Separating Beads and Cells in Multi-channel Microfluidic Devices Using Dielectrophoresis and Laminar Flow
Summary
Dielectrophoresis (DEP) is an effective method to manipulate cells. Printed circuit boards (PCB) can provide inexpensive, reusable and effective electrodes for contact-free cell manipulation within microfluidic devices. By combining PDMS-based microfluidic channels with coverslips on PCBs, we demonstrate bead and cell manipulation and separation within multichannel microfluidic devices.
Abstract
Microfluidic devices have advanced cell studies by providing a dynamic fluidic environment on the scale of the cell for studying, manipulating, sorting and counting cells. However, manipulating the cell within the fluidic domain remains a challenge and requires complicated fabrication protocols for forming valves and electrodes, or demands specialty equipment like optical tweezers. Here, we demonstrate that conventional printed circuit boards (PCB) can be used for the non-contact manipulation of cells by employing dielectrophoresis (DEP) for bead and cell manipulation in laminar flow fields for bioactuation, and for cell and bead separation in multichannel microfluidic devices. First, we present the protocol for assembling the DEP electrodes and microfluidic devices, and preparing the cells for DEP. Then, we characterize the DEP operation with polystyrene beads. Lastly, we show representative results of bead and cell separation in a multichannel microfluidic device. In summary, DEP is an effective method for manipulating particles (beads or cells) within microfluidic devices.
Protocol
A general diagram of the equipment setup is shown in Figure 1A. The PCB-sample assembly is further detailed in a cross-sectional schematic in Figure 1B.
1. Preparing PCB Electrodes:
- Design PCB electrodes to the desired geometry to generate a non-uniform electrical field. Customized PCB electrode chips can be ordered through commercial fabrication facilities (Figure 1C).
- Prepare pre-fabricated PCB electrodes by soldering 16-gauge wire to the end of each printed metal electrode (Figure 1D).
- Place the wire onto the end of the electrode. Hold the wire in place on the metal area of the PCB with the hot soldering iron to heat the wire.
- Feed a small amount of solder into the heated wire to fill the wire with solder.
- After the wire is filled with solder, remove the soldering iron, holding the wire in place while the solder cools.
- Repeat the soldering process for each electrical connection on the PCB (Figure 1D).
2. Prepare Microfluidic Channels:
- Polydimethylsiloxane (PDMS)-based microfluidic channels are prepared using the PDMS elastomer, Sylgard 184 from Dow Corning. A master mold which defines the channels is usually created through standard microfabrication processes using a silicon wafer and SU-8 photoresist.
- Mix base compound to curing agent at a 10:1 ratio for 5 minutes.
- Pour the liquid PDMS onto the prefabricated SU-8 master mold and remove air bubbles by exposing liquid PDMS to vacuum for a few minutes. Repeat vacuum process if needed to completely remove all bubbles.
- Cure the PDMS at 70 °C for 2 hours.
- Remove PDMS slab with microfluidic channels from the wafer with a razor blade, careful not to break wafer.
- Punch holes for introducing fluids and cells into the microfluidic device. (Note: Syringe pumping, gravity or surface-tension based flow can all be used with DEP.)
- Inspect the microfluidic device to ensure it is free of dust and debris. Cleaning the PDMS can be easily achieved using 3M Scotch Magic Tape.
- Plasma bond the PDMS microfluidic channels to a clean no.0 thickness (80-130 μm) coverslip. Heat the coverslip-microfluidic assembly at 100 °C for a minimum of 15 min.
3. Prepare Low-conductivity Media:
- Low-conductivity media is prepared by mixing 8.5% sucrose + 0.3% glucose (w/v) in deionized (DI) water.1
Note: We have previously demonstrated that cells can be cultured for days in conventional cell culture media after being exposed to low conductivity for short periods (approximately 30 min).11
4. Assemble Microfluidic Channels Onto PCB Electrodes:
- Place a small amount (approximately 10 μL) of mineral oil onto the PCB to ensure tight contact between the PCB and the coverslip.
Note: An optional step for decreasing electrode visualization is to coat the PCB surface with a very thin layer of black permanent marker or paint. - Place the coverslip-microfluidic channel assembly onto the oiled PCB, with coverslip forming contact with the oil (coverslip down). Gently press the coverslip-microfluidic assembly down to ensure a good contact and to minimize air bubbles that can detract from cell and bead visualization. An example of the completed device is shown in Figure 1D.
5. Sort and Concentrate Beads and Cells using DEP:
- Fill the microfluidic channels with DI water or low-conductivity media; the plasma bonding process facilitates the easy loading of aqueous solutions into the microfluidic channels by temporarily making the normally hydrophobic PDMS surface hydrophilic.
- Introduce cells and/or polystyrene beads into the channel reservoir (Figure 1C). Here we use human colon adenocarcinoma (HT-29) cells.
- Connect the output of a function generator to the input of an AC power amplifier, then connect the output of the amplifier to the electrode wires. Cover all electrical wires and surfaces of the setup with electrical tape to protect users from the potential exposure to shock. A schematic diagram of the equipment setup is shown in Figure 1A.
- Set the function generator to produce a sine wave output of 1.0-1.5 MHz. The amplitude of the output should be adjusted for the RF power amplifier to produce an output of 80-100V to the PCB. The RF power amplifier in this work requires an input voltage around 220-330 mV. To separate cells and beads, the laminar flow rate must be compliant with the DEP force to move the cells and beads within the main channel (width w = 100 μm, height h = 27 μm) into the target channels (width w = 100 μm, height h = 27 μm).
Caution: consult with PCB manufacturer to determine the maximum operating voltages to avoid issues such as PCB overheating or melting. Check the equipment manufacturers’ specifications for the function generator and power amplifier manufacturers to determine safe operating settings. - Initiate DEP to sort cells and beads. Cells experience a positive-DEP whereas polystyrene beads experience a negative-DEP in a non-uniform electric field within the specified frequencies. This enables DEP-force-activated bioactuation and separation of cells and other particles within microfluidic devices using affordable and reusable PCBs as electrodes.
6. Representative Results:
When DEP is effective at actuating cells or particles, a robust alignment within non-uniform electrical fields is observed for static baths or slow fluid velocities. Under conditions of higher fluid velocities, the cell or particle behavior depends on the relative orientation of the axial flow to the electrical field and the flow velocity. In addition, the cell and particle behaviors are also dependent upon the electrical field strength and the degree of non-uniformity within the electrical field. Typical behaviors of cells or particles include ‘pearl chains,’ cycling, stalling, or turning.
Conditions that compromise or abolish DEP include the presence of salts or other ionic molecules in the fluid, weak electric field strength, excessive flow velocities, coverslips that are too thick, or conductive solutions between the coverslip and PCB electrodes (e.g. a cracked coverslip can induce the mixing of water and mineral oil).
Here, when using no.0 thickness coverslips (80-130 μm) and an electrode spacing of (231 μm), the gradient of the square of the electric field intensity applied to the HT-29 cells and beads is estimated to be between 2-8 to 6-8 V2/μm3.1
The DEP force can be estimated by measuring the velocity of the particle, manipulated by DEP in static fluid (Figure 2A-B). Due to the small inertia of the particle and highly viscous environment of microfluidic channel, the hydrodynamic drag force will be equal to, but in opposite direction with the DEP force. The average bead velocity in the low-conductivity media is 34.5 μm/s with DEP voltage of 93 Vpp and 1.5 MHz. The hydrodynamic drag force can be calculated with following equation.1
Fdrag = 6πηRv
The average viscosity η of the low-conductivity media at 20 °C is 1.27 mPa•s and the bead radius R is 7.5 μm. The hydrodynamic drag force for the polystyrene microbead is estimated to be 6.19 pN. To demonstrate the ability to actuate beads within microfluidic channels (Figure 2C-F), we initiated the flow of the low-conductivity media by employing surface tension mediated flow. The average bead velocity in the single input channel was 1540 μm/sec, whereas the average velocity within the individual channels was reduced to 565 μm/sec (Figure 2C). By initiating DEP, the beads were retained within the central channel rather than being released into the side channel. Thus, under these conditions, the DEP forces were sufficient to overcome the drag force of the fluid into the two side channels.
The same principle was also used to divert the beads from the central cannel to a single side channel by simply changing the orientation of the channels and the bead flow to that of the DEP electrodes (Figure 2E-F). By angling the metal electrode on the side of the single inlet channel, when DEP was initiated, the beads were guided to the side of the channel as the beads approached the trifurcation point. There, the DEP forces were sufficient to pull the beads into one of the side channels where the laminar flow continued to propel them down the channel (Figure 2F).
Because cells and polystyrene beads have distinct differences in their ability to be polarized and actuated in non-uniform electrical fields, we use DEP to demonstrate the ability to perform the on-chip separation of cells and beads, simultaneously. We used the same microfluidic channel structure and PCB electrodes as configured in Figure 2E-F, but introduced a suspension of HT-29 cells and beads in low-conductivity media into the single inlet channel (Figure 3). When DEP is introduced, and under these conditions, the HT-29 cells were retained in the two left output channels whereas the beads were retained in the individual output channel on the right. Here the cells show a characteristic ‘pearl-chaining’ behavior as they are collected in the left output channel. Occasionally a bead and cell become attached together in which they are collected together, often in the cell output channels. From this experimental data, we determine the sorting rate to be 713 particles (cells and beads) per minute, or the equivalent of 14,260 cells and beads in 20 minutes. The number of cells and beads retrieved is relevant and useful for molecular biology, imaging, biochemical and lab-on-a-chip applications.
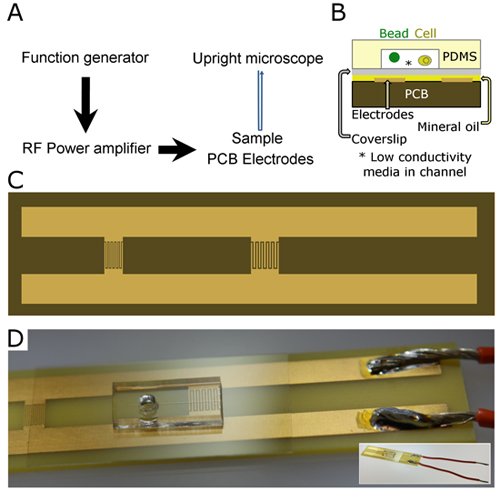
Figure 1. PCB-based DEP of cells and particles in microfluidic channels. (A) The equipment setup for DEP-based actuation starts with a function generator to define the frequency and amplitude of the electrical signal, then a power amplifier to boost the signal strength of the electrical field generated on the PCB. (B) The microfluidic device assembly for sample actuation consists of PDMS microfluidic channels irreversibly bound to a no. 0 thickness coverslip (80-130 μm) through oxygen plasma, non-conductive media to bathe the cells and/or beads. (C) The PCB electrodes used here consist of two regions where the electrodes are interdigitated to generate a strong non-uniform electric field. (D) The completed device: a PCB with a trifurcated microfluidic device on a single coverslip, inset shows the whole device with electrode wires. (C-D) For scale, PCB measurements are 8.4 cm (l), 2.1 cm (w), with 5 mm-wide electrodes.
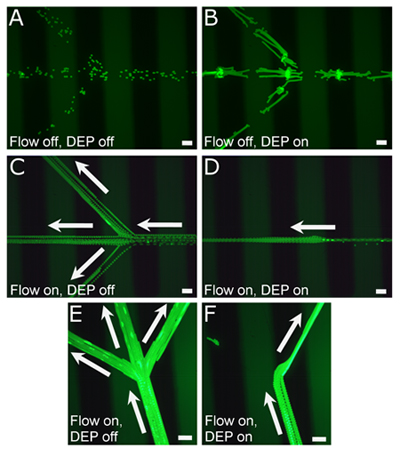
Figure 2. Representative images of cells and beads in channels before and during DEP actuation. (A) 15 μm fluorescent polystyrene beads in a low-conductivity media solution within a PDMS microfluidic channel, without laminar flow (time lapsed, 1.23 sec). (B) Upon DEP initiation, the beads migrate toward the PCB electrode patterns (black stripes) rather than between electrodes (time lapsed, 8.18 sec). (C) Beads in the same channels under laminar flow are divided between the three separate channels (time lapsed, 5.3 sec); when DEP is initiated (D), the beads are actuated to flow only in the central channel (time lapsed, 4.3 sec). (E) By changing the orientation of the channels to the PCB electrodes, beads can be differentially directed into the side channels (F) using DEP rather than the central channel as shown in (D). (E-F) Time lapsed is 8.23 sec and 5.16 sec, respectively. All scale bars = 100 μm.
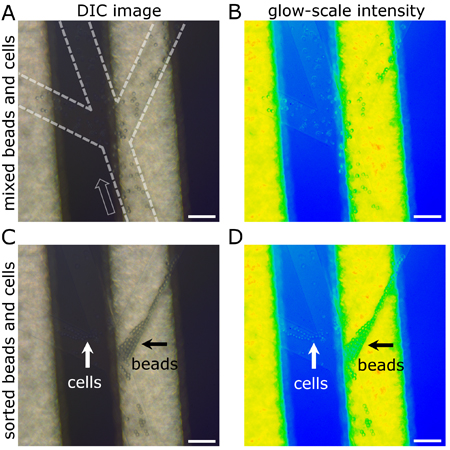
Figure 3. Representative images of cells and beads in channels before and during DEP actuation. (A-B) Prior to initiating DEP, a mixed solution of human colon adenocarcinoma (HT-29) cells and beads flow from one channel to 3 separate output channels. The open arrow identifies the direction of laminar flow and channels are outlined with dashed lines for orientation and clarification. (C-D) By inducing DEP, beads and cells are selectively actuated into separate channels as identified. HT-29 cells exit the central and left channel whereas beads exit the right channel. Characteristic pearl chaining of beads and cells is observed during DEP actuation. (A, C) Differential Interference Contrast imaging of cells and beads in microchannels renders the beads easily visible. Glow scale intensity images (B, D) of the same DIC images (A, C) improve the visualization of cells in channels. The reflective metal electrodes ((A, C) light stripe and (B, D) yellow and green) provide a prominent landmark for aligning microchannels to the electrodes for effective DEP-based actuation. Scale bars = 100 μm.
Discussion
Cell manipulation in microfluidic devices is desirable for the sorting or selective placement of single cells or for population studies.2 Laminar flow is used in conjunction with valves and pumps to manipulate cells within microfluidic devices. However, these methods alone are challenging and require detailed fabrication processes and skills.3 Centrifugation can simplify the demands for cell placement but simultaneous imaging is a challenge; furthermore, the channel architecture for centrifugation must be considering carefully when designing the desired manipulations and considering effects of centripetal forces.4 Laser tweezers can be used for cell placement but the method is expensive and not amenable for high-throughput cell sorting.5 Yet, DEP has been demonstrated as an effective system of “electrical tweezers” for effective cell placement, characterization and manipulation.6,7
Specifically, DEP has been used to selectively capture and sort cells for on-chip processing of living and dead cells,7-10 and for collecting cells on resonant sensors for the measurement of cell mass.11 We have previously demonstrated that by increasing the DEP force on bacteria or beads above the fluidic drag force, the on-chip concentration and trapping of polystyrene beads and Listeria monocytogenes V7 can be accomplished.12 Mixed populations of E. coli and L. monocytogenes bacteria can also be directed and released through DEP pulses. Furthermore, larger particles can be differentially captured and concentrated on the electrodes based on the large particle size, whereas smaller particles are not capture but are removed with the fluidic flow.13 When DEP forces do not overcome the fluidic dragging forces on the particles, the bead or cell is not captured, but rather moved within the fluidic stream. As shown in Figure 2C, beads can be focused into the central region of the fluid stream sufficient to retain the beads in the central channel. This could be due to the combined effect of particle-to-particle influences within the electrical field, fluid velocities greater than DEP dragging forces, the combination of these, or some other undefined effect.
Newer advances in contactless DEP allow for maximizing cell capture and manipulation with the minimum required field, thus protecting cell types, to the greatest extent, during DEP manipulation.1,9 Contactless DEP offers promise to the microfluidics community for sorting, collecting and positioning cells within microfluidic devices. We anticipate that with the increased demand for, and implementation of, DEP for manipulation within microfluidic devices, further discoveries and innovations will expand the understanding and influence of DEP forces.
PCBs can be manufactured through high-volume processes at an affordable price with a rapid fabrication turnaround time, making them good platforms for commercialization. In addition, PCBs are easy to use and accessible for scientists in a range of disciplines for on-chip cell manipulations and sorting.
When designing the layout of the PCB electrodes, consideration should be given to the desired particle/cell trajectory. Key factors for consideration include particle size and type (bead and/or cell), cell type, microchannel size, flow velocity, electrode spacing (which determines the electrical field strength), coverslip thickness, and the fluid conductivity. These factors influence the force required and available to manipulate the particle or cell, and ultimately the separation efficiency. Our protocol presented here demonstrates an effective setup for initiating DEP for microsphere and cell separation. For potential applications, users should match the architecture of the microfluidic channel design with the desired flow paths for cells and beads, as well as the electrode patterns to optimize efficiency for each application. Electrode spacing and coverslip thickness can be used according to the previously reported guidelines when designing the microfluidic channel layout.1
The conductivity and permittivity of the cell/particle and the surrounding media must be different enough to facilitate positive or negative DEP, while keeping the cells intact. The polarity of DEP for a spherical particle can be determined from the real part of a complex value of the following Clausius-Mosotti factor at the frequency ω of DEP voltage.
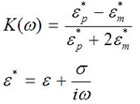
Equation 1
In this equation ε, is permittivity, σ is conductivity, and ε* is complex permittivity. The subscripts p and m denote the particle and media, respectively. When a cell has a higher permittivity than the media, or the real part of the Clausius-Mosotti factor becomes positive, the particle becomes more polarized than the surrounding media. Due to a non-uniform electric field, the particle’s polarization becomes non-uniform and this creates a positive DEP (+DEP) force that draws the cell toward the region with higher electric field intensity. If a cell has a lower permittivity than the surrounding media, or the real part of the Clausius-Mosotti factor becomes negative, it will undergo negative DEP (-DEP), and the cell is forced toward the minimal field region. If the cell and media have approximately the same complex permittivity, no force can be generated to manipulate the cell. For this reason, pure water is a preferred media for the DEP particle manipulation. However, to avoid the osmotic stress on the cell from pure water, low conductivity media was formulated to keep the conductivity unchanged, but to increase osmolarity to decrease osmotic stress to the cells. Conventional cell culture media or physiological buffers, such as DMEM or PBS, have high conductivity, which is not suitable for the DEP manipulation.
We also have previously demonstrated that cells can be captured on sensors with DEP using low-conductivity media. After a brief period for cell attachment, the low conductivity media can be replaced for the required cell culture media to support cell growth for days.11
From our experience, fluorescent beads are very bright with respect to the live cell fluorescence, thus it can be a challenge to match the bead intensity to the intensity of a living and fluorescing cell. To improve the visualization of both the cells and the beads, we used DIC microscopy on an upright microscope for imaging both. To display the cells and beads, we presented the data in an intensity glow-scale image, which retains the data in a wide color spectrum for easier viewing. Thus, when designing the study of interest, the imaging parameters and resources must be considered.
In summary, we demonstrate the ability to selectively actuate beads and cells into separate channels using DEP. With the increasing utility of microfluidic channels for cell biology, biochemistry and bioengineering applications, DEP is a desirable option for cell collection, placement and sorting. The PCB electrodes fabrication is inexpensive and convenient, the electrodes are easy to use, and the rapid fabrication times are ideal for the implementation of DEP.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We express appreciation to Woo-Jin Chang for providing the trifurcating channels for DEP actuation and to Mitchell Collens for his assistance in equipment setup. The work has been supported by US NSF through Grant OISE-0951647 and by Taiwan NSC through Grant 99-2911-1-002-007.
Materials
| Material Name | Type | Company | Catalogue Number | Comment |
|---|---|---|---|---|
| Sylgard 184 kit (PDMS) | Elsworth Adhesives | 184 SIL ELAST KIT 0.5KG | Dow Corning Sylgard | |
| 16 awg hook-up wire | Belden | 8521 | (Type MW) Mil-W-76C-PVC | |
| Gold seal Cover Glass | Ted Pella | 260320 | No. 0 thick, 24 x 60 mm | |
| Miltex Biopsy Punch | VWR Scientific | 95039-104 | Miltex, assorted sizes | |
| Custom PCB electrodes | Bay Area Circuits | Custom parts | ||
| Mineral oil | Fisher Scientific | O121-1 | ||
| Deionized water | House DI water supply | None | Use filtered DI water | |
| D-Glucose Anhydrous Granular AR | Mallinckrodt Chemicals | 4912-06 | ||
| Sucrose, Crystal | Mallinckrodt Chemicals | 8360-06 | ||
| Fluorescent polymer microspheres | Bangs Laboratories | FS07F/9277 | 15μm Dragon green microspheres | |
| HT-29 cell line | ATCC | HTB-38D | Human colon adenocarcinoma | |
| Typsin 0.05% EDTA | Invitrogen | 25300054 | ||
| Eclipse E600FN upright microscope | Nikon | Eclipse E600FN | ||
| Phantom V310 High-speed imaging camera | Phantom | V310 | ||
| BX51 upright research microscope | Olympus | BX51 | ||
| SpotFlex high resolution color camera | Diagnostic Instruments | FX1520 | ||
| Function generator | Agilent | 3325A | ||
| RF Power amplifier | EIN | 2100L |
References
- Park, K., Suk, H. J., Akin, D. Dielectrophoresis-based cell manipulation using electrodes on a reusable printed circuit board. Lab Chip. 9, 2224-2224 (2009).
- Nilsson, J., Evander, M., Hammarstrom, B. et al., Review of cell and particle trapping in microfluidic systems. Anal. Chim. Acta. 649, 141-141 (2009).
- Fu, A. Y., Chou, H. P., Spence, C. An integrated microfabricated cell sorter. Anal. Chem. 74, 2451-2451 (2002).
- Rhee, S. W., Taylor, A. M., Cribbs, D. H. External force-assisted cell positioning inside microfluidic devices. Biomed. Microdevices. 9, 15-15 (2007).
- Zhang, H., Liu, K. K. Optical tweezers for single cells. J. R. Soc. Interface. 5, 671-671 (2008).
- Hunt, T. P., Westervelt, R. M. Dielectrophoresis tweezers for single cell manipulation. Biomed. Microdevices. 8, 227-227 (2006).
- Vahey, M. D., Voldman, J. An equilibrium method for continuous-flow cell sorting using dielectrophoresis. Anal. Chem. 80, 3135-3135 (2008).
- Burgarella, S., Bianchessi, M., Merlo, S. A Modular Platform for Cell Characterization, Handling and Sorting by Dielectrophoresis. Cytometry A. 77A, 189-18 (2010).
- Shafiee, H., Sano, M. B., Henslee, E. A. Selective isolation of live/dead cells using contactless dielectrophoresis (cDEP). Lab Chip. 10, 438-438 (2010).
- Vahey, M. D., Voldman, J. High-Throughput Cell and Particle Characterization Using Isodielectric Separation. Anal. Chem. 81, 2446-2446 (2009).
- Park, K., Jang, J., Irimia, D. Living cantilever arrays’ for characterization of mass of single live cells in fluids. Lab Chip. 8, 1034-1034 (2008).
- Gomez-Sjoberg, R., Morisette, D. T., Bashir, R. Impedance microbiology-on-a-chip: Microfluidic bioprocessor for rapid detection of bacterial metabolism. J. Microelectromech. Syst. 14, 829-829 (2005).
- Li, H. B., Zheng, Y. N., Akin, D. Characterization and modeling of a microfluidic dielectrophoresis filter for biological species. J. Microelectromech. Syst. 14, 103-103 (2005).

