Evaluation of Amino Acid Consumption in Cultured Bone Cells and Isolated Bone Shafts
Summary
This protocol presents a radiolabeled amino acid uptake assay, which is useful for evaluating amino acid consumption either in primary cells or in isolated bones.
Abstract
Bone development and homeostasis is dependent upon the differentiation and activity of bone forming osteoblasts. Osteoblast differentiation is sequentially characterized by proliferation followed by protein synthesis and ultimately bone matrix secretion. Proliferation and protein synthesis require a constant supply of amino acids. Despite this, very little is known about amino acid consumption in osteoblasts. Here we describe a very sensitive protocol that is designed to measure amino acid consumption using radiolabeled amino acids. This method is optimized to quantify changes in amino acid uptake that are associated with osteoblast proliferation or differentiation, drug or growth factor treatments, or various genetic manipulations. Importantly, this method can be used interchangeably to quantify amino acid consumption in cultured cell lines or primary cells in vitro or in isolated bone shafts ex vivo. Finally, our method can be easily adapted to measure the transport of any of the amino acids as well as glucose and other radiolabeled nutrients.
Introduction
Amino acids are organic compounds that contain an amino (-NH2) and carboxyl (-COOH) functional groups with a variable side chain that is specific to each amino acid. In general, amino acids are well known as the basic constituent of protein. More recently, novel uses, and functions of amino acids have been elucidated. For example, individual amino acids can be metabolized to generate intermediate metabolites that contribute to bioenergetics, function as enzymatic cofactors, regulate reactive oxygen species or are used to synthesize other amino acids1,2,3,4,5,6,7,8,9,10. Many studies demonstrate that amino acid metabolism is critical for cell pluripotency, proliferation, and differentiation in various contexts3,6,11,12,13,14,15,16,17.
Osteoblasts are secretory cells that produce and secrete the Collagen Type 1 rich extracellular bone matrix. To sustain high rates of protein synthesis during bone formation, osteoblasts demand a constant supply of amino acids. To meet this demand, osteoblasts must actively acquire amino acids. Consistent with this, recent studies reveal the importance of amino acid uptake and metabolism in osteoblast activity and bone formation15,16,17,18,19,20.
Osteoblasts acquire cellular amino acids from three major sources: extracellular milieu, intracellular protein degradation and de novo amino acid biosynthesis. This protocol will focus on the evaluation of amino acid uptake from extracellular environment. The most common methods to measure amino acid uptake rely on either radiolabeled (e.g., 3H or 14C) or heavy isotope labeled (e.g., 13C) amino acids. Heavy isotopomer assays can analyze amino acid uptake and metabolism more thoroughly and safely but are more time consuming taking multiple days to complete as it takes a day to prepare and derivatize samples and multiple days to analyze on the mass spectrometer depending on the number of samples21,22. By comparison, radiolabeled amino acid uptake assays are not informative about downstream metabolism but are cheap and relatively fast, being able to be completed within 2-3 h from the start of the experiment23,24. Here, we describe an easily modifiable basic protocol designed to evaluate radiolabeled amino acid uptake in cultured primary cells or cell lines in vitro or individual bone shafts ex vivo. The application of these two protocols can be extended to other radiolabeled amino acids and other bone associated cell types and tissues.
Protocol
All mouse procedures described herein were approved by the Animal Studies Committees at the University of Texas Southwestern Medical Center at Dallas. The radiation protocol was approved by the Radiation Safety Advisory Committee at the University of Texas Southwestern Medical Center at Dallas.
1. Amino acid uptake in cells (Protocol I)
- Plate 5 x 104 ST2 cells in each well of a 12-well tissue culture plate. Plate cells in α-MEM containing 10% FBS, 100 U/mL penicillin and 0.1 µg/mL streptomycin (Pen/Strep). Plate extra wells of cells to quantify the cell number per condition for the normalizations in step 1.12. Incubate cells in a humidified cell culture incubator at 37 °C with 5% CO2.
- Culture the cells for 2-3 days until confluent.
- On the day of the experiment, prepare the following solutions: 1x Phosphate Buffered Saline (PBS), pH 7.4 and Krebs Ringers HEPES (KRH) buffer, pH 8.0: (120 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, NaHCO3, 5 mM HEPES, 1 mM D-Glucose). Prewarm to 37 °C.
- Aspirate the medium and wash the cells twice with 1 mL of 1x PBS, pH 7.4.
NOTE: This protocol is also appropriate for rapidly dividing non-confluent cells. In this case, it is important to normalize the radioactivity to either absolute cell number or DNA content. In addition, consider increasing the size of the culture plate or flask to increase the overall cell number and cpm values. This is important to determine empirically on an individual basis. - Aspirate 1x PBS and wash the cells once with 1 mL KRH.
- Make 4 µCi/mL L-[3,4-3H]-Glutamine working media by diluting 4 µL of [1 µCi µL-1] L-[3,4-3H]-Glutamine stock per 1 mL of KRH.
NOTE: Before using radioactive materials, please contact the Office of Radiation Safety at your home institution to obtain approvals. All the procedures related to radiation must be performed behind the plexiglass shield. - Incubate cells with 0.5 mL of KRH containing 4 µCi/mL L-[3,4-3H]-Glutamine working media for 5 min.
- Collect the radioactive medium and dispense into the liquid waste container. Wash the cells three times briefly with ice-cold KRH to terminate the reaction. Collect and discard all the washes in the radioactive liquid waste container.
- Add 1 mL of 1% SDS to each well and triturate 10x to lyse and homogenize the cells. Transfer the cell lysates to 1.5 mL tubes. Discard cell culture plates, serological pipettes, and pipette tips in the solid radioactive waste container.
- Centrifuge at >10,000 x g for 10 min. Transfer the supernatants to scintillation vials containing 8 mL of the scintillation solution. Mix by shaking the scintillation vials vigorously. Discard tubes and pipette tips in solid radioactive waste container.
- Read radioactivity in counts per minute (cpm) using a Scintillation counter. Discard scintillation vials in scintillation vial waste container.
- Trypsinize, resuspend, and count the cells from the remaining non-radioactive plates of cells (see step 1.1) to estimate the number of cells in the lysed, radioactive cultures. Using a hemocytometer, count the number of cells per non-radioactive well for each experimental condition. Normalize the cpm from step 1.11 to the estimated cell number from the non-radioactive plates.
- After completion of the experiment, decontaminate the cell culture hood, bench, and all instruments with a radioactivity decontaminant spray. Finally, perform wipe tests to ensure the working area is radiation-free.
2. Amino acid uptake in freshly isolated bone tissues (Protocol II)
- Prewarm KRH to 37 °C.
- Euthanize a 3-day-old mouse and remove the skin on the arms. Disarticulate the humeri from the shoulder using scissors and dissect out both humeri. Remove all extemporaneous tissues using a scalpel and forceps. Remove the epiphyses from the bone.
NOTE: In addition to humerii, this protocol can be adapted to neonatal femora, tibiae and calvariae as well as humerii, femora and tibiae isolated from 2- and 4-month-old mice (unpublished data). - Flush out marrow from the bone and weigh the bone shafts to normalize in step 2.14.
- Boil one humerus in 1x PBS at 100 °C for 10 min to decellularize the bone. The decellularized boiled bone is used as a negative control.
NOTE: As a quality control, paraffin embed the boiled and non-boiled bones for histological stains to visually confirm that boiling was efficacious in decellularizing the bone. - Equilibrate both humeri in 1 mL of KRH for 30 min in the cell culture incubator at 37 °C.
- Make a working solution of 4 µCi/mL L-[2,3,4-3H]-Arginine in KRH by diluting 4 µL of 1µCi/µL L-[2,3,4-3H]- Arginine stock per 1 mL KRH.
NOTE: This protocol is appropriate to evaluate the uptake of whichever radiolabeled nutrient is chosen. L-[2,3,4-3H]-glutamine, L-[14CU]-alanine, and L-[2,3-3H]-proline have been tested with similar results. Arginine data is shown to highlight the utility of this protocol.
CAUTION: All the procedures using radiation must be performed behind the plexiglass shield while using appropriate personal protective equipment (PPE). - Incubate both the experimental and boiled control bone separately in KRH containing 4 µCi/mL L-[2,3,4-3H]-Arginine for up to 90 min at 37 °C.
NOTE: It is important to empirically determine the incubation times for different conditions including the age of the mice, different skeletal elements and the types of radiolabeled amino acids by performing a time course. Saturation mostly occurs after approximately 3-4 h. Uptake should be evaluated at a timepoint in the linear rage of uptake. - Remove the radioactive medium and discard in the liquid radioactive waste container. Wash humeri three times using ice cold 1 mL of KRH to terminate the reaction. Discard all the washes in the liquid radioactive waste container.
- Transfer each bone into a 1.5 mL tube. Add 500 µL of RIPA buffer [150 mM NaCl; 5 mM EDTA; 50 mM Tris (pH 8.0); 0.5% NP40 (v/v); 0.5% DOX (w/v); 0.1% SDS (w/v)]. Discard the culture plates, serological pipettes, and pipette tips in a radioactive solid waste container.
- Homogenize the humeri by chopping 100 times with scissors in a RIPA buffer.
- Sonicate (Amplitude: 35%, Pulse 1 s) the resulting homogenized bone for 10 s.
NOTE: Sonication is also known to produce aerosols. Sonication steps can be performed in a biological safety cabinet. To reduce aerosol formation, it is important to not overfill the tubes. Sonication of small sample volumes can result in incomplete sonication or sample loss due to foaming. If foaming occurs, centrifuge the sample for 5 min to settle. - Clarify the lysate by centrifugation for 10 min at >10,000 x g at room temperature. Transfer 200 µL of the supernatant to scintillation vials containing 8 mL of the scintillation solution. Mix by shaking the scintillation vials vigorously. Discard all solid radioactive waste (e.g., used tubes, pipette tips, plates, etc.) in the solid radioactive waste container.
- Read radioactivity (cpm) using the Scintillation counter. Discard used scintillation vials in the radioactive waste container designated for scintillation vials.
- Subtract the cpm of boiled bone (basal value) from the cpm of experimental bone. Normalize the radioactivity with bone weight from step 2.3.
- Spray the cell culture hood, instruments, and bench with radioactivity decontaminant. Perform wipe tests to confirm the working area is radiation-free.
Representative Results
Amino acid transport is regulated by many membrane-bound amino acid transporters that have been categorized into distinct transport systems based on numerous characteristics, including substrate specificity, kinetics, as well as ion and pH dependence25. For example, glutamine uptake can be mediated by the Na+-dependent transport systems A, ASC, γ+L and N or the Na+-independent System L. The Na+-dependent systems are distinguished by the ability to substitute Li+ for Na+ (System N) or sensitivity to the amino acid analogue 2-(methylamino)isobutyric acid (MeAIB) (System A). The purpose of this experiment was to define the amount of glutamine consumption attributable to each transport system. L-[3,4-3H] glutamine uptake was measured in confluent ST2 cells in normal KRH containing 120 mM NaCl, Na+ free KRH containing 120 mM Choline chloride, normal KRH containing 5 mM MeAIB or Na+ free KRH containing 120 mM LiCl. Total radioactivity (counts per minute) was normalized to cell number. Na+ removal led to 90% reduction in glutamine uptake. This indicated that System L accounts for 10% of glutamine uptake whereas 90% of glutamine uptake is Na+ dependent in ST2 cells (Figure 2). The presence of Li+ only increased glutamine uptake by 2% compared to the Na+ free condition whereas 5 mM MEAIB did not affect glutamine uptake. These data indicate that the majority of glutamine uptake is mediated by Systems ASC and γ + L while System N and System A are responsible for roughly 2% and 0% of Na+-dependent glutamine uptake, respectively.
We modified Protocol I to characterize amino acid uptake in bone shafts ex vivo. In this experiment we followed Protocol II to characterize arginine uptake kinetics in isolated long bones from 3-day-old (p3) mice. To do this, we first dissected both humeri from p3 mice. The epiphyses were removed, and the marrow was spun out and each humerus was weighed for normalization. The contralateral humerus was designated as a negative control and was boiled to kill cellular activity. The experimental and boiled humeri were then incubated with radiolabeled 4µCi/mL L-[2,3,4-3H]-Arginine for up to 2 h. Arginine uptake increased in a linear manner over the course of this experiment (Figure 3A). For comparison, the boiled humeri did not exhibit a dynamic increase of arginine uptake in this experiment, but rather had a basal radioactivity that is attributed to adsorption of the probe in the bone matrix. The normalized and corrected arginine uptake is shown in Figure 3B.
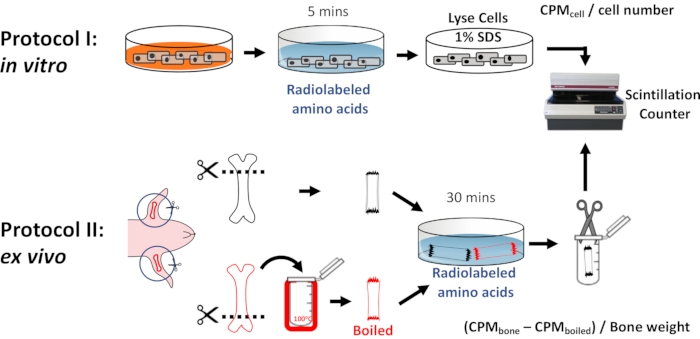
Figure 1: Workflow of radiolabeled amino acid uptake assays. Schematic overview of amino acid uptake assays in vitro (Protocol I) and in bones ex vivo (Protocol II). Please click here to view a larger version of this figure.
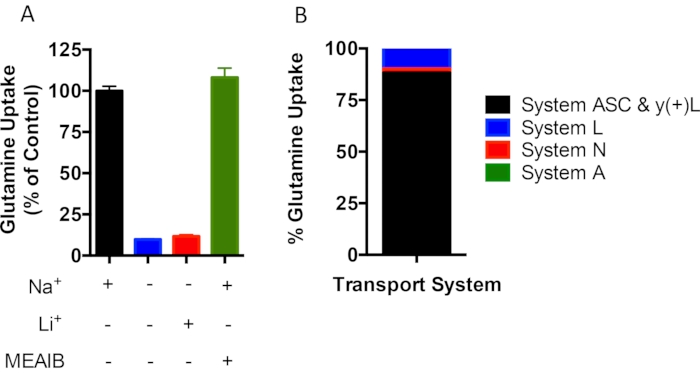
Figure 2: Kinetic properties of L-[3,4-3H]-Glutamine uptake in ST2 in vitro. (A) L-[3,4-3H]-Glutamine uptake assays performed in the presence (+) or absence (-) of sodium (Na+), MeAIB or lithium (Li+) in ST2 cells. (B) Percentage of estimated contributions of System A, N, L, or ASC and γ + L to glutamine uptake. Please click here to view a larger version of this figure.
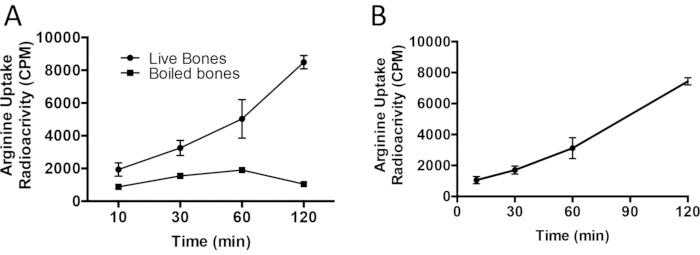
Figure 3: Ex vivo L-[2,3,4-3H]-Arginine uptake in neonatal humeri. (A) L-[2,3,4-3H]-Arginine uptake performed over a time course of 2 h in live and boiled humeri isolated from 3-day-old C57BL/6 mice. (B) Normalized L-[2,3,4-3H]-Arginine uptake with boiled humeri. Please click here to view a larger version of this figure.
Discussion
The protocol described herein provides a fast and sensitive approach to evaluate amino acid uptake in response to various experimental permutations either in vitro or ex vivo. When compared to commercially available kits (e.g., Glutamine and Glutamate Determination Kit), this method is much more sensitive, quicker, and less labor intensive16,17,25. In our protocol, we evaluate uptake in the Krebs Ringers HEPES buffer. We utilize this media as it is a basal formula that allows for rapid and easy modifications to test a diverse array of conditions to characterize amino acid uptake. For example, to test whether the transport system is Na+ dependent, we can simply replace NaCl with choline chloride to make Na+ free KRH. This flexibility is advantageous to interrogate the various transport systems mediating uptake of individual amino acids. Basal growth media such as alpha-MEM is also appropriate for this assay. In this case, we typically purchase alpha-MEM lacking the individual amino acid in question, which is replaced with the radiolabeled amino acid for the experiment. For example, to evaluate glutamine uptake we use glutamine-free alpha-MEM in primary bone marrow stromal cells15. KRH does have some limitations due to its simple composition. KRH is not an ideal option if the goal is to study secondary active transport systems such as symporters and antiporters. For instance, leucine uptake through Slc7a5/Slc3a2 occurs in exchange for glutamine. In this case, leucine uptake may be underestimated due to the lack of glutamine in KRH; rather, the use of leucine-free alpha-MEM medium would be preferable in this instance. It is also important to note that the use of a Geiger counter is not mandatory when working with isotope 3H. However, it is advised to keep the Geiger counter on as a courtesy to alert people that you are working with radiation.
A critical and mandatory control for the second protocol is the boiling of the contralateral bone. This is necessary as the bone matrix can absorb radioactive amino acids independent of facilitated transport by bone cells. By decellularizing the bone, we can estimate the amount of radiation trapped by bone matrix and use it as the baseline radioactivity to normalize uptake. Another important control is to weigh the bones after removal of the epiphyses and marrow. The weight of the bone is used to normalize uptake relative to the size of the bone shaft. This is important as the size of the bones may vary due to many factors ranging from variability during the removal of the epiphyses to genetic mutations that affect growth. An alternative method is to quantify DNA content of the bone shaft for normalization. In our experience, normalizing DNA content is comparable to bone weight but adds more steps dealing with radioactive materials. Thus, we prefer to normalize to bone weight.
These protocols can be adapted to cell lines, including ATDC5, MC3T3, 293, or other primary cells such as calvarial osteoblasts, chondrocytes, bone marrow stromal cells, bone marrow macrophages, and osteoclasts. In addition, the protocols are easily adaptable to evaluate other radioactive isotopes (e.g., 14C and 35S) as well as different nutrients, including amino acids, glucose, and fatty acids. Furthermore, protocol II can also be adapted to adult bones and pellet cultures, with the consideration of adjustment to the incubation time. Although this protocol is highly adaptable, it is important to characterize the kinetics of the new isotope labeled molecules or in new cell lines before conducting experiments. The transport kinetics may vary drastically between different molecules, different isotopes, or different cell types.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The Karner lab is supported by National Institute of Health R01 grants (AR076325 and AR071967) to C.M.K.
Materials
| 0.25% trypsin | Gibco | 25200 | |
| 12-well plate | Corning | 3513 | |
| 1mL syringe | BD precision | 309628 | |
| 30G Needle | BD precision | 305106 | |
| Arginine Monohydrochloride L-[2,3,4-3H]-, 1mCi | PerkinElmer | NET1123001MC | |
| Beckman LS6500 scintillation counter | |||
| Calcium chloride | Sigma | C1016 | |
| choline chloride | Sigma | C7077 | |
| D-(+)-Glucose solution | Sigma | G8769 | |
| Dissection Tool | Forceps, scissors, scapels | ||
| DPBS | Gibco | 14190 | |
| Ethylenediaminetetraacetic acid | Sigma | E9884 | |
| HEPES(1M) | Gibco | 15630 | |
| L-[3,4-3H(N)]-Glutamine | PerkinElmer | NET551250UC | |
| Liquid scintilation vials | Sigma | Z190535 | |
| lithium chloride solution, 8M | Sigma | L7026 | |
| Magnesium chloride | Sigma | M8266 | |
| MEMα | Gibco | 12561 | |
| Microcentrifuge tube, 15mL | Biotix | 89511-256 | |
| NP-40 | Sigma | 492016 | |
| Potassium chloride | Sigma | P3911 | |
| Sodium bicarbonate | Sigma | S6014 | |
| sodium chloride | Sigma | S9888 | |
| Sodium Deoxycholate | Sigma | D6750 | |
| Sodium dodecyl sulfate | Sigma | 436143 | |
| Sonicator | Sonic&Materials | VCX130 | |
| Tris Base | Sigma | 648311 | |
| Ultima Gold (Scintillation solution) | PerkinElmer | 6013329 | |
| α-(Methylamino)isobutyric acid | Sigma | M2383 |
References
- Xiao, M., et al. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes & Development. 26 (12), 1326-1338 (2012).
- Altman, B. J., Stine, Z. E., Dang, C. V. From Krebs to clinic: glutamine metabolism to cancer therapy. Nature Reviews Cancer. 16 (10), 619-634 (2016).
- Karner, C. M., Long, F. Wnt signaling and cellular metabolism in osteoblasts. Cell and Molecular Life Sciences. 74 (9), 1649-1657 (2017).
- Zarse, K., et al. Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metabolism. 15 (4), 451-465 (2012).
- Nagano, T., et al. Proline dehydrogenase promotes senescence through the generation of reactive oxygen species. Journal of Cell Science. 130 (8), 1413-1420 (2017).
- Comes, S., et al. L-Proline induces a mesenchymal-like invasive program in embryonic stem cells by remodeling H3K9 and H3K36 methylation. Stem Cell Reports. 1 (4), 307-321 (2013).
- Fan, J., et al. Glutamine-driven oxidative phosphorylation is a major ATP source in transformed mammalian cells in both normoxia and hypoxia. Molecular Systems Biology. 9, 712 (2013).
- Hosios, A. M., et al. Amino acids rather than glucose account for the majority of cell mass in proliferating mammalian cells. Developmental Cell. 36 (5), 540-549 (2016).
- Welbourne, T. C. Ammonia production and glutamine incorporation into glutathione in the functioning rat kidney. Canadian Journal of Biochemistry. 57 (3), 233-237 (1979).
- Sullivan, L. B., et al. Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell. 162 (3), 552-563 (2015).
- Nelsen, C. J., et al. Amino acids regulate hepatocyte proliferation through modulation of cyclin D1 expression. The Journal of Biological Chemistry. 278 (28), 25853-25858 (2003).
- Krall, A. S., Xu, S., Graeber, T. G., Braas, D., Christofk, H. R. Asparagine promotes cancer cell proliferation through use as an amino acid exchange factor. Nature Communications. 7, 11457 (2016).
- Green, C. R., et al. Branched-chain amino acid catabolism fuels adipocyte differentiation and lipogenesis. Nature Chemical Biology. 12 (1), 15-21 (2016).
- Shiraki, N., et al. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metabolism. 19 (5), 780-794 (2014).
- Yu, Y., et al. Glutamine metabolism regulates proliferation and lineage allocation in skeletal stem cells. Cell Metabolism. 29 (4), 966-978 (2019).
- Shen, L., Sharma, D., Yu, Y., Long, F., Karner, C. M. Biphasic regulation of glutamine consumption by WNT during osteoblast differentiation. Journal of Cell Science. 134 (1), (2021).
- Karner, C. M., Esen, E., Okunade, A. L., Patterson, B. W., Long, F. Increased glutamine catabolism mediates bone anabolism in response to WNT signaling. Journal of Clinical Investigation. 125 (2), 551-562 (2015).
- Hu, G., et al. The amino acid sensor Eif2ak4/GCN2 is required for proliferation of osteoblast progenitors in mice. Journal of Bone and Mineral Research. 35 (10), 2004-2014 (2020).
- Rached, M. T., et al. FoxO1 is a positive regulator of bone formation by favoring protein synthesis and resistance to oxidative stress in osteoblasts. Cell Metabolism. 11 (2), 147-160 (2010).
- Elefteriou, F., et al. ATF4 mediation of NF1 functions in osteoblast reveals a nutritional basis for congenital skeletal dysplasiae. Cell Metabolism. 4 (6), 441-451 (2006).
- Maleknia, S. D., Johnson, R. Mass spectrometry of amino acids and proteins. Amino Acids, Peptides and Proteins in Organic Chemistry. , 1-50 (2011).
- Rennie, M. J. An introduction to the use of tracers in nutrition and metabolism. The Proceedings of the Nutrition Society. 58 (4), 935-944 (1999).
- Hahn, T. J., Downing, S. J., Phang, J. M. Amino acid transport in adult diaphyseal bone: contrast with amino acid transport mechanisms in fetal membranous bone. Biochimica Biophysica Acta. 183 (1), 194-203 (1969).
- Rosenbusch, J. P., Flanagan, B., Nichols, G. Active transport of amino acids into bone cells. Biochimica Biophysica Acta. 135 (4), 732-740 (1967).
- Kandasamy, P., Gyimesi, G., Kanai, Y., Hediger, M. A. Amino acid transporters revisited: New views in health and disease. Trends in Biochemical Sciences. 43 (10), 752-789 (2018).

