Rapid Isolation of Viable Circulating Tumor Cells from Patient Blood Samples
Summary
Circulating tumor cells are isolated from the blood of cancer patients without inflicting cellular damage. Isolation of tumor cells is accomplished using a bimolecular surface of E-selectin in addition to antibodies against epithelial markers. A nanotube coating specifically promotes cancer cell adhesion resulting in high capture purities.
Abstract
Circulating tumor cells (CTC) are cells that disseminate from a primary tumor throughout the circulatory system and that can ultimately form secondary tumors at distant sites. CTC count can be used to follow disease progression based on the correlation between CTC concentration in blood and disease severity1. As a treatment tool, CTC could be studied in the laboratory to develop personalized therapies. To this end, CTC isolation must cause no cellular damage, and contamination by other cell types, particularly leukocytes, must be avoided as much as possible2. Many of the current techniques, including the sole FDA-approved device for CTC enumeration, destroy CTC as part of the isolation process (for more information see Ref. 2). A microfluidic device to capture viable CTC is described, consisting of a surface functionalized with E-selectin glycoprotein in addition to antibodies against epithelial markers3. To enhance device performance a nanoparticle coating was applied consisting of halloysite nanotubes, an aluminosilicate nanoparticle harvested from clay4. The E-selectin molecules provide a means to capture fast moving CTC that are pumped through the device, lending an advantage over alternative microfluidic devices wherein longer processing times are necessary to provide target cells with sufficient time to interact with a surface. The antibodies to epithelial targets provide CTC-specificity to the device, as well as provide a readily adjustable parameter to tune isolation. Finally, the halloysite nanotube coating allows significantly enhanced isolation compared to other techniques by helping to capture fast moving cells, providing increased surface area for protein adsorption, and repelling contaminating leukocytes3,4. This device is produced by a straightforward technique using off-the-shelf materials, and has been successfully used to capture cancer cells from the blood of metastatic cancer patients. Captured cells are maintained for up to 15 days in culture following isolation, and these samples typically consist of >50% viable primary cancer cells from each patient. This device has been used to capture viable CTC from both diluted whole blood and buffy coat samples. Ultimately, we present a technique with functionality in a clinical setting to develop personalized cancer therapies.
Protocol
The following protocol is for the production of a single microtube device.
1. Preparation of Halloysite Nanotube Solution
- Sonicate (10-13 W (rms)) 250 μL halloysite nanotube solution (6.6 wt% in water) 30 sec. Cool solution with cold water and repeat sonication once. Cool again.
- Draw sonicated solution into a syringe, attach a 0.45 μm syringe filter, and filter solution into clean microfuge tube. Vortex the solution occasionally to maintain homogeneity.
2. Coating of Microtube Inner Surface with Halloysite Nanotubes
- Obtain 50 cm long section of Micro-Renathane microtubing (300 μm inner diameter, 35.3 μL inner volume). Cut one end of the microtube at a diagonal and insert into a small IDEX adaptor piece. Insert the needle of a 3/10 cc 29G syringe into the other end of the microtube.
- To clean the microtube, place the open end of microtube (the end with the adaptor) into 70% ethanol and draw ~50 μL ethanol into the syringe to fill microtube.
- Rinse the ethanol out of the microtube by drawing a generous volume of distilled water into the syringe.
- Detach the syringe from the microtube, empty the syringe, and then reattach to the microtube.
- Prepare a solution of 0.02% w/v poly-L lysine in water. Draw 50 μL into the microtube and allow to sit for 5 min at room temperature (RT).
- Draw 100 μL filtered nanotube solution into the microtube and allow to incubate for 3 min at RT.
- Draw 100 μL water through the microtube to rinse out the nanotube solution. Allow coated microtube to sit, filled with water, overnight at RT.
3. Preparation of Microtubes for Cell Isolation
- Prepare a solution of 10 μg/mL protein G in 1X Dulbecco’s phosphate buffered saline (PBS, pH 7.0 – 7.2). Draw 50 μL PBS through microtube and then draw 50 μL of the protein-G solution and allow to incubate for 1.5 hr at RT.
- Prepare a solution containing 5 μg/mL E-selectin-IgG and 50 μg/mL antibody (anti-EpCAM for most cancer samples, anti-PSMA for prostate cancer samples) in PBS. Draw 50 μL of the E-selectin and antibody solution into the microtube and allow to incubate for 2 hr at RT.
- Nonspecific cellular adhesion is blocked with 5% milk protein. Prepare a solution of 5% (w/v) milk protein in PBS. Draw 50 μL milk protein solution into the microtube and allow to incubate for 1 hr at RT.
- Draw 50 μL PBS into tube and leave at RT until blood or buffy coat samples are ready for processing.
- 10 min prior to using the microtube for isolation, draw 50 μL PBS that has been saturated with Ca2+ (“PBS+”) to activate the selectin molecules.
4. Preparation of samples for cell isolation
- Draw or obtain 10 mL blood from patient into heparinized tube.
- Place 10 mL Ficoll-Paque lymphocyte isolation solution into 50 mL centrifuge tube. Gently layer 10 mL whole blood on top of Ficoll so as not to mix the blood and Ficoll.
- Centrifuge at 2000x g for 15 min at 4 °C with minimal deceleration.
- Remove the buffy coat layer and place in new tube.
- Wash buffy coat with PBS (centrifuge at 230x g for 10 min, discard the supernatant).
- Gently resuspend cells with 1 mL RBC lysis buffer and incubate for 10 min at RT to lyse erythrocytes.
- Add 10 mL PBS, mix gently, and centrifuge at 230x g for 10 min. Discard the supernatant and gently resuspend the pellet in 3 to 4 mL PBS+.
5. Cell isolation
- Attach one end of the functionalized microtube to a 5 mL syringe using IDEX adaptors.
- Insert the syringe onto a syringe pump.
- Submerge the open end of the functionalized microtube into the cell suspension.
- Process the cell suspension through the microtube at 1 to 4 mL/h.
- Transfer the open end of the microtube to a tube containing PBS+ and draw 300 μL PBS+ into syringe at 0.016 mL/min to remove unbound and loosely bound cells from the microtube.
- Place the open end of the microtube into a clean tube. Disconnect the syringe from the microtube and attach a syringe prefilled with Accutase. Gently perfuse enough Accutase into the microtube to fill (~50 μL) and allow to incubate at RT for 10 min.
- Attach a syringe prefilled with 1 mL of growth media (79% RPMI, 20% FBS, 1% penicillin streptomycin) and perfuse into microtube, collecting effluent into tissue culture treated well plate.
- Culture cells at 37 °C and 5% CO2 under humidified conditions.
6. Representative Results
The goal of this technique is to isolate viable cancer cells from the blood of cancer patients. Several methods exist to identify cancer cells in culture; a necessary verification of device success. We have chosen to stain cells in culture with antibodies against epithelial moieties such as EpCAM (epithelial cellular adhesion molecule) or PSMA (prostate specific membrane antigen), in addition to DAPI to identify intact cell nuclei (Fig. 2 and 3). The number of cancer cells captured using this technique is necessarily a function of the number of CTCs in the starting sample, and patient variability can be high. In processing samples taken from patients diagnosed with stage IV cancers, we routinely capture between 100 and 500 cancer cells per tube of blood, at purities >50%. Immediately following isolation, greater numbers of contaminating leukocytes may be present. However these numbers will be depleted following incubation in culture medium for up to 5 days.
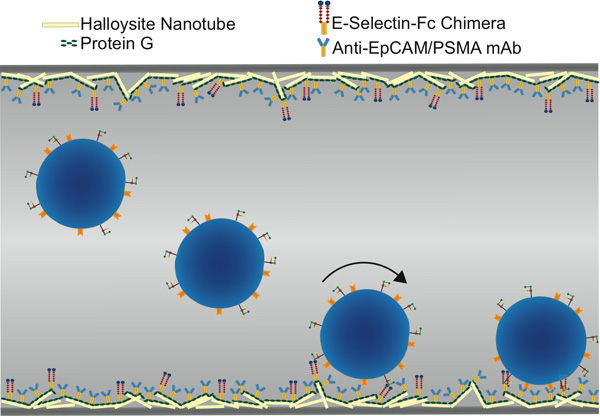
Figure 1. Schematic of functionalized microtube for CTC isolation, showing selectin-mediated rolling followed by antibody-mediated static adhesion.
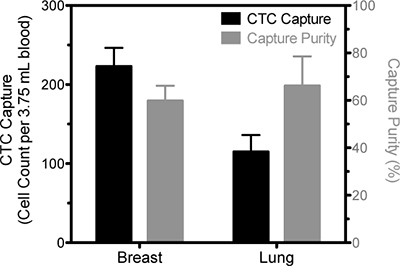
Figure 2. Representative data of CTC isolation from blood samples drawn from a breast cancer patient and a lung cancer patient. The number of viable cells positively identified as CTC based on EpCAM staining is represented by the solid bars and pertain to the left ordinate and the percentage of cells that were identified as CTC compared to the total number of captured cells is represented by the open bars and measured on the right ordinate. Results are following five days in culture.
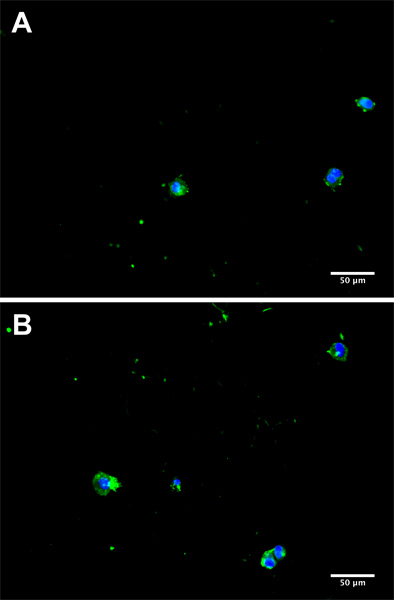
Figure 3. Representative micrographs from separate donors (A and B) of CTC in culture 5 days subsequent to isolation from a cancer patient blood sample. Cells were fluorescently stained for EpCAM with AlexaFluor 488 (green) and 4′,6-diamidino-2-phenylindole (DAPI) to visualize the nucleus (blue).
Discussion
It is often the case that the early steps in the discovery of new cancer therapeutics utilize cancer cell lines, which bear questionable resemblance to primary cancer cells yet remain in use due to their ease of use in the laboratory. Research into the development of new cancer therapies would be expedited if primary human cancer cells were utilized early in novel research. CTC are the most easily accessible type of cancer cell, due to their presence in blood and the ease of a standard blood draw. In addition, circulating tumor cells represent a necessary step in the process of metastasis5,6, so their relevance to the disease and utility for targeting new drugs is clear. Isolation of CTC from blood in vitro is complicated by their low concentrations: on the order of one per million leukocytes or one per billion erythrocytes7. The majority of current methods for detecting CTC in blood, including the only FDA-approved technique CellSearch(Veridex), damage or destroy cells in the detection process, precluding use beyond enumeration. The method described above does not compromise cell viability and thus opens the door for future clinical research on cancer. As the recovery of intact cells is the goal of this technique, it is imperative that cells be handled with care, especially during blood separation steps.
There are a number of tunable parameters in this system that could be altered to achieve improved yield depending on critical characteristics such as cancer type. In the steps described above, we had chosen to use EpCAM as a CTC-specific antibody for all cancer types except when processing prostate cancer samples, wherein anti-PSMA was used. Further substitutions of cancer-specific antibodies can certainly be used to improve performance for individual patients. Furthermore, selectin and antibody concentrations can be altered to enhance capture8.
An essential feature of the device is the incorporation of E-selectin molecules onto the surface. E-selectin is normally expressed on the luminal surface of endothelial cells and function to recruit fast moving leukocytes to sites of inflammation. Flowing leukocytes bind transiently to selectin molecules, resulting in a slower, rolling behavior that facilitates slower and stronger binding of the cell to the endothelium by integrins. Convincing experimental evidence exists that makes the case that CTC can extravasate by a similar mechanism9,10. The inclusion of selectin also allows the device to be operated at greater flow rates, rates that would otherwise prevent cells from binding to antibodies11.Thus our device physiologically mimics a venule to biomimetically capture flowing CTC without inflicting cellular injury.
Enhanced device performance can be attributed to the addition of the halloysite nanotube coating to the luminal surface of the device. Previous studies have shown that there are three major components of the nanotube coating that allows for improved functionality. First, the nanotube coating provides increased surface area, allowing for greater protein deposition onto the surface4. Second, in performing atomic force microscopy on the nanotube coating we have determined that individual nanotubes protrude from the surface into the flow. This allows selectin molecules to be presented as much as one micron above the surface so that cells can be captured and recruited to the surface earlier in their trajectory through the tube4. Finally, the halloysite nanotube coating is able to prevent leukocyte adhesion and spreading on the surface, allowing for a reduced number of leukocytes captured along with the CTC and thus greater subsequent CTC purities3.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
The work described was supported by the Cornell Center on the Microenvironment & Metastasis through Award Number U54CA143876 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. This work was additionally funded in part by a National Science Foundation Graduate Research Fellowship (A.D.H).
Materials
| Name of reagent | Company | Catalog Number |
| 300 um ID tubing | Braintree Scientific | MRE025 |
| Larger tubing for connection to syringe | IDEX Health & Science | 1507L |
| 5mL syringe for pump | BD | 309603 |
| Connector small to large tubing | IDEX Health & Science | P-770 |
| Connector large tubing to syringe | IDEX Health & Science | F-120 and P-659 |
| Syringe for microtube (3/10cc Insulin Syringe U-100 29G 1/2′) | BD | 309301 |
| Accutase | MP Biomedicals LLC | 1000449 |
| Ficol-Paque Plus | GE Healthcare Bio-Sciences AB | 17-1440-03 |
| RBC Lysis Buffer (Buffer EL, 1000mL) | Qiagen | 1014614 |
| Protein G, 5 mg | CalBiochem (calbiochem.com) | 539303 |
| Human EpCAM/TROP-1 Antibody | R&D Systems | MAB960 |
| PSMA Antibody (GCP-04) | abcam | ab66911 |
| 96 well plates (Microtest 96) | BD Falcon | 35-3072 |
| Blotting Grade Blocker Non-Fat Dry Milk, 300g | Bio-Rad Laboratories | 216005508 |
| Halloysite Nanotubes | NaturalNano | NN-HNT200 |
| Calcium Carbonate, 5g | Aldrich Chemistry | 481807 |
| rhE-Selectin/Fc Chimera, 100 ug | R&D Systems | 724-ES |
| RPMI Medium 1640 | Gibco | 22400-089 |
| DPBS | Gibco | 14190-095 |
| Poly-L lysine 0.1% w/v in water | Sigma-Aldrich | P8920-100ML |
| Sonic Dismembrator | Fisher Scientific | Model 100 |
| Fetal Bovine Serum | Atlanta Biochemicals | S11056H |
| Penicillin Streptomycin | Sigma Life Siiences | P0781 |
Referenzen
- Allard, W. J. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 10, 6897-6904 (2004).
- Hughes, A. D., King, M. R. Nanobiotechnology for the capture and manipulation of circulating tumor cells. Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology. , (2011).
- Hughes, A. D. Microtube Device for Selectin-Mediated Capture of Viable Circulating Tumor Cells from Blood. Clinical Chemistry. 58, 846-853 (2012).
- Hughes, A. D., King, M. R. Use of naturally occurring halloysite nanotubes for enhanced capture of flowing cells. Langmuir. 26, 12155-12164 (2010).
- Al-Mehdi, A. B. Intravascular origin of metastasis from the proliferation of endothelium-attached tumor cells: a new model for metastasis. Nat. Med. 6, 100-102 (2000).
- Gupta, G. P., Massague, J. Cancer metastasis: Building a framework. Cell. 127, 679-695 (2006).
- Lee, D., King, M. R. Microcontact Printing of P-Selectin Increases the Rate of Neutrophil Recruitment Under Shear Flow. Biotechnology Progress. 24, 1052-1059 (2008).
- Gout, S., Tremblay, P. L., Huot, J. Selectins and selectin ligands in extravasation of cancer cells and organ selectivity of metastasis. Clin. Exp. Metastasis. 25, 335-344 (2008).
- Geng, Y., Marshall, J., King, M. Glycomechanics of the Metastatic Cascade: Tumor Cell-Endothelial Cell Interactions in the Circulation. Annals of Biomedical Engineering. , 1-16 .
- Stott, S. L. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. U.S.A. 107, 18392-18397 (2010).

