Using Bioluminescent Imaging to Investigate Synergism Between Streptococcus pneumoniae and Influenza A Virus in Infant Mice
Summary
A concurrent infection with influenza A virus is one of the factors implicated in the induction of invasive pneumococcal disease during asymptomatic Streptococcus pneumoniae carriage. Here we describe a mixed infection method using infant mice to investigate the synergism between these two respiratory pathogens.
Abstract
During the 1918 influenza virus pandemic, which killed approximately 50 million people worldwide, the majority of fatalities were not the result of infection with influenza virus alone. Instead, most individuals are thought to have succumbed to a secondary bacterial infection, predominately caused by the bacterium Streptococcus pneumoniae (the pneumococcus). The synergistic relationship between infections caused by influenza virus and the pneumococcus has subsequently been observed during the 1957 Asian influenza virus pandemic, as well as during seasonal outbreaks of the virus (reviewed in 1, 2). Here, we describe a protocol used to investigate the mechanism(s) that may be involved in increased morbidity as a result of concurrent influenza A virus and S. pneumoniae infection. We have developed an infant murine model to reliably and reproducibly demonstrate the effects of influenza virus infection of mice colonised with S. pneumoniae. Using this protocol, we have provided the first insight into the kinetics of pneumococcal transmission between co-housed, neonatal mice using in vivo imaging 3.
Protocol
1. Preparation of Infectious Stock of Bioluminescent S. pneumoniae
- Inoculate a McCartney tube containing 10ml Todd-Hewitt broth supplemented with 0.5% (w/v) yeast extract and a small piece of blood agar with bioluminescent S. pneumoniae and grow statically at 37°C to optical density (600nm) of 0.40-0.45.
- Place the culture on wet ice for 5 min to arrest the cells in this growth phase.
- To the 10 ml of mid-log growth phase S. pneumoniae add 1 ml of 80% (v/v) glycerol and mix, aliquot into the desired volumes and store at -70°C.
- Determine the infectious dose of the bioluminescent S. pneumoniae stock by viable count of serial dilutions on blood agar plates.
2. Growth and Preparation of influenza A virus stock
- Influenza A virus is grown in allantoic fluid of 10-day old embryonated chicken eggs.
- Thaw a vial with influenza A virus in 37°C water bath.
- Using a light source placed over the airsac (candling), mark on the egg shell the location of the air sac at a point free of underlying veins.
- Disinfect the surface of the eggs by wiping with 70% ethanol and wipe clean.
- Pierce a small hole in the air sac just above the mark using a jeweler’s scribe.
- With the eggs positioned in a carton with the air sac uppermost, use a 13mm 1mL syringe and a 26G needle to inoculate each egg through the hole in the shell with 0.1mL of appropriately diluted influenza A virus. Point the needle directly downwards into the allantoic cavity, piercing the airsac.
- Seal the hole with melted wax and incubate the eggs for 2 days in an humidified egg incubator at 35°C. After 2 days incubation, candle the eggs to check for the presence of blood vessels to confirm the embryo is still alive. If the egg is black inside, the embryo has died and the egg should be discarded and not used for harvesting of influenza A virus.
- Place the eggs at 4°C overnight to kill the embryos and to constrict blood vessels in order to minimize disruption of the vessels, as contact between blood and virus would result in binding of virus to red blood cells.
- The next day, disinfect the surface of the eggs by wiping with 70% ethanol.
- Work in a class II Biosafety Cabinet to collect the allantoic fluid containing the influenza A virus from the eggs. Remove the wax and use curved scissors to cut around the top of the egg to remove the air sac above the allantoic membrane.
- Puncture and peel back the allantoic membrane using sterile forceps.
- Aspirate the allantoic fluid with a 10mL sterile pipette while holding the embryo to one side with another sterile pipette. Collect pooled allantoic fluid in 50mL tubes and keep on ice at all times. If the allantoic fluid is bloody or contains yolk do not add to the pool.
- Centrifuge allantoic fluid for 5min at 2,000rpm at room temperature to remove debris.
- Aliquot the infectious allantoic fluid in 1-2 ml cryotubes and store at -70°C.
- Determine the influenza A virus titre of the thawed allantoic fluid in 3 independently performed plaque forming assays in Madin-Darby canine kidney cells (see below) . Use a new aliquot of virus for each experiment. Repeated freeze-thawing will reduce the infectivity titre.
3. Intranasal infection of mice with bioluminescent S. pneumoniae and/or influenza A virus
- Thaw frozen stock of bioluminescent S. pneumoniae and dilute to desired dose in PBS.
- Gently pick up 5 day old mice between index finger and thumb and hold upright. Use a pipette to drop bioluminescent S. pneumoniae onto nares in a volume of 3μl. Wait until all of the inoculum is inhaled before placing mouse back in the cage. Use 3μl of PBS for mock infection.
- Three to nine days after colonization with bioluminescent S. pneumoniae, thaw an aliquot of allantoic fluid containing influenza A virus and dilute in PBS to desired concentration. Infect mice with influenza A virus in a volume of 3μl PBS as described at step 3.2. Use 3μl of diluted allantoic fluid from uninfected eggs for mock-virus infection.
- Monitor the welfare of the mice daily by observing their appearance, behaviour, body condition and, in mice that are ≤ 3 days old, the presence of milk spots. A guide for humane end points and intervention criteria should be developed in consultation with the Laboratory Animal Veterinarian (Table 3).
4. Bioluminescent imaging using the IVIS Spectrum (Caliper Life Sciences)
- Prepare a mixture of ketamine at 0.75 mg/ml and xylazine 1.76mg/ml in injectable water.
- To anaesthetize mice, inject 100μl/10 g body weight into peritoneal cavity.
- Place anaesthetized mice into imaging box. Careful attention should be paid to keeping the anatomical region of interest parallel to the camera when placing the mouse inside of the imaging box/ chamber.
- Place box with mice in IVIS and allow isoflurane entry into the box at 0.5 L/min to maintain anaesthesia.
- Set IVIS machine to correct imaging platform and binning and capture image for 7 minutes.
- Allow mice to recover from anaesthesia in a warmed box (eg. using a heat pad) before returning to home cage.
- Images are analyzed and adjusted to the same scale using the Living Image software package version 3.0 (Caliper Life Sciences).
- Bioluminescent signals in a selected region of interest can be quantified as either the total flux (photon/s) or the average radiance (photons/s/cm2/sr).
5. Processing of tissues to determine bacterial load and virus titer
- Mice are euthanased by CO2 asphyxiation and the fur disinfected using 70% ethanol.
- Immobilize the animal on a hard cutting board.
- Using a sterile scalpel blade, cut through the middle of the head of the animal, starting posterior, and expose the nasopharynx by bending each side of the head outwards.
- Collect the nasopharyngeal tissue of each animal by scraping the tissue from each half of the head using sterile forceps and place in 1.5mL ice-cold RPMI. Keep on ice.
- Open up the chest cavity using scissors and remove the lungs using sterile forceps and scissors. Rinse the lungs three times in PBS to remove any blood and collect in 1.5mL ice-cold RPMI. Keep on ice.
- Use a homogeniser to homogenize the tissues, place back on ice.
- Remove an aliquot of the homogenized tissue and use this to determine the bacterial load. Prepare serial dilutions of the tissue homogenate and plate on HBA. Culture plates at 37°C for 18-24 hrs and determine viable count. The bacterial titers in a particular organ can then be correlated with the number of photons observed in that region.
- Centrifuge the rest of the homogenized tissue at 3,500rpm for 10 min at 4°C. Collect the supernatant and transfer to clean, sterile tubes. Store supernatant at -70°C and determine viral titer by plaque assay.
6. Plaque assay to determine influenza A virus titer in tissue homogenates
- Virus infectivity titers are determined by plaque formation on confluent monolayers of Madin-Darby canine kidney (MDCK) cells. MDCK cells are routinely cultured in RF10 (Table 1) in tissue culture bottles (Nunc) and passaged when confluent.
- Detach MDCK cells from tissue culture bottles using trypsin. Wash the cells in RF10 and collect cells by centrifugation at 1200 rpm for 5 min. Discard the supernatant and resuspend the pelleted cells in RF10. Count the MDCK cells using a hemocytometer and vital dye. Seed each 35mm well of 6-well plates (Nunc) with 1.2×106 MDCK cells in 3ml RF10 and culture the cells at 37°C and 5% CO2 to achieve confluency (in ~24 hours).
- Prepare Leibovitz dsL15 media and 1.8% (w/v) agarose overlay media (Table 1). Aliquot dsL15 media and 1.8% (w/v) agarose in 50mL in sterile bottles. Keep dsL15 media at 4°C until use and agarose at 56°C until use.
- Check that the MDCK cell monolayers are confluent and wash the cells with RPMI+ by aspirating the RF10 and replacing it with ~1.5ml RPMI+. At no stage in the procedure should the plates be allowed to completely dry out. Incubate the plates at 37°C until ready to add the samples containing virus.
- Prepare serial dilutions of the virus samples in RPMI+. Keep samples on ice until use.
- Aspirate RPMI+ from the monolayers and add 135uL RPMI+ of appropriate dilutions of virus sample to each well. Test each sample in duplicate.
- Incubate the MDCK cells with the virus samples for 45 min at 37°C and 5% CO2. Gently shake the plates every 15 minutes to distribute the virus evenly and prevent drying of the MDCK cell monolayers.
- Move the agarose from 56°C to 46°C waterbath and warm dsL15 media to 46°C in waterbath.
- After 45min incubation of the virus on the MDCK monolayers, take plates out of 5% CO2 incubator. Remove agarose and L15 media from 46°C waterbath. Add 0.2mL of 0.1% trypsin to 50mL of dsL15 media, than add 50mL of dsL15+trypsin to 50mL of 1.8% (w/v) agarose. Mix the agarose and L15 gently and add 3mL of overlay medium to each well of the 6-well plates. Gently swirl to ensure the entire MDCK cell monolayer is covered. Once the overlay is set incubate plates at 37°C and 5% CO2 for 2-4 days.
- At the end of the incubation period, count the number of plaques as a measure of virus infectivity. If necessary, the plaques can be visualized by staining of the monolayers with crystal violet dissolved in methanol.
7. Section C – Secrets to success:
1 All experimental work with S. pneumoniae (eg. inoculation of liquid cultures, processing of tissues from mice infected with S. pneumoniae) is performed in a class II Biosafety Cabinet.
1.1 We create bioluminescent S. pneumoniae strains by introducing plasmid pPJTG28 3. However, a wide variety of different bioluminescent S. pneumoniae strains are available 4-6. Bioluminescent S. pneumoniae strains can also be purchased from Caliper Life Sciences Inc (MA, USA).
1.1 Growth kinetics of various S. pneumoniae strains may differ and should be determined for each strain. It may take between 3 to 4 hours to reach OD600nm=0.4 to 0.45.
1.4 We would normally expect the infectious stocks to reach concentration of 10e8 CFU/ml and recommend a dilution range from 10e-4 to 10e-8 to determine the concentration of viable bacteria.
2.6 The appropriate titre to use for inoculating the eggs, depends on the virus strain used and may range from 10e-3 to 10e-5. Influenza A virus strains obtained from tissue culture supernatants may sometimes be used neat.
3.2a We routinely use 2,000 CFU of S. pneumoniae strain EF3030 to colonize 5 day old mice 3. The precise infectious dose of the inoculum is determined retrospectively for each experiment by plating serial dilutions of the inoculum on horse blood agar plates. The ability of the S. pneumoniae strain to colonize mice is first determined by viable count of tissue homogenates (eg. nasopharynx, lungs) at several time points after inoculation of the mice.
3.2b It is not necessary to use anaestetics to administer the bacteria or virus to the animals via inhalation. Infant mice (as well as adult mice) should be restrained by hand and held upright by the investigator, the inoculum (3 microliter for infant mice, up to 10 microliter for adult mice) is then dropped onto the nares and inhalation all of the inoculum is confirmed by observation.
3.2c Remove neonatal mice one by one from the nest and inoculate with bacteria or virus as needed. Mark the tail of the animal using a permanent marker if necessary and return animal back to cage immediately. Rub the animal with some bedding material so that the dam will accept the pups back into the nest. Note: permanent marker rubs off the animals quickly and needs to be re-applied daily. Alternatively, a tattoo system may be used to identify individual animals.
3.3 We use 20 PFU influenza A virus Udorn/307/72 (H3N2) to infect 8 to 14 day old mice 3. Mortality and morbidity observed in the animals will depend on the strain of S. pneumoniae and influenza A virus used. Table 3 provides a useful set of humane end points and intervention criteria to use in mice younger than 21 days of age. We typically do not detect any mortality or morbidity when using S. pneumoniae EF3030 and influenza A virus strain Udorn/307/72 (H3N2) when mice are >10 days old.
4.2 To be accurate, mice can be weighed before injection of the anesthetic solution. As a guide, about 50-75 microliters of anesthetic solution is used for 20 day old mice. The mouse is fully anesthetized when not responding to a pinch in the tail or hind limb. While in the IVIS, the mice will also be exposed to isoflurane which will add anesthesia. The timing of imaging following infection of the mice depends on the strain(s) of S. pneumoniae and influenza A virus used as well as on the symptoms of interest. Performing a few pilot experiments to optimize the timing of bioluminescent imaging may be necessary.
4.3 Mice can either be placed into an isolation box (which prevents contaminants from entering or exiting the box) or alternatively, can be placed directly into the instrument for imaging. We use the isolation box for experiments with infant mice, in order to ensure the mice remain anaesthetized during the imaging procedure and to prevent contamination of the IVIS with S. pneumoniae and/or influenza A virus.
4.3 Use a ventral image of the mice to obtain an image of respiratory tract. A lateral image of the mouse provides a more sensitive image of the ears.
4.4 When mice are placed directly into the chamber, isoflurane is delivered to the mice via nose cones. For experiments in which infant mice are used, the isolation box is preferred since the nose cones do not fit on infant mice.
4.5 The total exposure time needed to capture an image may vary between experiments according to the strength of the bioluminescent signal, location of the signal, bacterial strain and the mouse strain used. Regarding the latter, both pigment and fur can significantly attenuate the total detectable signal. The camera on the IVIS Spectrum is a back-thinned, back illuminated CCD offering a broad dynamic range (65536 grey levels). Ideally, the exposure time should be set to capture between 60 and 60,000 counts. In our experience with C57BL/6 mice, increasing the imaging time beyond 7 minutes does not markedly improve the signal to noise ratio. Alternative options for increasing the bioluminescent signal include increasing the degree of binning (ie. to large) and using a smaller field of view. Importantly, quantification should not be performed on areas inside the image containing saturated pixels, as this provides an underestimated value for the number of photons detected. If unsure as to the optimal conditions, the auto-exposure feature can be used as a guideline.
4.5 If a strong bioluminescent signal in one part of the mouse is obstructing the detection of bioluminescence in a nearby region, selected regions of the mouse can be covered with black paper and imaging can proceed as normal
4.7 If the luminescent signal is quantified in photons (as opposed to counts) variations in imaging conditions (e.g. imaging time) between experiments will not affect the measurement of the bioluminescent signal.
4.8 If using the total flux to measure the photons in a specified region, ensure that the same ‘region of interest’ shape is used for all samples.
5.1a Mice can be euthanized at any time point after infection with influenza A virus, depending on interest of the researcher. We typically determine bacterial and viral titre at 6 days after infection with influenza A virus Udorn/307/72 (H3N2) and do not observe any mortality/morbidity. However, morbidity and mortality observed in these experiments will vary depending on the S.pneumoniae and influenza A virus strain used and the time point chosed should take mortality and morbidity rates into account to minimize the impact on animal welfare. Table 3 provides a useful guide to intervention criteria and humane endpoints for use in these experiments. Generally, mice are euthanized and tissue processed as described within 3 hours after in vivo bioluminescent imaging.
5.1b Mice younger than 10 days old are resistant to hypoxia and are euthanized by decapitation. In the described experimental setup, mice are at least 14 days old and may be euthanized by CO2 asphyxiation.
5.7 Blood-agar plates may be supplemented with 5μg/ml gentamicin to prevent growth of commensal bacteria in homogenates of nasal tissues. The range of dilutions used will depend on the S. pneumoniae strain used. For experiments in 20 day old mice with S. pneumoniae strain EF3030, we use neat, 10e-1, 10e-2 and 10e-3.
5.8 Store each sample of homogenized tissue in at least two aliquots, so that a backup sample is available in case the virus plaque assay fails! An accurate viral titer can only be obtained from a sample thawed once after freezing but not from samples repeatedly freeze-thawed.
6.1 When grown 90-100% confluent, a single 150cm2 tissue culture flask will yield enough MDCK cells for five 6-well plates.
6.5 The optimal serial dilution will depend on the virus strain used and the time after infection. Typically, we would use neat, 10e-1 and 10e-2 dilutions for organ homogenates taken 6 days after infection with influenza A virus.
6.6 Use of serial dilutions of virus stock is an appropriate positive control for use in each plaque assay. As a negative control, RPMI+ can be used.
8. Representative results:
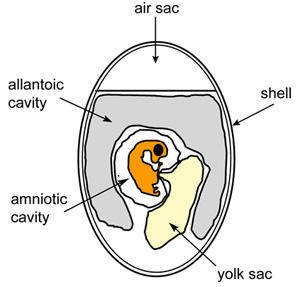
Figure 1. Schematic diagram of embryonated chicken egg.
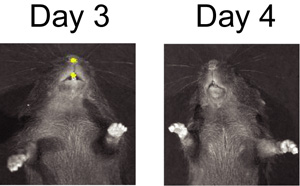
Figure 2. BLI of S. pneumoniae infected mouse on day 3 and day 4 after mock infection with influenza virus.
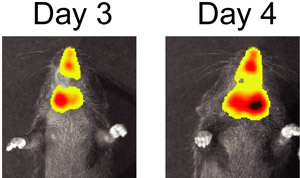
Figure 3. BLI of S. pneumoniae and influenza virus co-infected mouse on day 3 and day 4 after infection with influenza virus.
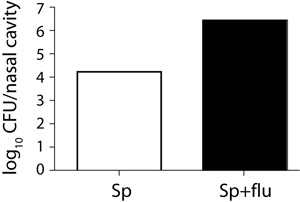
Figure 4. Bacterial load in the nose of a single and a co-infected mouse.
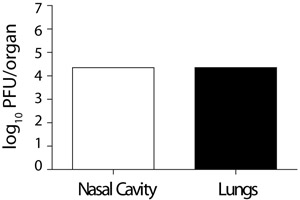
Figure 5. Viral titres in the nose and lung of a co-infected mouse.
| Media | Contents |
| 1.8% (w/v) Agarose | Add 0.9 gram of agarose to 50mL distilled H2O, sterilize by autoclaving at 121°C for 10min. Transfer to 56°C waterbath to cool. |
| Double strength Leibovitz’s L15 medium (ds L15) | 1 satchel of L15 powder (to make up 1L) is dissolved in 450 mL sterile distilled H2O and stirred until dissolved. The pH is adjusted to pH 6.8 with 1M HCl. Then supplement with 4mL of 7% (w/v) NaHCO3, 0.4mL HEPES (0.5M, pH 6.8), 200 U/ml penicillin, 200 μg/ml streptomycin, 60 μg/ml gentamicin. Adjust final volume to 500mL and filter sterilize. |
| Horse Blood Agar (HBA) | 4% HBA (Oxoid, Basingstoke, UK), dH2O, 7% (v/v) Horse Blood |
| RF10 | RPMI-1640 containing 2mM glutamine, 2nM sodium pyruvate, 24 μg/ml gentamicin, 100 U/ml penicillin, 100 μg/ml streptomycin, 10% (v/v) heat-inactivated Fetal Calf Serum. |
| RPMI+ | RPMI-1640 containing 2mM glutamine, 2nM sodium pyruvate, 24 μg/ml gentamicin, 100 U/ml penicillin, 100 μg/ml streptomycin |
| Todd-Hewitt Broth + 0.5% yeast extract | Todd Hewitt Broth (Oxoid, Basingstoke, UK) in dH2O supplemented with 2% (w/v) yeast extract |
Table 1. Specific media used in this protocol.
| Material Name | Typ | Company | Catalogue Number | Comment |
| RPMI medium 1640 | Reagent | Gibco | 21870-076 | |
| Trypsin | Reagent | Worthington Biochemical Corporation | LS003740 | TPCK treated |
| Leibovitz’s L-15 medium | Reagent | Gibco | 41300-039 | |
| Agarose, type I | Reagent | Sigma-Aldrich | A6013 | Use low melting temperature agarose, such as SeaPlaque agarose |
| Madin-Darby Canine Kidney cells | Reagent | ATCC | CCL-34 |
Table 2. Specific reagents and equipment:
| Effect on animal welfare | ||
| Mild to Moderate signsa | Severe signsb | |
| Non-specific sign(s): | ||
| Appearance | Slight to moderate piloerection with no dehydration (skin tenting) | Piloerection, with dehydration (skin tenting) |
| Body Condition | reduced weight gain | No weight gain or reduction in weight |
| Behaviour | Subdued but responsive, decreased interaction with peers. | Unresponsive to activity and provocation. Isolated from other littermates |
| Specific conditions or abnormal clinical sign(s): | ||
| Absence of milk spot (<2-3 days old) |
Try to encourage suckling if this has been observed for <24h. | If no evidence of milk spot even with encouragement after 24-48h, or if obviously has not suckled, consider euthanasia. |
| Other signs | Seek Animal Facility Manager/Laboratory Animal Veterinarian advice re: appropriate action or euthanise if animal is in moderate or severe pain or distress | |
Table 3. Intervention criteria for infant mice (<21 days old):
a When one or more ‘mild to moderate’ signs are observed increase frequency of observations to twice daily and seek advice from animal welfare officer where appropriate so that treatment and care can be given.
b When one or more ‘severe’ signs are observed, euthanise the mice using appropriate method. Mice < 10 days old are euthanized by decapitation, mice > 10 days old are euthanized by CO2 asphyxiation.
Discussion
In vivo imaging with the IVIS machine is a unique methodology that allows the real time visualization of microbial pathogenesis 4, 6-9. Here, we show the application of this technology to understanding the synergism between S. pneumoniae and influenza virus in infant mice. Due to the non-invasive nature of this technique, we have been able to monitor the progression of infection in each individual mouse over time. This has enabled us to document the kinetics of pneumococcal transmission amongst co-housed infant mice 3. The non-invasive nature of this technique enables researchers to use fewer animals per experiment and hence reduce animal usage. It is also possible to use the IVIS machine to visualize the dissemination of the infection to previously unknown sites of infection in the body, as well as sites not easily sampled by dissection (such as the middle ear).
Before commencing the imaging experiments, we confirmed that colonization levels of the bioluminescent EF3030 strain were comparable to colonization levels of the parent EF3030 strain by enumerating viable count in tissue homogenates. In addition, we first confirmed that infection with influenza virus resulted in increased load of bioluminescent EF3030 in the nasopharynx of colonized animals as demonstrated with the parent EF3030 strain (results not shown).
While the IVIS technology is easy to operate and generates reproducible and easy to understand data, experiments must be carefully designed so that the sensitivity and utility of the technology is maximised. For example, the sensitivity of the IVIS camera is affected by the depth and opacity of the tissue 8, which in our experience, increases the detection limit for bioluminescent signals originating from the lungs. In addition, dark fur and pigmented skin reduces light transmission by as much as 10-fold (compared to hairless mice) 8. While developing this method, we have optimized this protocol for use with 5 to 14 day old C57BL/6 mice and have since demonstrated that IVIS can be used to detect colonization of 20 day old C57BL/6 mice with bioluminescent S. pneumoniae EF3030. However, signal detection may be increased when these experiments are performed in nude or BALB/c mice. Nevertheless, the IVIS camera represents a novel approach to monitor, characterize and ultimately understand the in vivo development of pneumococcal disease following influenza A virus infection.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
The authors would like to acknowledge the technical advise from David Briles (University of Alabama at Birmingham, AL, USA). We would like to thank Yvette Chen, the Animal Welfare Officer of The University of Melbourne, and David Taylor, the Animal Facility Manager, for helpful suggestions and discussions concerning animal welfare and ethics and development of intervention criteria and humane end points for the experiments described in this manuscript.
Odilia Wijburg and Patrick Reading are supported by a NHMRC R.D. Wright Fellowship, Kirsty Short is supported by a GSK Postgraduate Support grant and a Puzey Scholarship.
Referenzen
- Brundage, J. F., Shanks, G. D. Deaths from bacterial pneumonia during 1918-19 influenza pandemic. Emerg Infect Dis. 14, 1193-1199 (2008).
- Petersdorf, R. G., Fusco, J. J., Harter, D. H., Albrink, W. S. Pulmonary infections complicating Asian influenza. AMA Arch Intern Med. 103, 262-272 (1959).
- Diavatopoulos, D. A. Influenza A virus facilitates Streptococcus pneumoniae transmission and disease. Faseb J. , (2010).
- Orihuela, C. J., Gao, G., Francis, K. P., Yu, J., Tuomanen, E. I. Tissue-specific contributions of pneumococcal virulence factors to pathogenesis. J Infect Dis. 190, 1661-1669 (2004).
- Kadurugamuwa, J. L. Noninvasive monitoring of pneumococcal meningitis and evaluation of treatment efficacy in an experimental mouse model. Mol Imaging. 4, 137-142 (2005).
- Smith, M. W., Schmidt, J. E., Rehg, J. E., Orihuela, C. J., McCullers, J. A. Induction of pro- and anti-inflammatory molecules in a mouse model of pneumococcal pneumonia after influenza. Comp Med. 57, 82-89 (2007).
- Owen, S. J. Nasal-associated lymphoid tissue and olfactory epithelium as portals of entry for Burkholderia pseudomallei in murine melioidosis. J Infect Dis. 199, 1761-1770 (2009).
- Luker, G. D., Leib, D. A. Luciferase real-time bioluminescence imaging for the study of viral pathogenesis. Methods Mol Biol. 292, 285-296 (2005).
- Hutchens, M., Luker, G. D. Applications of bioluminescence imaging to the study of infectious diseases. Cell Microbiol. 9, 2315-2322 (2007).

