In Vitro Ubiquitination and Deubiquitination Assays of Nucleosomal Histones
Summary
Ubiquitination is a post-translational modification that plays important roles in cellular processes and is tightly coordinated by deubiquitination. Defects in both reactions underlie human pathologies. We provide protocols for conducting ubiquitination and deubiquitination reaction in vitro using purified components.
Abstract
Ubiquitination is a post-translational modification that plays important roles in various signaling pathways and is notably involved in the coordination of chromatin function and DNA-associated processes. This modification involves a sequential action of several enzymes including E1 ubiquitin-activating, E2 ubiquitin-conjugating and E3 ubiquitin-ligase and is reversed by deubiquitinases (DUBs). Ubiquitination induces degradation of proteins or alteration of protein function including modulation of enzymatic activity, protein-protein interaction and subcellular localization. A critical step in demonstrating protein ubiquitination or deubiquitination is to perform in vitro reactions with purified components. Effective ubiquitination and deubiquitination reactions could be greatly impacted by the different components used, enzyme co-factors, buffer conditions, and the nature of the substrate. Here, we provide step-by-step protocols for conducting ubiquitination and deubiquitination reactions. We illustrate these reactions using minimal components of the mouse Polycomb Repressive Complex 1 (PRC1), BMI1, and RING1B, an E3 ubiquitin ligase that monoubiquitinates histone H2A on lysine 119. Deubiquitination of nucleosomal H2A is performed using a minimal Polycomb Repressive Deubiquitinase (PR-DUB) complex formed by the human deubiquitinase BAP1 and the DEUBiquitinase ADaptor (DEUBAD) domain of its co-factor ASXL2. These ubiquitination/deubiquitination assays can be conducted in the context of either recombinant nucleosomes reconstituted with bacteria-purified proteins or native nucleosomes purified from mammalian cells. We highlight the intricacies that can have a significant impact on these reactions and we propose that the general principles of these protocols can be swiftly adapted to other E3 ubiquitin ligases and deubiquitinases.
Introduction
Ubiquitination is one of the most conserved post-translational modifications and is critical for a wide variety of organisms including yeast, plants and vertebrates. Ubiquitination consists of the covalent attachment of ubiquitin, a highly conserved 76 amino acid polypeptide, to target proteins and occurs in three sequential steps involving three enzymes, i.e., E1-activating, E2-conjugating and E3 ligase1,2,3. This post-translational modification plays central roles in a wide spectrum of biological processes. Indeed, the E3 ligases, which provide the specificity of the reaction, constitute a large superfamily of enzymes and are the most abundant enzymes of the ubiquitin system4,5,6. The downstream effects of protein ubiquitination depend on nature of the modification: monoubiquitination, multi-monoubiquitination, and linear or branched polyubiquitination. Monoubiquitination is rarely associated with proteasomal degradation, but instead this modification is involved in mediating various signaling events. Polyubiquitination involves the N-terminal or the lysine residues in ubiquitin molecule itself, and the destiny of a polyubiquitinated protein depends on which residue is involved in ubiquitin chain extension. It has long been known that polyubiquitination mediated by lysine 48 of ubiquitin induces proteasomal degradation. On the contrary, polyubiquitination via lysine 63 of ubiquitin is often associated with protein activation7,8,9. Similar to other important post-translational modifications, ubiquitination is reversible and ubiquitin removal from proteins is ensured by specific proteases termed deubiquitinases (DUBs), which have emerged as important regulators of cellular processes2,10. Importantly, many DUBs are highly specialized, and regulate, through deubiquitination, specific substrates, indicating that a fine balance between ubiquitination and deubiquitination is critical for protein function. E3s and DUBs, along with the proteasome degradation machinery and accessory factors, form the Ubiquitin Proteasome System (UPS, with >1200 genes) which regulates major signaling pathways, several of which are associated with cell growth and proliferation, cell fate determination, differentiation, cell migration, and cell death. Importantly, deregulation of several signaling cascades involving ubiquitination promotes tumorigenesis and neurodegeneration diseases5,11,12,13,14.
Ubiquitination plays pervasive roles in chromatin biology and DNA-dependent processes15,16,17. For instance, monoubiquitination of histone H2A on lysine 119 (hereafter H2A K119ub) is a critical post-translational modification involved in transcriptional repression and DNA repair18,19,20,21,22. H2A K119ub is catalyzed by the Polycomb Repressive Complex 1 (PRC1), which plays a key role in the maintenance of epigenetic information and is highly conserved from Drosophila to human. Canonical PRC1 is constituted notably by the RING1B and BMI1, which are the core E3 ubiquitin ligase complex responsible for the above-mentioned ubiquitination event22,23. In Drosophila, H2A monoubiquitination (H2A K118ub which corresponds to H2A K119ub in mammalians) is reversed by the DUB Calypso, which interacts with Additional Sex Comb (ASX) forming the Polycomb-repressive DUB (PR-DUB) complex24. The mammalian ortholog of calypso, BAP1, is a tumor suppressor deleted or inactivated in various human malignancies25,26,27,28,29,30,31,32,33. BAP1 regulates DNA-dependent processes in the nucleus and Calcium-signaling-mediated apoptosis at the endoplasmic reticulum33,34,35,36,37,38,39,40,41,42. BAP1 assembles multi-subunit protein complexes containing transcription regulators notably ASXL1, ASXL2 and ASXL3 (ASXLs), three orthologues of ASX38,43. ASXLs use the DEUBiquitinase ADaptor (DEUBAD) domain, also termed ASXM domain, to stimulate BAP1 DUB activity35,36,44. Hence, ASXLs play important roles in coordinating BAP1 DUB activity at chromatin and more broadly its tumor suppressor function.
Several methods exist to study ubiquitination and deubiquitination processes. Notably, biochemical assays using proteins purified from bacteria remain very powerful in demonstrating direct ubiquitination of, or removal of ubiquitin from, specific substrates. These experiments can be conducted to investigate a range of parameters such as the determining the requirement of minimal complexes, determining reactions kinetics, defining structure/function relationships, and understanding the impact of pathological gene mutations. Here, we provide protocols to conduct ubiquitination and deubiquitination reactions on chromatin substrates with purified components. As a model system, in vitro ubiquitination and deubiquitination of nucleosomal H2A protein is presented. Bacteria-purified proteins assembled in minimal complexes of RING1B/BMI1 and BAP1/DEUBAD are used for ubiquitination or deubiquitination of nucleosomal H2A, respectively.
Protocol
1. GSH-agarose Affinity Purification of the GST-RING1B(1-159)-BMI1(1-109) E3 Ubiquitin Ligase Complex
- Use the pGEX6p2rbs-GST-RING1B (1-159 aa) – BMI1 (1 -109aa) bacteria expression construct to transform BL21 (DE3) bacteria (see Table of Materials)23. This construct allows the expression of murine RING1B domain 1-159 fused to BMI1 domain 1-109 with GST tag in pGEX-6P-2 backbone.
- Perform an overnight starter culture by inoculating RIL-bacteria expressing GST-RING1B (1-159 aa) – BMI1(1 -109 aa) in 20 mL of LB broth medium in presence of 100 µg/mL ampicillin and 50 µg/mL chloramphenicol. Incubate by shaking at 37 °C overnight. On the next day, add the starter culture to a 1 L flask containing 500 mL of fresh LB broth medium (1/26 dilution) with ampicillin and incubate the culture in the shaker at 37 °C for 2 to 4 h.
- During the incubation time, measure the OD at 600 nm with a spectrophotometer. When bacteria culture reaches 0.6 OD units at 600 nm, take a 1 mL aliquot in an 1,5 mL tube as the non-induced sample. Add 400 µM of Isopropyl β-D-1-thiogalactopyranoside (IPTG) to the 1 L culture to induce protein expression. Incubate bacteria at 25 °C for 6 h to overnight.
- Centrifuge the 1 mL aliquots at 14,000 x g for 30 s, discard the supernatant, resuspend the pellet in 200 μL of Laemmli sample buffer, and keep for SDS-PAGE and Coomassie blue analysis of protein induction.
- Transfer the induced bacteria culture from the flask into a clean centrifugation bottle and spin down at 3,500 x g for 15 min at 4 °C. Discard the supernatant and resuspend the pellet with 40 mL of cold PBS and spin down at 3,500 x g for 15 min at 4 °C.
- Discard the supernatant and freeze the pellet at -80 °C or proceed to the next step. If the proteins are induced at the expected molecular weight, proceed to the next step.
- Resuspend the bacteria pellet in 25 mL of ice-cold lysis buffer A (50 mM Tris-HCl pH 7.5, 100 mM NaCl, 1 mM EDTA, 1% NP40, 1 mM PMSF, 0.5 mM DTT, and 1/100 anti-protease cocktail containing AEBSF, Aprotinin, Bestatin, E-64, Leupeptin, and Pepstatin A). Make sure that all the bacteria pellet is resuspended. PMSF and anti-protease cocktail must be freshly added to the lysis buffer, only at the time of bacteria lysis.
CAUTION: Always keep the sample on ice.
NOTE: Lysozyme at 100 µg/mL can be added to enhance bacteria lysis. - Incubate the bacteria lysate on ice for 15 min. Use a probe sonicator and sonicate at 70% output amplitude for 30 s (4–5x), and then centrifuge at 21,000 x g for 20 min at 4 °C.
CAUTION: During sonication, keep always the samples on ice. - During the time of centrifugation, take 500 µL packed GSH-agarose beads (50% slurry) in a 15 mL tube and add 10 mL of buffer A without PMSF and anti-proteases. Mix briefly and spin at 2,500 g for 3 min at 4 °C, repeat this wash step two more times and keep the beads on ice.
- Following bacterial lysate centrifugation, collect the supernatant and transfer it into a 50 mL conical tube. Take an aliquot of 100 μL as a total lysate and add 100 μL of 2x Laemmli sample buffer for SDS-PAGE and Coomassie blue analysis.
- Filter the lysate through 0.45 μm pore syringe filter and mix it with the GSH beads. Incubate at 4 °C with shaking for 5 h. Spin down the beads at 2,500 x g for 3 min, remove, and save the supernatant as the flow through. Wash the proteins-GSH bound beads 6 times (5 mL each) with buffer A containing anti-protease cocktail and PMSF.
- After the last wash, transfer the beads along with 10 mL of buffer into an empty chromatography column, add 1.5 mL buffer A containing 25 mM glutathione, and collect the elution by gravity into 1.5 mL microtubes. Repeat the elution procedure 4 times.
NOTE: Glutathione stock solution is prepared at 200 mM concentration in 50 mM Tris-HCl, pH 7.5 - Take an aliquot of 10 µL from each one of the 4 elutions and add 10 µL of 2x Laemmli sample buffer. These samples will be used for SDS-PAGE and Coomassie blue analysis to determine the presence and the purity of the purified proteins.
NOTE: Following the last elution, keep the beads in buffer at 4 °C for recycling and future use. - Choose the eluted fractions showing reasonable amounts of purified proteins for further analysis. Make small aliquots of the preparation. Store the purified proteins in -80°C. Usually, protein concentration will range from 0.5 µg to 2 µg per µL and samples can be stored in this state.
- OPTIONAL: Concentrate the sample or change buffer using a 10K centrifugal filter unit
2. Purification of the BAP1/DEUBAD (ASXL2) Deubiquitinase Complex
- Use pET30a+-His-BAP138 and pDEST-MBP-DEUBAD (ASXL2)35 bacterial expression constructs to transform BL21 (DE3) RIL bacteria for the production of His-BAP1 and MBP-DEUBAD respectively.
- Perform two separate overnight starter cultures by incubating His-BAP1 and MBP-DEUBAD (ASXL2) expressing bacteria in 20 mL of LB broth of ampicillin medium containing 50 µg/mL chloramphenicol and the corresponding antibiotic for each plasmid, 100 µg/mL of kanamycin or 100 µg/mL of ampicillin respectively. Place on shaker at 37°C.
- Perform steps 1.3–1.6.
- Resuspend the bacteria pellets in 25 mL of ice-cold lysis buffer B (50 mM Tris pH 7.3, 500 mM NaCl, 3 mM β-Mercaptoethanol, 0.2% Triton X-100, 1 mM PMSF and 1/100 anti-protease cocktail). Mix equal volumes of the His-BAP1 with the MBP-DEUBAD lysates and incubate on ice for 15 min. Sonicate and then centrifuge the lysate as indicated in protocol 1 (step 9).
- During the centrifugation, prepare 500 µL packed Ni-NTA agarose beads (50% slurry) by washing them three times with buffer B without PMSF and anti-protease cocktail.
- Following bacterial lysate centrifugation, collect the supernatant and transfer it into a 50 mL conical tube. Take an aliquot of 100 μL as a total lysate and add 100 μL of 2x Laemmli sample buffer for SDS-PAGE and Coomassie blue analysis.
- Filter the lysate through 0.45 μm pore syringe filter and mix it with the Ni-NTA beads. Incubate at 4°C with shaking for 5 h. Spin down the beads at 2,500 x g for 3 min, remove, and save the supernatant as the flow through.
- Wash the beads 6 times (5 mL each) with buffer B containing 20 mM 1,3-Diaza-2,4-cyclopentadiene (Imidazole).
NOTE: Make a stock solution of 2 M imidazole at pH 7.3.
- Wash the beads 6 times (5 mL each) with buffer B containing 20 mM 1,3-Diaza-2,4-cyclopentadiene (Imidazole).
- After the last wash, transfer the beads with 10 mL of buffer into an empty chromatography column, add 1 mL buffer B containing 200 mM Imidazole and collect the elution by gravity into 1.5 mL microtubes containing 10 µL DTT (500 mM) and 2 µL EDTA (500 mM, pH: 8). Repeat the elution procedure 4 times. Take 10 µL of each elution fraction and add 10 µL of 2x Laemmli sample buffer for SDS-PAGE and Coomassie blue analysis. Pool the desired eluted fractions.
NOTE: Keep the beads in buffer at 4 °C for recycling and future use. - Dilute the eluted complex 3 times with buffer C (50 mM Tris pH 7.3, 300 mM NaCl, 1% Triton X-100, 5 mM DTT, 1 mM EDTA, 1 mM PMSF, and 1/100 protease inhibitor cocktail) prior to incubation with the amylose agarose beads.
- Prepare amylose agarose beads (500 µL packed) by washing them 2 times with the buffer C without adding PMSF and anti-protease cocktail. Add the beads to the diluted elution and incubate overnight at 4 °C.
- Centrifuge at 2,500 x g for 3 min and keep the flow through. Wash the proteins-bounds beads with 5 mL of buffer C (5–6x).
- Keep the purified His-BAP1/MBP-DEUBAD complex on the amylose agarose beads in buffer D (50 mM HEPES pH 8.0, 50 mM NaCl, 10% Glycerol, and 1 mM DTT) and store at -80 °C. Take 20 µL of the beads fraction (50% beads solution) and add 20 µL of 2x Laemmli sample buffer for SDS-PAGE and Coomassie blue analysis.
3. Purification of the Nucleosomes from HEK293T Cells
- Culture HEK293T cells in complete Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% newborn calf serum (NBS), 2 mM L-glutamine, and 1% penicillin/streptomycin.
- Plate around 12 x 106 HEK293T cells in 20 mL of complete media per 15 cm culture dish one day before the transfection (10 dishes were used). Before transfection, change the cell’s medium to 12 mL of serum-free medium and transfect the cells with 21 µg of pCDNA-Flag-H2A using 63 µl of polyethylenimine (PEI) at 1 mg/mL36. Change to complete medium 12 h later.
- Three days post-transfection, wash the cells with 15 mL ice-cold PBS (twice) and harvest them in 3 mL of ice-cold PBS using a cell scraper. Centrifuge at 2,100 x g for 8 min at 4 °C. Discard the PBS and proceed to the lysis step or freeze the cell pellet at -80 °C.
- Resuspend the cell pellet in about 10 volumes of buffer E (50 mM Tris-HCl pH 7.3, 420 mM NaCl, 1% NP-40, 1 mM MgCl2, 1 mM PMSF, 1/100 protease inhibitor cocktail, and 20 mM of N-methylmaleimide (NEM)) and incubate on ice for 20 min. Adding N-methylmaleimide (NEM) is critical to inhibit DUBs that co-purify with nucleosomes.
- After centrifugation at 3,000 x g for 5 min, discard the supernatant and resuspend the chromatin pellet in 10 volumes of buffer E. Mix by inversion and centrifuge at 3,000 x g for 5 min.
- Repeat the wash step another time using buffer E and two times using buffer F "MNase Buffer" (20 mM Tris-HCl pH 7.5, 100 mM KCl, 2 mM MgCl2, 1 mM CaCl2, 0.3 M sucrose, 0.1% NP-40, 1 mM PMSF, and 1/100 protease inhibitor cocktail).
CAUTION: Pellet will float during washes and centrifugation.
- Repeat the wash step another time using buffer E and two times using buffer F "MNase Buffer" (20 mM Tris-HCl pH 7.5, 100 mM KCl, 2 mM MgCl2, 1 mM CaCl2, 0.3 M sucrose, 0.1% NP-40, 1 mM PMSF, and 1/100 protease inhibitor cocktail).
- Resuspend the chromatin in 5 mL buffer F and treat the pellet with Micrococcal nuclease (MNase) (3 U/mL for 10 min at room temperature).
NOTE: The reaction can be aided by mixing using a Dounce Homogenizer. - After incubation, take a 40 μL aliquot of the mixture for analysis of nucleosomal DNA fragments. Mix this aliquot with 40 μL of Phenol/chloroform and 20 μL of 6x DNA loading buffer, vortex, spin at 18,000 x g for 2 min, and load the DNA on a 2% agarose gel.
- When the DNA is predominantly a 147 bp fragment corresponding to mononucleosomes, stop the reaction by adding 5 mM EDTA at final concentration.
- Following centrifugation at 21,000 x g for 20 min at 4°C, incubate the soluble chromatin fraction with anti-Flag resin overnight at 4 °C. Wash the beads with 6x Buffer G (50 mM Tris-HCl, pH 7.3, 5 mM EDTA, 150 mM NaCl, 1% NP-40, 1 mM PMSF, 1 mM DTT, 1/100 protease inhibitors cocktail).
- Transfer the beads with 5 mL buffer G into an empty chromatography column. Elute the beads-bound nucleosomes with 200 μg/mL of Flag elution peptide. Use an elution buffer composed of buffer G containing 200 μg/mL Flag elution peptides (see Table of Materials) and 1/5 (vol:vol) 1 M Tris, pH 8.0. Elute the nucleosomes 3x with 260 μL Flag elution Buffer (2 h each elution).
- Test the 3 elutions by taking an aliquot for phenol-chloroform extraction and load the DNA in a 2% agarose gel. Take 10 µL of each elution fraction and add 10 µL of Laemmli sample buffer 2x for SDS-PAGE and Coomassie blue analysis and load each elution for Western Blot detection of ubiquitinated H2A. Store the elution at -80 °C. In general, purification from human cells yields less amount of proteins than that from bacteria, with concentrations ranging from 0.1 to 0.5 μg/μL.
4. Nucleosome Ubiquitination Assay Using BMI1/RING1B E3 Ubiquitin Ligase Complex
- Centrifuge the nucleosomes through 10K pore 0.5 mL centrifugal filters to change the substrate from the elution buffer to the reaction buffer H (25 mM Tris, pH 7.5; 10 mM MgCl2, and 5 mM ATP). Alternatively, commercially available nucleosomes can be used for the assay.
NOTE: Changing substrate suspension solution to reaction buffer ensures reproducibility and prevents potential E3 ligase inhibition by compounds present in the Flag elution mixture. - Incubate 1 μg of the nucleosomes in 40 μL total volume of buffer H. Add Ub Activating Enzyme (UBE1) (250 ng), Ub (50 ng/μL) and E2 Ub-conjugating enzymes (672 ng), and 1 μg of BMI1/RING1B E3 ubiquitin ligase complex. Incubate the reaction for 3 h at 37 °C with occasional shaking.
NOTE: Several controls can be conducted in parallel including omitting, E1, E2, E3, ATP and ubiquitin. This ensures the specificity for the ubiquitination reaction. - Stop the reaction by adding 40 μL of 2x Laemmli sample buffer and analyze histone H2A K119 ubiquitination by SDS-PAGE and western blotting, according to standard procedures. Use either anti-H2A or anti-H2A K119ub antibodies (see Table of Materials).
5. In Vitro Nucleosomal H2A DUB Assay Using BAP1/DEUBAD
- Use the purified nucleosomes for the in vitro deubiquitination assay. Resuspend 100–500 ng of nucleosomes in 40 μL buffer I (50 mM Tris-HCl, pH 7.3, 1 mM MgCl2, 50 mM NaCl, and 1 mM DTT). Add 1 μg of BAP1 bacteria-purified His-BAP1 and MBP-DEUBAD. Carry out the deubiquitination reaction for 3 h at 37 °C with occasional shaking.
- Stop the in vitro reaction by adding 40 μL of 2x Laemmli buffer and analyze by immunoblotting.
Representative Results
GST-BMI1 and RING1B proteins are well produced in bacteria and can be readily extracted in the soluble fraction. Figure 1A shows a Coomassie blue staining for a typical purification of the GST-BMI1-RING1B complex. The GST-BMI1 and RING1B bands migrate at the expected molecular weight, ~45 kDa and ~13 kDa respectively. Notably the E3 ligase complex is highly homogenous with very low levels of bacteria proteins contaminants and/or degradation products. Moreover, the stoichiometry of the purified complex is optimal, as with a molar ratio of 1:1, the band intensity of GST-RING1B protein is expected to be two to three times stronger than that of BMI1. Similarly, the purification of His-BAP1/MBP-DEUBAD complex also resulted in relatively highly homogenous preparations with bands at ~90 kDa and ~55 kDa, respectively (Figure 1B). The tandem affinity purification of BAP1 and DEUBAD, through nickel-agarose and amylose-agarose respectively ensures and adequate stoichiometry and the removal of free BAP1 from the purified complex. On the other hand, the purified nucleosomal fraction is essentially composed by 147bp band indicating the presence of highly enriched mononucleosomal fraction (Figure 1C). Coomassie blue staining of these purified nucleosomes shows a typical band pattern of the four histone subunits with stoichiometric amounts (Figure 1D). Commercially available recombinant mononucleosomes also display similar protein and DNA patterns when migrated on SDS-PAGE and agarose gels respectively. Of note, monoubiquitinated forms of H2A and Flag-H2A can be readily distinguished on HEK293T-purified nucleosomes. We used recombinant nucleosomes along with E1/E2/Ubiquitin/ATP and observed that ubiquitination of histone H2A with the BMI1-RING1B complex shows a time-dependent increase of the ubiquitinated form of the protein, while the levels of the non-modified form are concomitantly decreased. The ubiquitination reaction is total, as virtually all H2A proteins are transformed to H2A K119ub (Figure 1E). On the other hand, deubiquitination assay was conducted on native nucleosomes. Following incubation of these nucleosomes with BAP1/DEUBAD, we observed a time-dependent-reduction of H2A K119ub signal (Figure 1F). Note that two bands of ubiquitinated H2A observed with anti-H2A K119ub correspond to transfected Flag-H2A and endogenous H2A migrating at ~30 kDa and ~25 kDa bands respectively.
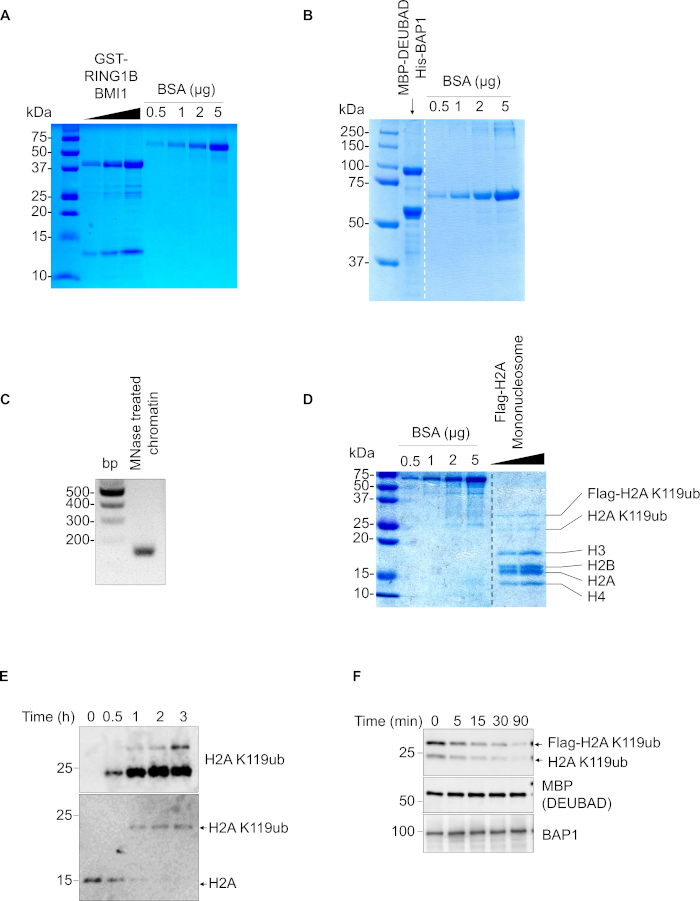
Figure 1: Purification, ubiquitination, and deubiquitination. (A) Purification of GST-RING1B-BMI1 proteins and analysis using Coomassie blue staining. GST-RING1B-BMI1 were produced in bacteria and purified using GST affinity beads. To confirm purification purity and proteins size, elutions were loaded on 15% SDS-PAGE gel and stained with Coomassie blue. Different amount of BSA are used to determine protein quantity. The stained gel was scanned to generate the figure. (B) Purification of MBP-DEUBAD/His-BAP1 complex. MBP-DEUBAD and HIS-BAP1 were produced separately in bacteria. After lysis, MBP-DEUBAD and HIS-BAP1 were mixed and purified using nickel and maltose beads. The purified complex was loaded on 8% SDS-PAGE gel and stained with Coomassie blue. (C) Purification of mononucleosome following MNase treatment. Chromatin from HEK293T expressing pcDNA.3 Flag-H2A was extracted and treated with MNase to generate and purify mononucleosomes. To confirm the generation of mononucleosome, a fraction of chromatin was taken for phenol chloroform extraction and detection under UV light. Loading on 2% agarose gel reveals a typical 147 base-pair DNA fragment indicative of mononucleosome. (D) Purified mononucleosomes were used for Coomassie blue staining. (E) In vitro ubiquitination of nucleosomes. Recombinant nucleosomes were incubated at 37 °C with the bacterial purified E3 ubiquitin ligase dimer, GST-RING1B/BMI1 and E1/E2/ATP/Ubiquitin, for the indicated times. H2A monoubiquitination (H2A K119ub) was analysed by western blotting. (F) Deubiquitination assay of H2A K119ub using the BAP1/DEUBAD deubiquitinase complex. In vitro deubiquitination assays were conducted using bacteria purified His-BAP1/MBP-DEUBAD and mammalian purified nucleosomes. Reactions were done for the indicted times and analyzed by western blotting. Please click here to view a larger version of this figure.
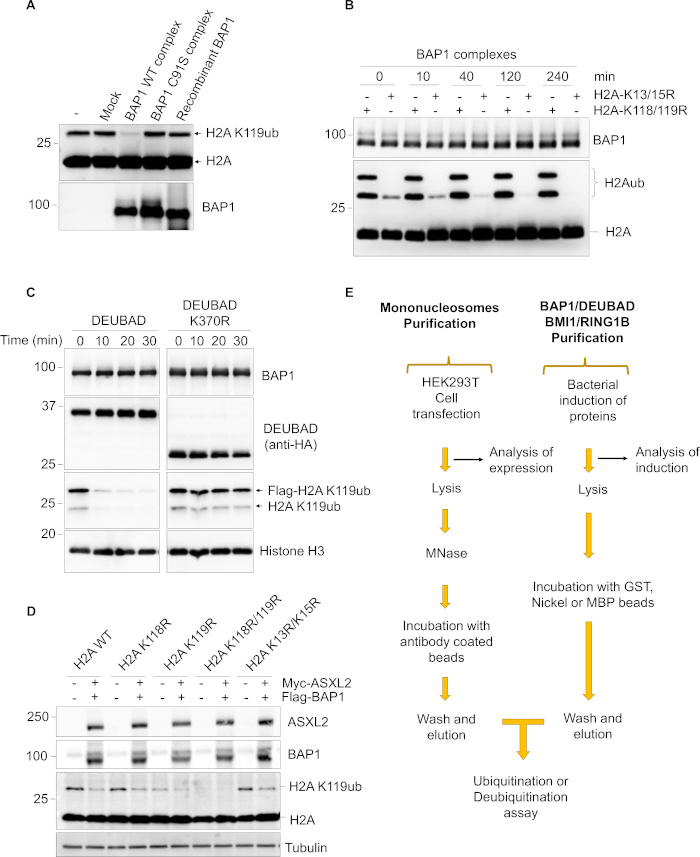
Figure 2: Controls and overview of the protocol. (A) Purified mammalian BAP1 or its catalytic dead mutant (C91S) as well as bacteria-purified recombinant BAP1 were used for H2A deubiquitination on nucleosomes. Nucleosomes incubated with buffer alone or with elutions obtained from the mock purification were also included as controls. (B) BAP1 does not deubiquitinate H2A K13/K15 whose ubiquitination is mediated by RNF168 E3 ligase. HEK293T cells were co-transfected with either H2A K13R/K15R or H2A K118R/K119R along with RNF168 to ensure ubiquitination on K13/K15 residues. The purified nucleosomes were used for deubiquitination by the BAP1 complexes. The reaction was stopped at different time points and subjected to immunoblotting. (C) Purification of non-ubiquitinated and monoubiquitinated DEUBAD in complex with BAP1 from mammalian cells. The purified complex was used for deubiquitination assay on nucleosomal H2A K119ub. Reactions were done for the indicated time points and analyzed by western blotting. (D) BAP1 deubiquitinates H2A K119ub in vivo. HEK293T cells were co-transfected with H2A or H2A mutants (H2A K118R, K119R, K118R/K119R or K13R/K15R) along with BAP1 and ASXL2 constructs. Two days post transfection, cells were harvested for immunoblotting using the indicated antibodies. (E) Schematic representation of the experimental workflow. Please click here to view a larger version of this figure.
Discussion
There are several advantages of establishing robust in vitro ubiquitination and deubiquitination assays for proteins of interest. These assays can be used to: (i) establish optimal conditions and define minimal requirement for these reactions, (ii) determine enzymatic kinetic and biochemical constants, (iii) define the roles of cofactors or inhibitors that can impact these reactions, (iv) identify interaction interfaces, (v) test the impact of artificial or disease-associated mutations and (vi) establish assay conditions that can be further used to develop high throughput chemical screening assays. Here, we exemplify these assays by describing how we perform BMI1/RING1B-mediated H2A ubiquitination and BAP1/ASXL-mediated H2A deubiquitination on nucleosomal substrates.
The ubiquitination of histone H2A can be recapitulated in vitro using minimal components, as almost complete ubiquitination of H2A can be observed using BMI1-RING1B complex. Of note, this reaction is conducted on recombinant nucleosomes. These have the advantage of being free from other histones post-translational modifications that occur in mammalian cells. Nonetheless, the ubiquitination reaction can also be efficiently conducted on native nucleosomes. If the ubiquitination reaction is weak, the ratio of enzymes versus substrates can be increased to observe optimal ubiquitin ligation. Of note, if deubiquitination reactions or interaction assays are to be conducted following substrate ubiquitination, then ubiquitination can be directly conducted on beads-bound substrates. The ubiquitinated substrate can be recovered by pull-down and the supernatant containing the components of the ubiquitination reaction can be discarded. However, when enzymatic characteristics are to be established, enzymes need to be eluted and the stoichiometry of components evaluated by size exclusion chromatography.
The deubiquitination reaction requires less components than the ubiquitination reaction. However, purification of the DEUBAD alone generally lead to protein precipitation. Importantly, when the DEUBAD is purified with BAP1, this greatly increases its solubility. Of note, we suggest conducting deubiquitination assays with various amounts of the substrate and enzyme complex. The deubiquitination reaction can be inhibited by excess amounts of chromatin, which can be probably explained by increased concentrations of inhibitors associated with nucleosomes. We also observed deubiquitination with excess of free BAP1. Thus, several controls have to be included such as incubating nucleosomes with BAP1 alone (Figure 2A). It is also important to note that some DUBs might co-purify with chromatin, and even though we treat chromatin with NEM, DUBs might be partially reactivated following addition of DTT which reduces the catalytic cysteine. In addition, metalloproteinase DUBs might also be present, and these enzymes are insensitive to NEM and are thus not inhibited. Therefore, additional controls should be included consisting of only nucleosomes without adding enzymes and DUB catalytic dead mutant (Figure 2A). Of note, the specificity of BAP1 DUB activity toward H2A K119ub is well documented and was validated in our assay conditions. We purified from HEK293T cells nucleosomes expressing either H2A K118/K119R or H2A K13/K15R. Expression of RNF168 leads to an important ubiquitination on K13/K15. BAP1 DUB activity is observed only on nucleosome containing H2A K13/K15R corresponding to H2A K119ub (Figure 2B). Another consideration is that post-translational modifications that can be critical for enzymatic activity are likely not to be present in bacterial proteins. Thus in vitro reactions using components purified from mammalian cells could also be considered35. For instance, monoubiquitination of DEUBAD, a process observed in higher eukaryotes, greatly promote the deubiquitinase activity of BAP135 (Figure 2C). Thus, purification of components from mammalian cells could also be considered. In addition to in vitro reactions, it is possible to monitor BAP1 DUB activity directly in cells following transfection. For example, transfection of HEK293T cells with H2A construct can give a clear signal of H2A K119ub because of its abundance in cells. In these conditions, most of the ubiquitination happens on K118/K119 residues, as mutation of these sites to arginine lead to complete absence of H2A ubiquitination (Figure 2D). Co-expressing BAP1 and ASXL2 along with H2A allows to conclude on the BAP1 DUB activity in cells. However, several points need to be considered as overexpression can lead to erroneous results. Thus, these experiments need to be optimized and well controlled.
In summary, the protocols described here, recapitulated in Figure 2E, are straightforward, do not require a specialized infrastructure or experimental setup, and can be scaled-up to produce substantial amounts of modified nucleosomes for multiple applications downstream including enzymatic assays, mass spectrometry and protein-protein interaction assays.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Diana Adjaoud for technical assistance. This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (2015-2020), Genome Quebec (2016-2019) and Genome Canada (2016-2019) to E.B.A. E.B.A. is a senior scholar of the Fonds de la Recherche du Québec-Santé (FRQ-S). L.M and N.S.K. have a PhD scholarship from the FRQ-S. H.B had a PhD scholarship from the Ministry of Higher Education and from Scientific Research of Tunisia and the Cole Foundation.
Materials
| Amylose agarose beads | New England Biolabs | #E8021 | |
| Amicon Ultra 0.5 ml centrifugal filters 10K | Sigma-Aldrich | #UFC501096 | |
| Anti-H2AK119ub (H2Aub) | Cell Signaling Technology | #8240 | |
| Anti-Flag-agarose beads | Sigma-Aldrich | #A4596 | |
| Anti-protease cocktail | Sigma-Aldrich | #P8340 | |
| BL21 (DE3) CodonPlus-RIL bacteria | Agilent technologies | #230240 | |
| DMEM | Wisent | #319-005-CL | |
| Empty chromatography column | Biorad | #731-1550 | |
| Flag peptide | Sigma-Aldrich | #F3290 | |
| GSH-agarose beads | Sigma-Aldrich | #G4510 | |
| HEK293T | ATCC | #CRL-3216 | |
| Imidazole | Sigma-Aldrich | #I5513 | |
| Micrococcal nuclease (MNase) | Sigma-Aldrich | #N3755 | |
| Ni-NTA agarose beads | ThermoFisher Scientific | #88221 | |
| N-methylmaleimide (NEM) | Bioshop | #ETM222 | |
| Pore syringe filter 0.45 μm | Sarstedt | #83.1826 | |
| Polyethylenimine (PEI) | Polysciences Inc | #23966-1 | |
| pGEX6p2rbs-GST-RING1B(1-159)-Bmi1(1-109) | Addgene | #63139 | |
| Ub Activating Enzyme (UBE1) | Boston Biochem | #E-305 | |
| UBCH5C (UBE2D3) | Boston Biochem | #E2-627 |
References
- Ye, Y., Rape, M. Building ubiquitin chains: E2 enzymes at work. Nature Reviews Molecular Cell Biology. 10, 755-764 (2009).
- Komander, D., Clague, M. J., Urbe, S. Breaking the chains: structure and function of the deubiquitinases. Nature Reviews Molecular Cell Biology. 10, 550-563 (2009).
- Ciechanover, A. The unravelling of the ubiquitin system. Nature Reviews Molecular Cell Biology. 16, 322-324 (2015).
- Nakayama, K. I., Nakayama, K. Ubiquitin ligases: cell-cycle control and cancer. Nature Reviews Cancer. 6, 369-381 (2006).
- Senft, D., Qi, J., Ronai, Z. A. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nature Reviews Cancer. 18, 69-88 (2018).
- Zheng, N., Shabek, N. Ubiquitin Ligases: Structure, Function, and Regulation. Annual Review of Biochemistry. 86, 129-157 (2017).
- Iwai, K., Fujita, H., Sasaki, Y. Linear ubiquitin chains. NF-kappaB signalling, cell death and beyond. Nature Reviews Molecular Cell Biology. 15, 503-508 (2014).
- Kulathu, Y., Komander, D. Atypical ubiquitylation – the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nature Reviews Molecular Cell Biology. 13, 508-523 (2012).
- Yau, R., Rape, M. The increasing complexity of the ubiquitin code. Nature Cell Biology. 18, 579-586 (2016).
- Mevissen, T. E. T., Komander, D. Mechanisms of Deubiquitinase Specificity and Regulation. Annual Review of Biochemistry. 86, 159-192 (2017).
- Bedford, L., Lowe, J., Dick, L. R., Mayer, R. J., Brownell, J. E. Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nature Reviews Drug Discovery. 10, 29-46 (2011).
- Minton, K. Inflammasomes: Ubiquitin lines up for inflammasome activity. Nature Reviews Immunology. 14, 580-581 (2014).
- Popovic, D., Vucic, D., Dikic, I. Ubiquitination in disease pathogenesis and treatment. Nature Medicine. 20, 1242-1253 (2014).
- Upadhyay, A., Amanullah, A., Chhangani, D., Mishra, R., Mishra, A. Selective multifaceted E3 ubiquitin ligases barricade extreme defense: Potential therapeutic targets for neurodegeneration and ageing. Ageing Research Reviews. 24, 138-159 (2015).
- Hammond-Martel, I., Yu, H., Affar el, B. Roles of ubiquitin signaling in transcription regulation. Cellular Signalling. 24, 410-421 (2012).
- Schwertman, P., Bekker-Jensen, S., Mailand, N. Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nature Reviews Molecular Cell Biology. 17, 379-394 (2016).
- Uckelmann, M., Sixma, T. K. Histone ubiquitination in the DNA damage response. DNA Repair. 56, 92-101 (2017).
- Robzyk, K., Recht, J., Osley, M. A. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 287, 501-504 (2000).
- Hwang, W. W., et al. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Molecular Cell. 11, 261-266 (2003).
- Kim, J., Hake, S. B., Roeder, R. G. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Molecular Cell. 20, 759-770 (2005).
- Wood, A., et al. an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Molecular Cell. 11, 267-274 (2003).
- Wang, H., et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 431, 873-878 (2004).
- Buchwald, G., et al. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1B. The EMBO Journal. 25, 2465-2474 (2006).
- Scheuermann, J. C., et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 465, 243-247 (2010).
- Jensen, D. E., et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 16, 1097-1112 (1998).
- Harbour, J. W., et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 330, 1410-1413 (2010).
- Abdel-Rahman, M. H., et al. GermLine BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. Journal of Medical Genetics. , (2011).
- Bott, M., et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nature Genetics. 43, 668-672 (2011).
- Goldstein, A. M. GermLine BAP1 mutations and tumor susceptibility. Nature Genetics. 43, 925-926 (2011).
- Testa, J. R., et al. GermLine BAP1 mutations predispose to malignant mesothelioma. Nature Genetics. 43, 1022-1025 (2011).
- Wiesner, T., et al. GermLine mutations in BAP1 predispose to melanocytic tumors. Nature Genetics. 43, 1018-1021 (2011).
- Dey, A., et al. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science. 337, 1541-1546 (2012).
- Pena-Llopis, S., et al. BAP1 loss defines a new class of renal cell carcinoma. Nature Genetics. 44, 751-759 (2012).
- Bononi, A., et al. BAP1 regulates IP3R3-mediated Ca2+ flux to mitochondria suppressing cell transformation. Nature. 546, 549-553 (2017).
- Daou, S., et al. Monoubiquitination of ASXLs controls the deubiquitinase activity of the tumor suppressor BAP1. Nature Communications. 9, 4385 (2018).
- Daou, S., et al. The BAP1/ASXL2 Histone H2A Deubiquitinase Complex Regulates Cell Proliferation and Is Disrupted in Cancer. The Journal of Biological Chemistry. 290, 28643-28663 (2015).
- Mashtalir, N., et al. Autodeubiquitination protects the tumor suppressor BAP1 from cytoplasmic sequestration mediated by the atypical ubiquitin ligase UBE2O. Molecular Cell. 54, 392-406 (2014).
- Yu, H., et al. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Molecular and Cellular Biology. 30, 5071-5085 (2010).
- Yu, H., et al. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proceedings of the National Academy of Sciences of the United States of America. 111, 285-290 (2014).
- Dai, F., et al. BAP1 inhibits the ER stress gene regulatory network and modulates metabolic stress response. Proceedings of the National Academy of Sciences of the United States of America. 114, 3192-3197 (2017).
- Zhang, Y., et al. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nature Cell Biology. 20, 1181-1192 (2018).
- Kolluri, K. K., et al. Loss of functional BAP1 augments sensitivity to TRAIL in cancer cells. eLife. 7, (2018).
- Machida, Y. J., Machida, Y., Vashisht, A. A., Wohlschlegel, J. A., Dutta, A. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. The Journal of Biological Chemistry. , (2009).
- Sahtoe, D. D., van Dijk, W. J., Ekkebus, R., Ovaa, H., Sixma, T. K. BAP1/ASXL1 recruitment and activation for H2A deubiquitination. Nature Communications. 7, 10292 (2016).

