Graphene Biosensor-Based Field-Effect Biosensing to Detect Protein-Protein Interactions
Abstract
Source: Qvit, N., et al. Exploring Biomolecular Interaction Between the Molecular Chaperone Hsp90 and Its Client Protein Kinase Cdc37 using Field-Effect Biosensing Technology. J. Vis. Exp. (2022).
In this video, we describe the field-effect biosensing (FEB) technology to detect and quantify protein-protein interactions. The target proteins immobilized on the graphene biosensor interact with analyte proteins in solution, resulting in electrical current alterations in the FEB system. This change in the electrical response indicates the binding affinity between the proteins.
Protocol
NOTE: The recombinant proteins used in this study, Hsp90, and Cdc37, were commercially obtained (see Table of Materials).
1. Chip Activation
NOTE: All materials to be used in the experiment are listed in the Table of Materials. Filter all prepared solutions through a sterile 0.2 μm filter.
- Prepare 1-Ethyl-3-(3-dimethylamino propyl) carbodiimide (EDC) solution by adding 2 mg of EDC to 2.5 mL of 1 M 2-(N-morpholino) ethane sulfonic acid (MES) buffer (pH = 6.0) in a 15 mL tube. Prepare N-Hydroxysulfosuccinimide (sulfo-NHS) solutions by adding 6 mg of sulfo-NHS to 2.5 mL of 1 M MES buffer (pH = 6.0) in a separate 15 mL tube. Aliquot 50 µL of each solution in independent tubes and store at -20 °C for future use.
- Mix equal volumes of EDC and sulfo-NHS solution (50 µL of EDC + 50 µL of sulfo-NHS) by pipetting up and down (do not vortex).
NOTE: The mixed solution of EDC/sulfo-NHS must be used within 30 min to maintain effective cross-linking for proper chip functionalization. - Place the biosensor chip (5.7 cm x 2.4 cm; see Table of Materials) supplied by the company in a glass Petri dish with a fitted lid. All the functionalization steps involved in chip activation are suggested to be done within the Petri dish. Apply 50 µL of 1 M MES buffer (pH = 6.0) to the biosensor chip, incubate for 1 min at room temperature, and then aspirate the buffer.
- Apply 50 µL of EDC/sulfo-NHS solution immediately to the sensor chip. Cover the Petri dish and incubate for 15 min at room temperature. Aspirate EDC/sulfo-NHS solution from the chip.
- Rinse the chip once with 50 µL of 1 M MES buffer (pH = 6.0); aspirate the MES buffer.
2. Target protein immobilization
- Rinse the chip 2x with 50 µL of 1x PBS (pH = 7.4). Aspirate the PBS from the chip and add the target molecule, Hsp90 (50 µL; 500 nM).
NOTE: Buffer mismatch may ruin the entire experiment; hence, before the experiment, it is important to make sure that the target molecule is in the same buffer as that used for calibration (e.g., 1x PBS (pH = 7.4)). If needed, perform a buffer exchange by overnight dialysis before the experiment. In this experiment, overnight dialysis was performed for both recombinant proteins, Hsp90 and Cdc37, against 1x PBS (pH = 7.4; see Table of Materials) with proper buffer exchange at 4 °C. The concentration of the target material (Hsp90 in this case) may vary according to different experimental protocols and the nature of the target materials (protein/peptide/ligands). - Cover the glass Petri dish and incubate for 30 min at room temperature. Aspirate the solution containing the target molecule and rinse 3x with 50 µL of 1x PBS (pH = 7.4). Aspirate the 1x PBS (pH = 7.4) solution from the chip.
- Add 50 µL of Quench 1 (3.9 mM amino-PEG5-alcohol in 1x PBS (pH = 7.4)) solution to the chip. Cover the glass Petri dish and incubate for 15 min at room temperature. Aspirate the Quench 1 solution from the chip.
- Add 50 µL of Quench 2 (1 M ethanolamine (pH = 8.5)) solution to the chip. Cover the glass Petri dish and incubate for 15 min at room temperature. Aspirate Quench 2 solution from the chip and rinse the chip 5x using 50 µL of 1x PBS, leaving the last PBS droplet on the sensor.
3. Preparing analyte samples
- Prepare analyte dilution series for Cdc37 in the desired concentration range. For the first experiment, the following concentrations were used: 25 nM, 50 nM, 100 nM, 200 nM, 400 nM, 800 nM, 1,000 nM, 2,000 nM, 3,000 nM, and 5,000 nM. For the second experiment, a different set of concentrations ranging from 0.4 nM to 200 nM was used.
- Design the experiment to include at least eight different analyte concentrations to obtain a reliable KD value. Prepare the different dilutions of the analyte protein in the same buffer as that used for calibration and target protein; here it is 1x PBS (pH = 7.4).
4. Loading of the activated biosensor chip into the FEB device
NOTE: The FEB device consists of a reader featured with LED light indications and a cartridge to insert the biosensor chip.
- After target protein immobilization, insert the activated chip into the cartridge of the device, which is connected via USB to a computer. After the chip insertion, a green LED light will be displayed on the reader indicating that the FEB device is ready for the experiment. Install the automated software (see Table of Materials) supplied by the company on the computer, to which the FEB device is connected, to monitor the experiment step by step as described below.
5. Run the experiment
- Press the Run Experiment module on the automated software and choose 10 Points with Regeneration or any other desired protocol. Fill in the following details: operator name, experiment name, date (e.g., Yana, Hsp90 + Cdc37, 14.03.2021); regeneration buffer (e.g., PBS buffer); the immobilized target (e.g., Hsp90); analyte in solution (e.g., Cdc37).
- Press the Begin the Experiment button displayed on the software and follow the instructions shown by the automated software as described below.
NOTE: The software is fully automated, user-friendly, and guides the user throughout the experiment step by step. A pop-up window will appear on the screen with instructions to proceed further at each step of the experiment. The software will provide instructions for each repetitive step consecutively from calibration, analyte association, dissociation, regeneration, and wash (5x) for each analyte concentration throughout the experiment. - Perform instrument calibration. To do so, aspirate the remaining PBS solution from the chip and apply 50 µL of calibration buffer (1x PBS; pH = 7.4). Press the 继续 button and wait for 5 min until the calibration step is finished. The software displays the endpoint determined for the calibration step (5 min) with a warning alarm to follow up.
- Next, perform an analyte association. To do so, aspirate the calibration buffer from the chip and apply 50 µL of the lowest analyte concentration (25 nM of Cdc37). Press the 继续 button and wait for 5 min until the association step is finished. The software displays the endpoint for the association step (5 min) with a warning alarm to proceed.
- Perform an analyte dissociation. To do so, aspirate the analyte solution from the chip and apply 50 µL of the dissociation buffer (1x PBS; pH = 7.4). Press the 继续 button and wait for 5 min until the dissociation step duration (5 min) is finished. The software displays the endpoint for the dissociation step (5 min) with a warning alarm to follow up.
- Next, perform chip regeneration. Aspirate the dissociation solution from the chip and apply 50 µL of regeneration buffer (1x PBS; pH = 7.4). Press the 继续 button and wait for 30 s until the regeneration step duration (30 s) is finished. The software displays the endpoint for the regeneration step (30 s) with a warning alarm to follow up.
- Finally, wash the chip. Aspirate the regeneration solution from the chip and apply 50 µL of wash buffer (1x PBS; pH = 7.4) to the chip. Aspirate the solution from the chip and repeat this 5x. Leave the last drop of wash buffer on the chip and press the 继续 button and wait for 30 s until the wash step duration is finished in the software display.
NOTE: The software displays the endpoint for the wash step (30 s) with a warning alarm to proceed with the next cycle of the experiment. - Repeat the steps for each analyte concentration used; the five steps of calibration, analyte association, dissociation, regeneration, and wash (5x) constitute one cycle. For the experiment shown here, we performed 10 cycles for 10 analyte concentrations (ranging from 25 nM to 5,000 nM or 0.4 nM to 200 nM; Figure 1).
Representative Results
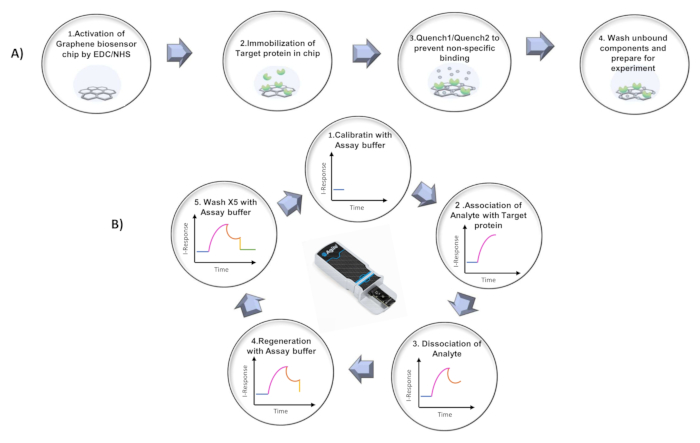
Figure 1: Protocol summary. (A) Summary of the steps for the chip activation process. (B) Graphical representation of the five repeating steps in the protocol.
Disclosures
The authors have nothing to disclose.
Materials
| Automated analysis software | Agile plus software, Cardea (Nanomed) | NA CAS number: NA |
Referred to in the text as the automated analysis software supplied with the instrument. Generates automated analysis. |
| COOH-BPU (Biosensing Processing Unit) | Agile plus software, Cardea (Nanomed) | NA CAS number: NA |
Biosensor chip |
| Data review software | Datalign 1.0, Cardea (Nanomed) | NA CAS number: NA |
Referred to as the supplied data review software in the text. Supplied with the instrument and allows to review and export the information data points. |
| Dialysis bag | CelluSep, Membrane filtration products | T2-10-15 CAS number: NA |
T2 tubings (6,000-8,000 MWCO), (10 mm fw, 6.4mm Ø, 0.32ml/cm, 15m) |
| EDC (1-Ethyl-3-(3-dimethylamino propyl) carbodiimide) | Cardea (Nanomed) | EDC160322-02 CAS number: 25952-53-8 |
White powder |
| MES (2-(N-morpholino) ethane sulfonic acid) buffer | Merck | M3671-50G CAS number: 4432-31-9 |
White powder |
| NHS (N-Hydroxysulfosuccinimide) chips | Cardea (Nanomed) | NA CAS number: NA |
Graphene-based chip |
| PBS (Phosphate-buffered saline) X 10 | Bio-Lab | 001623237500 CAS number: 7758-11-4 |
Liquid transparent solution |
| Pipette | Thermo Scientific | 11855231 CAS number: NA |
Finnpipette F3 5-50 µL, yellow |
| Quench 1 (3.9 mM amino-PEG5- alcohol in 1 X PBS) | Cardea (Nanomed) | 0105-001-002-001 CAS number: NA |
Liquid, transparent solution |
| Quench 2 (1 M ethanolamine (pH=8.5)) | Cardea (Nanomed) | 0105-001-003-001 CAS number: NA |
Liquid, transparent solution |
| Recombinant protein Cdc37 | Abcam | ab256157 CAS number: NA |
|
| Recombinant protein Hsp90 beta | Abcam | ab80033 CAS number: NA |
|
| Spreadsheet | Excel, Microsoft office | NA CAS number: NA |
|
| Statistical software | GraphPad, Prism | NA CAS number: NA |
Referred to as the other statistical software. Sigma plot, phyton or other statistical programes may also be used |
| Sulfo-NHS | Cardea (Nanomed) | NHS160321-07 CAS number: 106627-54-7 |
White powder |
| Tips | Alex red | LC 1093-800-000 CAS number: NA |
Tip 1-200 µl, in bulk, 1,000 pcs |

