Agroinfiltration and PVX Agroinfection in Potato and Nicotiana benthamiana
Summary
Agroinfiltration and PVX agroinfection are routine functional assays for transient ectopic expression of genes in plants. These methods are efficient assays in effectoromics strategies (rapid resistance and avirulence gene discovery) and crucial to modern research in molecular plant pathology. They meet the demand for robust high-throughput functional analysis in plants.
Abstract
Agroinfiltration and PVX agroinfection are two efficient transient expression assays for functional analysis of candidate genes in plants. The most commonly used agent for agroinfiltration is Agrobacterium tumefaciens, a pathogen of many dicot plant species. This implies that agroinfiltration can be applied to many plant species. Here, we present our protocols and expected results when applying these methods to the potato (Solanum tuberosum), its related wild tuber-bearing Solanum species (Solanum section Petota) and the model plant Nicotiana benthamiana. In addition to functional analysis of single genes, such as resistance (R) or avirulence (Avr) genes, the agroinfiltration assay is very suitable for recapitulating the R-AVR interactions associated with specific host pathogen interactions by simply delivering R and Avr transgenes into the same cell. However, some plant genotypes can raise nonspecific defense responses to Agrobacterium, as we observed for example for several potato genotypes. Compared to agroinfiltration, detection of AVR activity with PVX agroinfection is more sensitive, more high-throughput in functional screens and less sensitive to nonspecific defense responses to Agrobacterium. However, nonspecific defense to PVX can occur and there is a risk to miss responses due to virus-induced extreme resistance. Despite such limitations, in our experience, agroinfiltration and PVX agroinfection are both suitable and complementary assays that can be used simultaneously to confirm each other's results.
Introduction
Effectoromics, a high-throughput functional genomics approach has recently emerged as a powerful tool to identify resistance (R) genes in crop plants and matching avirulence (Avr) genes of pathogens1-4. In contrast to the more time-consuming stable transformation with R genes, the effectoromics strategy is based on transient assays of pathogen gene sequences.
Since the genomics era, genomes of plant pathogens have widely been explored. For example for oomycetes, which include the most devastating plant pathogens, large collections of sequences have been generated and analyzed for genes that play a role during the interaction with the plant5-10. One class of pathogen proteins represents effectors, which manipulate host cell structure and function either to facilitate infection (virulence factors) or to trigger defense responses (avirulence factors)11-13. Expression of Avr genes in plant cells containing R genes usually results in the hypersensitive cell death response (HR)14,15. In planta expression of R and Avr genes can be accomplished using transient expression systems such as Agrobacterium tumefaciens –based transient transformation (agroinfiltration)16. This transient transformation can also be applied in combination with viral expression systems (agroinfection)17,18.
For agroinfiltration, the most commonly used agent is A . tumefaciens, a broad host range pathogen of dicot plants. A . tumefaciens contains a tumor-inducing (Ti) plasmid. Transfer DNA (T-DNA) from a Ti plasmid will translocate into the plant cells after the virulence machinery of the bacterium is activated. This can be triggered in wounded plant cells, by the released low-molecular-weight phenolic compounds and monosaccharaides in a slightly acidic environment19. The virulence gene is activated after the infiltration of Agrobacterium suspensions into leaf panels defined by major veins. Then plant cells in the leaf panels will be transformed and express the transgene(s) contained in the T-DNA region.
Agroinfection is based on wound-inoculated Agrobacterium, which mediates translocation of a virus to plant cells. The virus then further spreads to adjacent plant tissues, in the absence of Agrobacterium. For agroinfection, several plant viruses can be used. RNA viruses are ideal vectors for gene expression because they can multiply to very high levels in infected plants. Among plant RNA viruses, Potato Virus X (PVX) is widely used for effectoromics screens. To facilitate functional tests for an inserted gene, binary vectors that contain the PVX genome flanked by the Cauliflower mosaic virus 35S promoter and the nopaline synthase terminator, were cloned into the T-DNA of A. tumefaciens20. After the T-DNA is transferred into plant cells, the PVX genome contained in the T-DNA is transcribed from the 35S promoter. Then virus particles spread systemically in the infected plants, resulting in the expression of the inserted gene. This method based on both Agrobacterium and PVX is called PVX agroinfection.
Here we show examples for both the agroinfiltration and PVX agroinfection assays. As host plants we use potato germplasm (Solanum section Petota), for which effectoromics approaches have been pioneered and proven successful3,4. We also use Nicotiana benthamiana, which is renowned as a model plant in Solanaceous plants14,21,22.
Protocol
1. Plant Growing and Testing Conditions
- Grow and maintain plants in controlled greenhouses or climate chambers within the temperature range of 18-22 °C and under natural light regime or with a 16 hr/8 hr day/night regime. Remove axillary branches in order to make the plants more manageable.
- For potato, maintain in vitro plantlets in sterile plastic jars containing Murashige-Skoog (MS) medium (20 g/L sucrose, 5 g/L MS salts with vitamins, 8 g/L agar, pH 5.8) under controlled conditions in climate chambers at 18 °C with a 16 hr/8 hr day/night regime for two weeks and then transfer them into pots of sterilized soil in regulated greenhouse compartments.
2. Agroinfiltration
- Plant material:
For Nicotiana benthamiana, use around 4-5 week old seed-grown plants. For potato, use around 4-5 week old transplants from in vitro. Choose young, healthy and fully developed leaves for infiltrations. - Agrobacterium culture:
- Prepare the YEB medium in advance (Table 1). Fill the 50 ml tubes with 10 ml YEB medium supplemented with 1 μl acetosyringone (200 mM), 100 μl MES buffer (2-(N-morpholino)-ethane sulfonic acid, 1 M) and the appropriate antibiotics. Pipette 20 μl glycerol stock of the desired strains (Table 2) (containing the gene of interest) into the YEB. Initiate cultures for all the Agrobacterium strains in this assay at the same time. Incubate cells by shaking for 1-2 days at 28 °C and 200 rpm to an OD600 of approximately 1.0.
- Harvest cells by centrifugation at 3,000 x g for 10 min. Pour off the supernatant and resuspend the pellet in freshly made MMA medium (Table 3) to an OD600 of 0.3. Gently vortex cells to resuspend them. For coinfiltration of two bacterial strains, mix the culture in a 1:1 ratio.
- Leave the culture on the bench at room temperature for 1-6 hr before infiltrations. In the meantime, label the plants to be infiltrated and the date of the experiment.
- Infiltrations:
Use a 1 ml needleless syringe to infiltrate the Agrobacterium suspensions. Carefully and slowly inject leaf panels with the suspensions from the syringe (for health and safety reasons eye protection should be worn during this process). Infiltrate at least three plants for each strain. Use three leaves per plant to serve as triplicates.
Note: Normally, 1 ml of Agrobacterium suspension s could be enough for 3 plants of N. benthamiana. For potato, more suspension is needed because the leaves of Solanum species are more difficult to infiltrate. Required volumes of suspensions depend on the Solanum species. Avoid cross-contamination by changing gloves or sterilizing gloves with ethanol between infiltrations and by not watering the plants until the day after the inoculation. - Scoring:
Score macroscopic responses about 3 days after infiltration when the cell death phenotype is clear (Figure 1). If the cell death phenotype is quantitative, use the given criteria (Figure 2). Briefly, the scales for macroscopic scoring of cell death mainly depend on the cell death percentages of the infiltrated area. Percentages of cell death are depicted on a scale from 0% (no symptoms) to 100% (confluent cell death). Intermediate responses range from weak responses such as chlorosis to increasing levels of cell death. If desired, the macroscopic scoring can also be quantitatively assessed using modern photo-imaging equipment.
3. PVX Agroinfection
- Plant material:
For N. benthamiana, use around 2-3 week old seed-grown plants. For potato, use around 2-3 week old transplants from in vitro. For large-scale tests, use slightly older (4-5 weeks) plants, as these plants have more and larger leaves to accommodate higher numbers of Agrobacterium inoculation spots. - Agrobacterium culturing:
- Prepare the YEB medium in advance. Fill the 10 ml tubes with 3 ml YEB supplemented with the appropriate antibiotics. Pipette 20 μl glycerol stock of the desired strains (Table 2) (containing the gene of interest) into the YEB. Initiate cultures for all the Agrobacterium strains in this assay at the same time. Incubate cells by shaking for 1-2 days at 28 °C and 200 rpm to an OD600 of approximately 1.0.
- Pipette about 300 μl of each Agrobacterium strain and spread them onto LB solid agar medium plates supplemented with the appropriate antibiotics and incubate cells at 28 °C for 1-2 days.
- Infections:
Use a spatula to collect the Agrobacterium culture in the plate. Dip a wooden toothpick in the Agrobacterium culture on the spatula and pierce the leaves to inoculate large amount of bacteria. Inoculate at least three plants for each strain. Use three leaves per plant to serve as triplicates. In each leaf, make multiple inoculation sites for each strain. - Scoring:
Score macroscopic responses about two weeks after inoculation (Figure 3). For high-throughput screens, record the qualitative responses (yes/no) for each inoculation spot. Then calculate the percentage of responding sites and compare them with controls.
Representative Results
Figure 1 shows a representative experiment of agroinfiltration with 7 different effectors (E1-E7) in potato and N. benthamiana. Cell death appears in the infiltrated leaf panels about 3 days after infiltration. The extent of the cell death phenotype needs to be compared with the controls. A mixture of the Agrobacterium strain AGL1(pVirG)23 containing pBINplus-R3a and pK7WG2-AVR3a was used as positive control24,25, while AGL1(pVirG) was used as negative control. Figures 1A and 1B show good examples of agroinfiltration in the potato genotype MaR8 (Mastenbroek R8)26 and N. benthamiana, respectively, which are very amenable for agroinfiltration. There is confluent cell death in the leaf panel coinfiltrated with positive control, while no cell death response occurs with negative control or pBINplus-R3a. In MaR8, two effectors AVR3a (E2) and AVR4 (E3) induce a cell death, while the other effectors (E1 and E4-E7) do not. In N. benthamiana, none of the tested effectors induce a cell death response. Figures 1C and 1D show examples of agroinfiltration in the wild potato Solanum berthaultii 483-1 and Solanum rechei 210-5, which are not well amenable for the agroinfiltration technique. In S. berthaultii 483-1, the leaf tissue shows a nonspecific necrosis to negative controls as well as to all tested effectors. In S. rechei 210-5, infiltrated leaf panels show very weak cell death response to positive control.
Figure 2 shows a range of scoring scales that can be used to quantify the response to agroinfiltrated Agrobacterium. Percentages of cell death are depicted on a scale 0-100%. Observed phenotypes range from macroscopically not visible symptoms (0%), through a range of intermediate responses displaying chlorosis and increasing levels of cell death, up to confluent cell death (100%).
Figure 3 shows a representative experiment after PVX agroinfection in potato. Normally, expanding cell death can be found at the sites about two weeks after tooth-pick inoculation. As shown in both figure panels, expanding cell death is present at the sites that were tooth-pick inoculated with pGR106-CRN2 (positive control using the effector crinkler Crn2), which is a general cell death inducing gene from P. infestans27. Apart from minor response to wounding, no expanding cell death is noted at the sites that were tooth-pick inoculated with the pGR106-empty (negative control). In Figure 3A, representing the potato genotype MaR3 (Mastenbroek R3), two pGR106-effectors (E1 and E2) did not induce cell death. In Figure 3B, a positive result of effector screening in the wild Solanum huancabambense 354-1 is presented; cell death response was observed to two pGR106-effectors (E1-E3, representing elicitins)28.
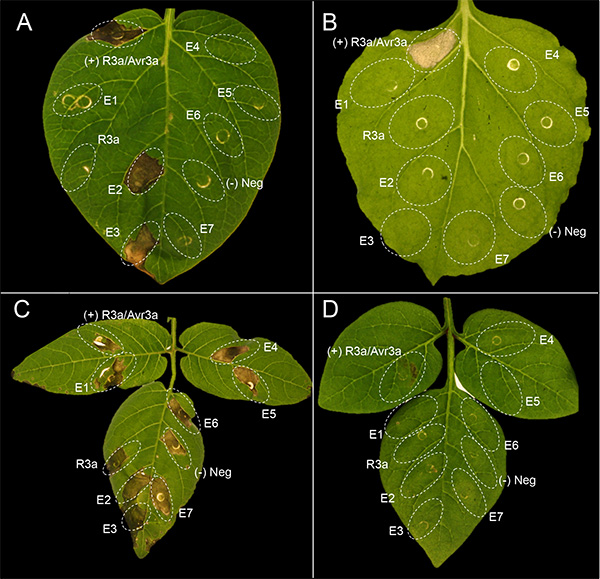
Figure 1. Examples of agroinfiltration in potato and N. benthamiana. Plants including (A) a potato genotype MaR8 (Mastenbroek R8), (B) N. benthamiana, (C) Solanum berthaultii 483-1 and (D) Solanum rechei 210-5 were infiltrated with a mixture of the Agrobacterium strain AGL1(pVirG) 23 containing pBINplus-R3a and pK7WG2-AVR3a (positive control), AGL1(pvirG) (negative control), pBINplus-R3a, and seven effectors (E1-E7). Click here to view larger image.
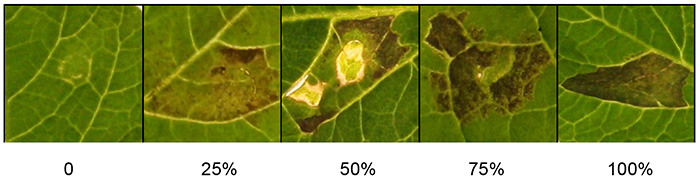
Figure 2. Quantification of agroinfiltration responses. The photograph shows representative scoring scales for cell death, ranging from 0% (no symptoms) to 100% (confluent cell death). Intermediate responses range from weak responses such as chlorosis to increasing levels of cell death. Click here to view larger image.
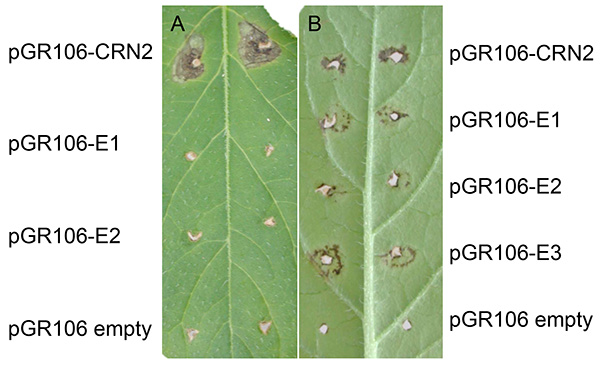
Figure 3. Examples of PVX agroinfection in potato. Potato genotype MaR3 (Mastenbroek R3) (A) and Solanum huancabambense 354-1 (B) tooth-pick inoculated with pGR106-CRN2 (positive control), pGR106-empty (negative control) and pGR106-E1-2 (effectors), or pGR106-E1-E3, respectively. Click here to view larger image.
Table 1. YEB medium
| 1 L | distilled H2O |
| 5 g | sucrose |
| 5 g | beef extract |
| 5 g | bacteriological peptone |
| 1 g | yeast extract |
| 2 ml | MgSO4 (1 M) |
Table 2. Vectors and strains used for agroinfiltration and PVX agroinfection. Several binary vectors can be used. Vectors in the list below allow high expression of the candidate genes and have worked well in our hands. We prefer using the Agrobacterium strain GV3101(pMP90)29 in N. benthamiana and find AGL130 containing the helper plasmid pVirG (pBBR1MCS-5.virGN54D)23 more suitable in potato31. Additional strains have been analyzed in other groups on various model plants including N. benthamiana but not in potato32.
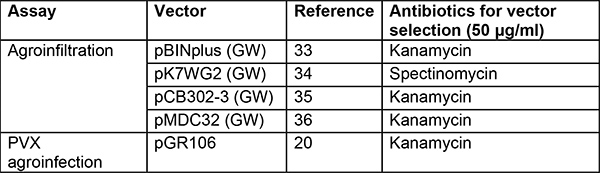
GW: gateway version36
Table 3. MMA medium.

Adjust the pH to 5.6.
Discussion
Transient assays like agroinfiltration and agroinfection are efficient methods that are vital to modern molecular plant pathology research. Despite some limitations, these methods meet the demand for efficient and robust high-throughput functional analysis in plants.
The agroinfiltration system is a widely used functional assay in a range of plant species. Agroinfiltration facilitates the delivery of several transgenes into the same cell with simultaneous expression of interacting proteins. This is advantageous for recapitulating R-AVR relationships, by coinfiltrating Agrobacterium strains that express Avr genes with strains that express the matching R genes. Also, for known R-AVR pairs, such coinfiltrations can be used as positive controls. Including such controls is important because in some plant genotypes, the transformation efficiency can be below the threshold to detect responses. Including negative controls, e.g. an Agrobacterium strain containing a vector without a gene insert, is also essential to determine whether a certain plant genotype raises nonspecific defense responses to the Agrobacterium. This feature occurs at a certain frequency in potato germplasm, and not all Solanum species are well suitable for this Agrobacterium-based expression system. Generally, the agroinfiltration assay works very well in N. benthamiana and most potato genotypes. In addition to effectoromics, there are various other potential applications for the agroinfiltration technique, such as production of proteins from transgenes and protein localization in plant cells by confocal microscopy.
PVX agroinfection is a highly sensitive screening system and typically more suitable for high-throughput screenings. Since the Agrobacterium is now only locally present, nonspecific responses to this bacterium are now not very disturbing, as the PVX virus takes over further spread of the transgene. However, plants may be resistant to PVX, or mount extreme resistance (ER) responses, and in that case the agroinfection method is not suitable. Another limitation of the PVX agroinfection method is the insert size of the gene of interest. Observed phenotypes of responses may vary from an intense black necrosis surrounding the wound to faint necrosis near the inoculation spot. In both N. benthamiana and Solanum species, PVX agroinfection is recognized as more sensitive than agroinfiltration.
Keeping into account that the genetic background of the diverse tested plant genotypes can have some restrictions (see above), we generally obtain similar conclusions by PVX agroinfection and agroinfiltration. These results are also comparable as obtained in other assays, such as protein infiltrations29 and ELISA3. Considering the advantages and limitations of both systems, we recommend using both methods to either complement each other or confirm independent results.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
The work is partially supported by Wageningen University Fund (WUF), China Scholarship Council Program for Graduate Students, and a NWO-VIDI grant 12378.
Materials
| Beef extract | Sigma-Aldrich | B4888 | |
| Bacteriological peptone | Oxoid | LP0037 | |
| Yeast extract | Oxoid | LP0021 | |
| MgSO4 | Sigma-Aldrich | 208094 | |
| MS salts (without vitamins) | Duchefa Biochemie | M0221 | |
| MES | Duchefa Biochemie | M1503 | |
| LB broth powder | Sigma-Aldrich | L3022 | |
| Acetosyringone | Sigma-Aldrich | D134406 | |
| Syringe (1 ml) | BD Plastipak | 300013 | |
| Incubator | Infors HT | Multitron II | |
| Centrifuge | Heraeus | Multifuge 3S-R | |
| Spectrophotometer | Eppendorf | Biophotometer 6131 |
Referenzen
- Vleeshouwers, V. G. A. A., et al. Understanding and exploiting late blight resistance in the age of effectors. Ann. Rev. Phytopathol. 49, 507-531 (2011).
- Ellis, J. G., Rafiqi, M., Gan, P., Chakrabarti, A., Dodds, P. N. Recent progress in discovery and functional analysis of effector proteins of fungal and oomycete plant pathogens. Curr. Opin. Plant Biol. 12, 399-405 (2009).
- Vleeshouwers, V. G. A. A., et al. Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. Plos One. 3, 1-10 (2008).
- Oh, S. K., et al. In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell. 21, 2928-2947 (2009).
- Tyler, B. M., et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 313, 1261-1266 (2006).
- Haas, B. J., et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 461, 393-398 (2009).
- Levesque, C. A., et al. Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol. 11, 1-22 (2010).
- Lamour, K. H., Win, J., Kamoun, S. Oomycete genomics: new insights and future directions. FEMS Microbiol. Lett. 274, 1-8 (2007).
- Stassen, J. H. M., et al. Effector identification in the lettuce downy mildew Bremia lactucae by massively parallel transcriptome sequencing. Mol. Plant Pathol. 13, 719-731 (2012).
- Kemen, E., et al. Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PLoS Biol. 9, 1-21 (2011).
- Win, J., et al. . Effector biology of plant-associated organisms: concepts and perspectives. , 1-13 (2012).
- Hogenhout, S. A., van der Hoorn, R. A. L., Terauchi, R., Kamoun, S. Emerging concepts in effector biology of plant-associated organisms. Mol. Plant Microbe Interact. 22, 115-122 (2009).
- Kamoun, S. A catalogue of the effector secretome of plant pathogenic oomycetes. Ann. Rev. Phytopathol. 44, 41-60 (2006).
- van der Hoorn, R. A. L., Laurent, F., Roth, R., De Wit, P. J. G. M. Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf-9-induced and Avr4/Cf-4-induced necrosis. Mol. Plant Microbe Interact. 13, 439-446 (2000).
- Dangl, J. L., Jones, J. D. G. Plant pathogens and integrated defence responses to infection. Nature. 411, 826-833 (2001).
- Kapila, J., de Rycke, R., van Montagu, M., Angenon, G. An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci. 122, 101-108 (1997).
- Kanneganti, T. D., Huitema, E., Kamoun, S. In planta expression of oomycete and fungal genes. Methods Mol. Biol. 354, 35-43 (2007).
- Vleeshouwers, V. G. G. A., Rietman, H., Lamour, K., Kamoun, S. In planta expression systems. Oomycete Genetics and Genomics. Diversity, Interactions, and Research Tools. 23, 455-475 (2009).
- Peng, W. T., Lee, Y. W., Nester, E. W. The phenolic recognition profiles of the Agrobacterium tumefaciens VirA protein are broadened by a high level of the sugar binding protein ChvE. J. Bacteriol. 180, 5632-5638 (1998).
- Lu, R., et al. High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 22, 5690-5699 (2003).
- Ma, L., Lukasik, E., Gawehns, F., Takken, F. L. W. The use of agroinfiltration for transient expression of plant resistance and fungal effector proteins in Nicotiana benthamiana leaves. Methods Mol. Biol. 835, 61-74 (2012).
- Bhaskar, P. B., Venkateshwaran, M., Wu, L., Ané, J. M., Jiang, J. Agrobacterium-mediated transient gene expression and silencing: A rapid tool for functional gene assay in potato. Plos One. 4, 1-8 (2009).
- van der Fits, L., Deakin, E. A., Hoge, J. H. C., Memelink, J. The ternary transformation system: constitutive virG on a compatible plasmid dramatically increases Agrobacterium-mediated plant transformation. Plant Mol. Biol. 43, 495-502 (2000).
- Huang, S., et al. Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato. Plant J. 42, 251-261 (2005).
- Armstrong, M. R., et al. An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl. Acad. Sci. U.S.A. 102, 7766-7771 (2005).
- Kim, H. J., et al. Broad spectrum late blight resistance in potato differential set plants MaR8 and MaR9 is conferred by multiple stacked R genes. Theor. Appl. Genet. 124, 923-935 (2012).
- Torto, T. A., et al. EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora. Genome Res. 13, 1675-1685 (2003).
- Vleeshouwers, V. G. A. A., et al. Agroinfection-based high-throughput screening reveals specific recognition of INF elicitins in Solanum. Mol. Plant Pathol. 7, 499-510 (2006).
- Koncz, C., Schell, J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of agrobacterium binary. Mol. Gen. Genet. 204, 383-396 (1986).
- Lazo, G. R., Stein, P. A., Ludwig, R. A. A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Bio-Technol. 9, 963-967 (1991).
- Rietman, H. . Putting the Phytophthora infestans genome sequence at work; multiple novel avirulence and potato resistance gene candidates revealed [PhD thesis]. , (2011).
- Wroblewski, T., Tomczak, A., Michelmore, R. Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol. J. 3, 259-273 (2005).
- van engelen, F. A., et al. pBINPLUS – an improved plant transformation vector based on pBIN19. Transgenic Res. 4, 288-290 (1995).
- Karimi, M., Inze, D., Depicker, A. GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193-195 (2002).
- Xiang, C. B., Han, P., Lutziger, I., Wang, K., Oliver, D. J. A mini binary vector series for plant transformation. Plant Mol. Biol. 40, 711-717 (1999).
- Curtis, M. D., Grossniklaus, U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462-469 (2003).

