass slides to remove dust and particulates.
OPTIONAL: Cover glass slides can be washed by spraying each side with 100% isopropanol followed by water. Repeat the isopropanol-water wash 3x and then dry with an N2 stream. - Arrange dry cover glass slides into a ceramic cradle. Place the cradle at the back of the plasma chamber so that the coverslips are parallel with the long edge of the chamber (this keeps them in place during plasma cleaning). Run the plasma cleaner for 15 min with oxygen at maximum power.
- Chamber Preparation
NOTE: The method described here uses homemade chambers from plastic PCR tubes to support the reaction volumes, but other low-adhesion materials, such as silicone wells, may also be suitable.- Using a razor blade or scissors, cut off the cap just below the frosted part of a 0.2 mL PCR tube (Figure 1).
- Paint the rim of the PCR tube with UV-activated adhesive, avoiding the inside of the tube. Gently place the PCR tube glue-down in the center of a plasma-cleaned coverslip.
NOTE: To avoid glue inside the chamber, use the minimum amount of glue to get a seal, and promptly treat with UV without bumping or disturbing the chamber. - Place the chamber under long-wavelength UV light for 5-7 min to cure the adhesive.
- Bilayer Formation
NOTE: Monodispersed SUVs should be used in this step for efficient bilayer formation. Table 1 provides recipes for all buffers used in the bilayer formation, including stock and final buffer concentrations.- Add 40 µL of supported lipid bilayer buffer (SLBB: 300 mM KCl, 20 mM HEPES, pH 7.4, 1 mM MgCl2; Table 1), 10 µL of 5 mM SUVs, and 1 µL of 100 mM CaCl2 to the well. Gently shake the chambers from side to side to disrupt the SUVs, and then incubate at 37 °C for 20 min.
NOTE: To limit evaporation, incubate in a covered Petri dish with a wet wipe.
- Add 40 µL of supported lipid bilayer buffer (SLBB: 300 mM KCl, 20 mM HEPES, pH 7.4, 1 mM MgCl2; Table 1), 10 µL of 5 mM SUVs, and 1 µL of 100 mM CaCl2 to the well. Gently shake the chambers from side to side to disrupt the SUVs, and then incubate at 37 °C for 20 min.
- Washing away excess lipids
NOTE: This step removes excess SUVs and acts as a buffer exchange step. It is very important not to scrape the bilayer with the pipette tip while washing; if the glass is exposed, septins and many other proteins will bind irreversibly, reducing the effective protein concentration.- After incubation, rinse the bilayer 6x with 150 µL of SLBB, pipetting up and down to mix each time while avoiding bubbles.
NOTE: Add 150 µL directly to the 50 µL of buffer and SUVs already present for a total of 200 µL in the reaction well. - When ready to start incubating with the septins or a different protein of choice, wash the bilayer 6x with 150 µL of reaction buffer (RXN: 33.3 mM KCl, 50 mM HEPES, pH 7.4, 1.4 mg/mL BSA, 0.14% methylcellulose, 1 mM BME).
NOTE: For best results, make the reaction buffer fresh and be very careful while mixing in the reaction well to avoid bubbles. - On the last wash step, remove 125 µL of reaction buffer, leaving 75 µL in the well. Add 25 µL of septins diluted in septin storage buffer (SSB: 300 mM KCl, 50 mM HEPES, pH 7.4, 1 mM BME) at the desired concentration and image by TIRF microscopy.
NOTE: When setting up this assay for the first time or incorporating a new lipid composition, it is good practice to ensure lipids are freely diffusing on the surface using fluorescence recovery after photobleaching (FRAP). While the rate of recovery will be different for different lipid compositions and inherently lower than for free-standing systems, the lipids should not be immobile.
- After incubation, rinse the bilayer 6x with 150 µL of SLBB, pipetting up and down to mix each time while avoiding bubbles.
Table 1: Buffer components for preparation of supported lipid bilayer and reactions. Volumes of stock solutions that are incorporated into buffers and the final concentrations of each component are shown. SLB and PRB can be stored at room temperature and reused between experiments. Reaction buffer and SSB are made fresh for each experiment.
| Supported Lipid Bilayer Buffer (SLBB) | ||
| Stock | Volume | Final concentration |
| 2 M KCl | 1.5 mL | 300 mM |
| 1 M HEPES | 200 µL | 20 mM |
| 500 mM MgCl2 | 20 µL | 1 mM |
| Water | 8 mL | |
| Pre-Reaction Buffer (PRB) | ||
| Stock | Volume | Final concentration |
| 2 M KCl | 166 µL | 33.3 mM |
| 1 M HEPES | 500 µL | 50 mM |
| Water | 9.33 mL | |
| Reaction Buffer | ||
| Stock | Volume | Final concentration |
| 2 M KCl | 166 µL | 33.3 mM |
| 1 M HEPES | 300 µL | 50 mM |
| 10 mg/mL BSA | 1.39 mL | 1.39 mg/mL |
| 1% Methylcellulose | 1.39 mL | 0.0014 |
| Water | Up to 10 mL | |
| BME | 0.7 µL | 1 mM |
| Septin Storage Buffer (SSB) | ||
| Stock | Volume | Final concentration |
| 2 M KCl | 1.5 mL | 300 mM |
| 1 M HEPES | 500 µL | 50 mM |
| Water | Up to 10 mL | |
| BME | 0.7 µL | 1 mM |
Representative Results
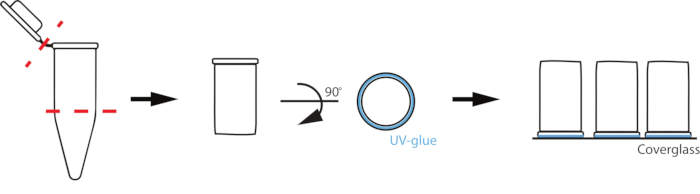
Figure 1: Schematic of reaction chamber set up. To prepare custom chambers, a 0.2 mL PCR tube is cut where the tube begins to taper and at the cap (red dashed lines). The uncut rim of the cut tube is then coated with a thin layer of UV-activated glue (blue) and placed glue-down on a coverslip.
Disclosures
The authors have nothing to disclose.
Materials
| 0.2 mL PCR Tubes with flat cap, Natural | Watson | 137-211C(EX) | |
| HEPES | Sigma Aldrich | H3375-1KG | |
| Magnesium chloride | VWR | 7791-18-6 | |
| Methyl cellulose 4000cp | Sigma-Aldrich | M052-100G | |
| Microglass coverslips for planar bilayers | Matsunami | Discontinued | 22×22 |
| Optical Adhesive | Norland Thorlabs | NOA 68 | Flexible adhesive for glass or plastics |
| Plasma Cleaner | Plasma Etch | PE-25 | Voltage: 120V, 60Hz. Current: 15 AMPS |
| Potassium chloride | VWR | 0395-1kg | |
| UV Lamp | Spectroline | ENF-260C | 115 Volts, 60 Hz, 0.20 AMPS |
| Bovine Serum Albumin (BSA) | Sigma-Aldrich | A4612-25G | |
| Beta mercaptoethanol (BME) | Sigma-Aldrich | M6250-100ML | |
| DOPC | Avanti Polar Lipids | 850375 | |
| Egg Liss Rhodamine PE | Avanti Polar Lipids | 810146 |

