An Optimized Enrichment Technique for the Isolation of Arthrobacter Bacteriophage Species from Soil Sample Isolates
Summary
We present an enrichment protocol for the isolation of bacteriophages infecting bacteria in the Arthrobacter genus. This enrichment protocol produces fast and reproducible results for the isolation and amplification of Arthrobacter phages from soil isolates.
Abstract
Bacteriophage isolation from environmental samples has been performed for decades using principles set forth by pioneers in microbiology. The isolation of phages infecting Arthrobacter hosts has been limited, perhaps due to the low success rate of many previous isolation techniques, resulting in an underrepresented group of Arthrobacter phages available for study. The enrichment technique described here, unlike many others, uses a filtered extract free of contaminating bacteria as the base for indicator bacteria growth, Arthrobactersp. KY3901, specifically. By first removing soil bacteria the target phages are not hindered by competition with native soil bacteria present in initial soil samples. This enrichment method has resulted in dozens of unique phages from several different soil types and even produced different types of phages from the same enriched soil sample isolate. The use of this procedure can be expanded to most nutrient rich aerobic media for the isolation of phages in a vast diversity of interesting host bacteria.
Introduction
The ubiquity of Arthrobacter species in soil environments offers a vast number and diversity of phages capable of being isolated from this species of host bacteria. Bacterial members of the Acintobacteriaceae family are most notable for their catabolic pathways of degrading recalcitrant compounds like atrazine and various other pesticides and herbicides1,2,3. Though most research has been done using environmental strains of Arthrobacter, clinical isolates of this genus is found in blood, urine, eyes, and many other human sources all displaying phylogenetic heterogeneity4.
While there is a rather extensive body of research on Arthrobacter bacteria, only a few studies report on the phages capable of infecting members of this diverse genus. Interestingly though, work done previously on Arthrobacter phages touches on several key distinct topics such as the typing of soil Arthrobacter species5, industrial uses with the purpose of reducing deleterious foam in activated sludge treatment plants 6, and work highlighting site specific recombination and integrase genes7.
Various enrichment technique protocols have been employed to generate pure phage isolates in Arthrobacter species. Early procedures include incubations of soil with added toxic agents like nicotine salts for periods of over one year8 giving rise to phages capable of only infecting A. globiformis. Studies done using soil percolated with labile organics appeared to produce detectable phages via plaque assay techniques, omitting lengthy incubation periods8. Interestingly though, a technique resembling direct plating was used in the past giving rise to several phages while still having a notably low success rate by the investigators5, citing past studies with low success rates8.
Overall, the isolation techniques used in the past were notable for having little efficacy in practice despite the Arthrobacter genus representing the most common aerobic soil isolate in nature4,9,,Van Twest and Kropinski10 present enrichment methods for isolating phages from water and soil adapted from earlier techniques used to enrich environmental bacterial isolates but these enrichment techniques proved inefficient in isolating Arthrobacter phages. The purpose of the method described here is to show “proof of concept” that the early enrichment methods can be adapted to consistently and effectively isolate Arthrobacter phages, overcoming previous technical challenges associated with isolating phages from this bacterial genus.
Protocol
1. Preparation of Arthobacter Cells for Phage Isolation
- Culture Arthrobactersp. KY3901 colonies streaked on an Luria Bertaini (LB) agar plate incubated at 30 °C for 2-3 days. Pick a colony and use a sterile loop to add it to 250 ml of LB broth in a baffled culture flask and incubate in a shaking incubator at 225 rpm at 30 °C.
- Allow approximately 24 hr of growth to obtain late exponential/early stationary phase cells for phage infection experiments. Monitor the state of bacterial growth closely to prevent cells from entering the middle to late stationary growth state.
NOTE: Cell growth should consist of an optical density OD600 between 0.5-.0.7 for the phage isolation and purification procedures/steps described below.
2. Collection of Phage from Soil Samples
- Gather 200-400 g of soil from desired location. Note: Since bacteria in the Arthrobacter genus are common soil bacteria they and the phages that infect them can be found in and are pervasive in most types of soil.
- Add at least 400 ml of Phage Buffer (PB) [68 mM NaCl, 10 mM MgSO4,10 mM Tris-CL (pH 7.5)] to the soil and mix in a large enough flask so that at least 200 ml of PB is in the supernatant and is able to be extracted.
- Mix by gently stirring or swirling until the soil is suspended and allow the soil sediment to settle, typically 30 min.
- Pass the “soil extract” through filter paper by gravity filtration to remove soil sediment debris.
- Add LB powder and mix to make a 2% solution (4 g LB powder in 200 ml soil extract).
- Pass the solution through a 0.22 µm filter by vacuum filtration to obtain a sterile filtrate. If possible, continue on to step 2.7 immediately. If need be, store filtered samples at 4 °C for up to a week before proceeding, however, the number of viable phage particles will be reduced.
- Aliquot multiple 50 ml portions of the filter-sterilized LB/soil extract mixture into individual 250 ml sterilized shaking baffled culture flasks using aseptic techniques. Add sterile 2.0 M CaCl2 solution to the final desired concentrations (0-50 mM) since different Arthrobacter phages grow optimally under different CaCl2 conditions.
NOTE: We find that the majority of the Arthrobacter phages identified by us are within the range of 1 mM-2.5 mM CaCl2. However, several of the isolated phages grew by us exclusively at 0 mM CaCl2.or at very high CaCl2 concentrations. Proceed immediately to step 3.1.
3. Phage Isolation
- Add 1 to 2.5 ml of late exponential/early stationary phase bacteria culture to each of the flasks. For an OD600 from 0.5-0.7, use 1 ml of cells. For an OD600 higher than an OD of 0.7 use 2.5 ml of cells.
NOTE: Cells in exponential phase growth typically yield more phages and higher titers than cells in stationary phase growth. - Shake flasks at 250 rpm at 30 °C for approximately 24 hr in a shaking incubator.
- After a 24 hr incubation period remove enrichment flasks from the shaking incubator and dilute the enrichment samples ten-fold in phage buffer.
- Set up culture tubes with 0.5 ml of late exponential/early stationary phase bacteria culture and add various amounts of the enrichment culture (5 µl to 500 µl of a 10-1 dilution in PB) to the culture tubes.
NOTE: It is advisable to test a wide range of concentrations since each soil sample generates different phage titers after enrichment. - Add CaCl2 to culture tubes to make a final concentration equal to the concentration present in the original enrichment flask. Use a range of different CaCl2 concentrations to select for many phages with varying CaCl2 dependencies.
NOTE: Most phages will be isolated within the CaCl2 range of 1-2.5 mM but there are phages isolated most optimally anywhere from 0-50 mM CaCl2 concentrations. Because of this it is best to check a range of CaCl2 concentrations to maximize the number of unique isolated Arthrobacter phages. - Mix 4.5 ml of LB top agar in the culture tube and pour the mixture onto an LB agar plate. Swirl gently to distribute across the plate evenly and allow for top agar to solidify for approximately 15 min.
NOTE: Top agar can be prepared in larger batches (250 ml) and extra top agar can be stored at RT and reused for subsequent experiments for at least two weeks. Although there are different formulas used to make top agar, we make top agar using 7g/L of agar mixed with LB. Place top agar in a 60 °C water bath to retain the liquid state during the course of an experiment. - Invert the plate and incubate at desired temperature (temperature to 30 °C) O/N to 48 hr. Vary these conditions to optimize for the phages present. Most isolated phages come from plates incubated at 30 °C at 1-2.5 mM CaCl2 concentration after a 24 hr incubation period.
4. Phage Purification
- After a 24 hr incubation check for plaque formation on plates. Continue incubation for up to 72 hr if no plaques are evident or to determine if additional plaques will appear. If needed, store plates with putative plaques for up to a week at 4 °C prior to proceeding.
NOTE: It is not uncommon to find a variety of different plaque morphologies on one plate. Finding plaques after 3 days of incubation is possible but rare. - Purify desired phages from clearly isolated plaques using the streak plaque method. Touch a plaque with a sterile wooden stick. Streak the plaque by running a wooden stick through a flame, allowing the stick to cool briefly, and then rubbing the stick on the agar plate in the gentle motion as shown in Figure 1. Repeat this using a clean stick for each pass.
- Alternatively, use the traditional plaque titer assay to isolate individual phage plaques. The plaque titer assay does require the use of more reagents such as LB agar and LB top agar and materials such as petri plates for phage plaque isolation. We have been highly successful using the streak plaque method but it is technically easier to isolate individual plaques using the plaque titer assay.
- Once the LB agar plate is streaked at least three times, mark the area with the lowest concentration of putative phage particles and GENTLY apply 0.5 ml of Arthrobacter and 4.5 ml of molten LB top agar (containing the same concentration of calcium chloride as the parent plate) and allow it to spread evenly across the plate.
- Invert and incubate plate O/N. Repeat the streak plaque technique for at least three iterations to ensure a pure phage species is isolated. Store plates at 4 °C for up to a week as needed between streaks without significant loss of phage viability.
- Once desired plaques have been grown and isolated on the final streak plate, touch one well isolated plaque with a micropipette tip and re-suspend in 100 µl PB. Serially dilute this 100 solution out to the 10-8 dilution by passing 20 µl of the sample into 180 µl of fresh PB.
- Add 10 µl of each dilution to 0.5 ml of grown Arthrobacter in a culture tube for 5 to 10 min at RT and plate by mixing the infected bacteria with 4.5 ml of molten LB top agar containing the same concentration of CaCl2 used to isolate the phage from the original soil sample. Save all dilutions made at 4 °C until the next day.
- Determine the phage serial dilution needed to make a web pattern using the traditional plaque titer assay. The purpose of the plaque titer assay is to determine the phage titer needed to obtain a web pattern plate. Identify a web pattern plate as mostly devoid of bacteria but containing remnants of bacteria that have not yet been lysed by phages. Add 5 ml of late exponential/ early stationary Arthrobacter culture to a sterile 50 ml culture flask.
- Add 100 µl of the dilution (that gave a web pattern to the Arthrobacter culture) in the culture flask and allow the phages to infect the bacteria cells for 5 to 10 min at RT. Mix with molten LB top agar containing the same concentration of CaCl2 as used to initially identify the phage and plate 5 ml of this mixture onto 10 fresh LB plates. Allow to solidify and incubate inverted plates at 30 °C.
- The next day, flood the web pattern plates with 5 ml of PB and store O/N at 4 °C or 4 hr at RT.
- Filter the phage lysate through a 0.22 µm syringe filter and store at 4 °C. Use this final phage stock for additional experiments such as electron microscopy images of phage particles, phage genomic DNA isolation for DNA restriction enzyme analysis and genomic sequencing using standard procedures. For long term storage of the phage stock, add an equal amount of the phage lysate to sterile glycerol in small vials for stock archiving at -80 °C.
NOTE: For those not experienced with these phage techniques, a great resource is the online accessible www.phagesdb.org website.
Representative Results
To demonstrate reproducibility of the improved enrichment technique for Arthrobacter phages, 30 different soil samples were used at different times and locations during the spring and summer of 2014. Of these 30 soil samples unique Arthrobacter phages were obtained from 22 of collected soil samples using this enrichment procedure. The standard enrichment procedure yielded unique phages from 3 of the same soil samples. The enrichment samples can have a very high phage titer needing initial dilution to isolate plaques or a relatively low phage titer requiring a larger volume of enriched Arthrobacter for successful detection of plaques. We use the plaque streak method to isolate pure phage populations after initial enrichment and plating on LB agar (Figure 2). Alternatively, a traditional standard plaque titer assay may be used to isolate a pure phage isolate though this method takes more time, reagents, and materials. As mentioned earlier, we do use the plaque titer assay to empirically determine phage titer numbers and the number of plaque forming units (PFUs) to form a webbing pattern on a plate. Figure 3 depicts a set of serial dilutions for phage Banana and demonstrates for one of the plates what a good web pattern should look like. Figure 4 depicts electron micrographic images of 20 Arthrobacter phages isolated using the enrichment technique. Of the 25 phages currently isolated, 23 of them are siphoviruses with rather long tails. Two of the isolated phages are myoviruses containing head diameters similar in length to that of their tails.
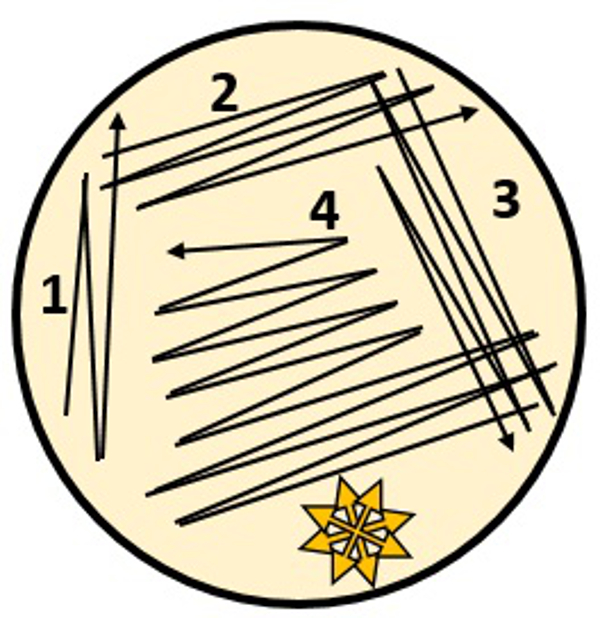
Figure 1. Streak plate technique schematic used for the isolation of individual phage plaques. The yellow star represents the area on the plate that the LB top agar plus bacteria solution should be carefully poured onto the plate after the phage plaque streaking has been completed.
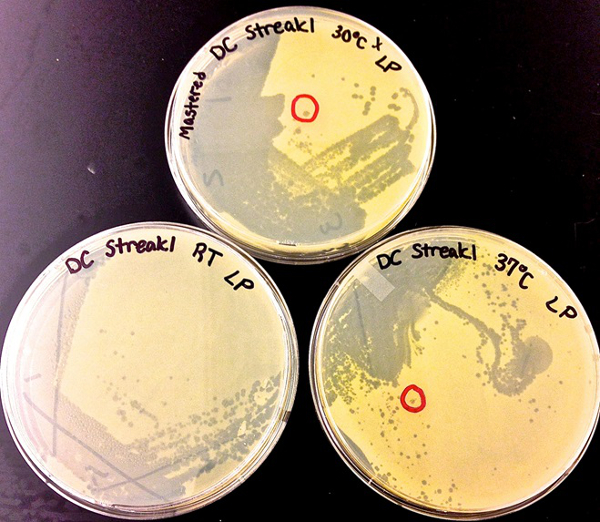
Figure 2. Examples of the streak plate technique used for the isolation of plaques for one phage (Dylan) at three different temperatures. Note that phage Dylan produces similar plaques at RT, 30 °C and 37 °C. The red circles on two of the plates surround clearly isolated plaques.
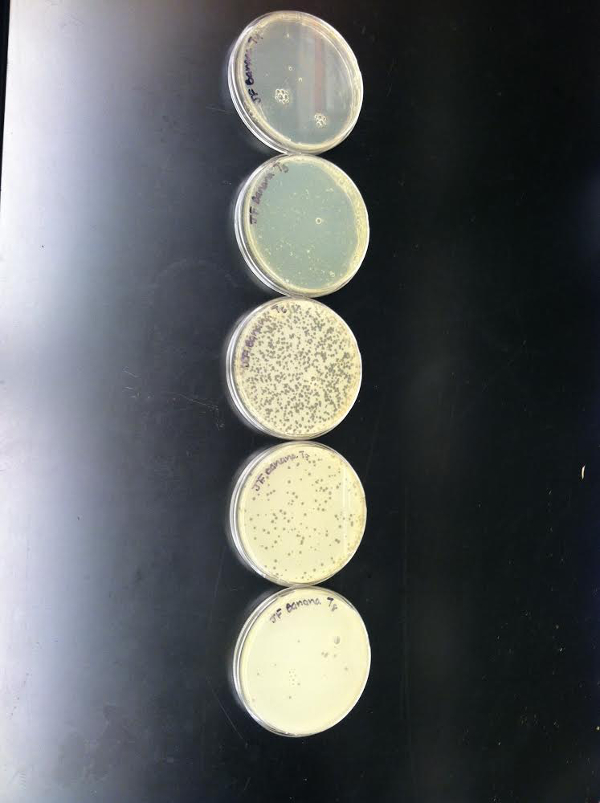
Figure 3. Plaque titer assay. Arthrobacter phage Banana was serial diluted in PB and plated on agar plates as described in sections 4.6 & 4.7 of the Protocol Text. Shown are serial dilutions T-4 to T-8 (from the top). Note that the T-5 dilution (from bottom to top) gave a web pattern with just a small amount of the bacterial lawn still remaining. Lower serial dilutions (T-1 to T-3) created clear plates with all of the bacteria lysed by phage particles demonstrated by the lack of any bacterial growth.
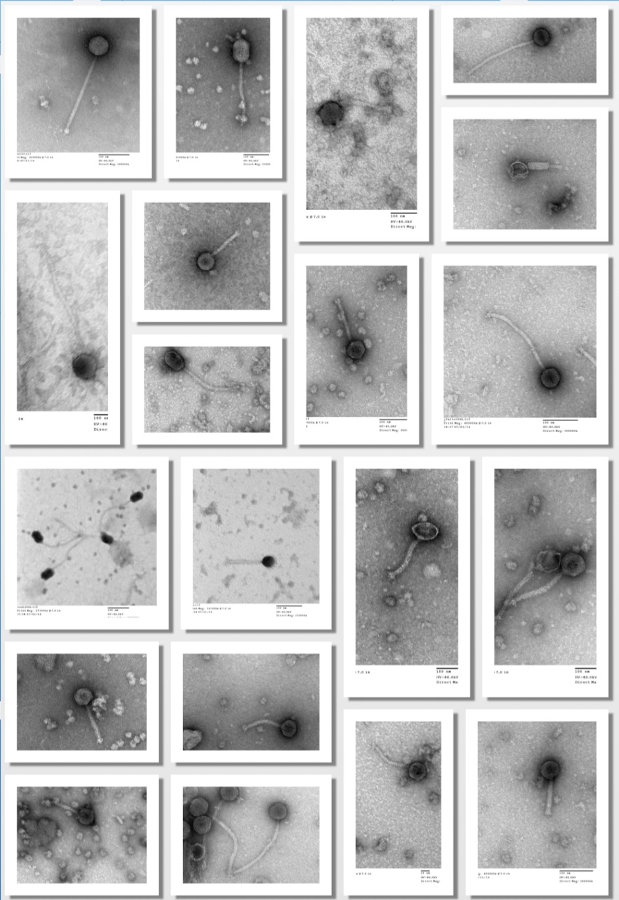
Figure 4. Electron micrographic images of 20 Arthrobacter phages isolated using the enrichment technique. The images were taken using a FEI Morgagni TEM. Please click here to view a larger version of this figure.
Discussion
Despite many previous attempts to isolate phages capable of infecting Arthrobacter hosts, we had little success using standard enrichment procedures. The generalized method of bacterial enrichment developed and adapted by van Twest and Kropinski10 to enrich phages from environmental samples remains the basis for the majority of enrichment procedures. Evidence from previous studies suggests that methods of direct plating have produced detectable plaques on strains of Arthrobacter albeit with very low success rates of isolation5. With the direct plating method, phages are extracted from soil samples in a phage buffer and filtered to remove contaminating bacteria. The filtered extract containing phage particles are added to the desired bacteria host to allow host cell infection and plated without additional rounds of bacteria growth and phage amplification.
Our enrichment technique is robust and optimized for the relatively easy isolation of Arthrobacter phages from Arthrobactersp. KY3901. With traditional enrichment procedures, phages, along with many different types of bacteria present in the soil samples, are extracted from soil in phage buffer and added to fresh growth media with desired bacterial cells used as the host for phage infection. With the enrichment method, all soil bacteria are removed from the soil extracted phage samples by filtration through a 0.22 µM filter. The sterile filtered lysate containing phage particles, but not soil bacteria, are diluted in 2x LB broth with the desired bacteria used as the phage propagation host. The main difference between our enrichment procedure and others is that there are NO contaminating soil bacteria competing for growth with the host bacteria during the enrichment incubation time period. The only host the soil extracted phage can infect is that of the desired host bacteria and therefore maximizes the growth potential of the host bacteria and amplification of the desired host-specific phages.
In the development of the procedure described here we attempted to account for selective bias inherent in any enrichment procedure by testing a range of CaCl2 concentrations11. What became apparent is that some phage isolates show physiological differences dependent on parameters like calcium ion concentration and temperature. For future Arthrobacter phage isolations we plan on testing key factors such as like pH, temperature, salinity, nutrients, divalent metal ions to isolate phages capable of reproducing in their hosts optimally under various environmental conditions.
We use freshly grown Arthrobacter cells in this experiment. We do not use cells that reach middle to late stationary phase of growth since we have had difficulty isolating phage or having decent titers with known phages at this growth period. Others have shown an inhibition of phage infection in stationary phase Achromobacter cells12 which may explain the lack of success of isolating phages using bacterial cells in middle to late stationary phase of growth. We have had success using cultures for these experiments with a cell density anywhere from an OD600 of 0.5 to 0.9. Grown cultures are stored at 4 °C and can be used for phage isolation up to seven days. However, the longer the Arthrobacter cells are stored at 4 °C the lower the titer and burst size of infectious phage particles. Similar results were found with using Escherichia coli for the infection of phageT413. Therefore using bacterial cells as soon as possible after they reach the desired growth maximizes chances of obtaining a novel phage from a soil sample.
Broadly speaking, the use of this enrichment technique with LB media should prove to be hardly exclusive to the isolation of Arthrobacter phages. While LB has been the industry standard for culturing E. coli strains and other members of the Enterobacteriaceae, it supports the growth of a wide variety of bacteria in aerobic conditions14. The nutrient rich composition of LB media can likely be used in this method to propagate phages in an incredibly diverse group of aerobic bacteria preventing technical challenges from hindering phage discovery.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
Funding for the development of this protocol was provided by the Southeastern Pennsylvania Consortium for Higher Education and the Cabrini College Science Department. Additional funding and support came from Arcadia University and Immaculata University. We especially thank Dr. Karen Snetselaar at St. Joseph University for kindly taking the electron microscopic images of our isolated phages. Additional support was provided by the Howard Hughes Medical Institute Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) program.
Materials
| LB Broth powder | Fisher | BP9722-2 | It's best to order these in bulk. |
| Granulated Agar | Fisher | BP1423-2 | It's best to order these in bulk. |
| 0.22 um syringe filters | Fisher | 09-719A | |
| .22 um buchner filters | Fisher | 430320 | More than 50 mL of liquid can be obtained by carefully swapping the receiving tube. |
| Eppendorf Tubes | Fisher | 05-408-129 | |
| 5 mL pipets individual | Fisher | 13-678-11D | |
| 50 mL conical tubes | Fisher | 76002844 | |
| 15 mL conical tubes | Fisher | 76002845 | |
| 10 mL pipets individual | Fisher | 13-676-10J | |
| 25 mL pipets individual | Fisher | 13-676-10K | |
| Whatman qualitative filter paper | Fisher | 1001-824 |
Referenzen
- Shapir, N., Mongodin, E. F., Sadowsky, M. J., Daugherty, S. C., Nelson, K. E., Wackett, L. P. Evolution of Catabolic Pathways: Genomic Insights into Microbial s-Triazine Metabolism. J Bacteriol. 189 (3), 674-682 (2007).
- Qingyan, L., Ying, L., Xikun, Z., Baoli, C. Isolation and Characterization of Atrazine Degrading Arthrobacter sp. AD26 and Use of this Strain in Bioremediation of Contaminated Soil. J Environ Sci. 20, 1226-1230 (2008).
- Wang, J., Zhu, L., Liu, A., Ma, T., Wang, Q., Xie, H., Wang, J., Jiang, T., Zhao, T. Isolation and characterization of an Arthrobacter sp. Strain HB-5 that Transforms Atrazine. Environ Geochem Hlth. 33, 259-266 (2011).
- Mages, I. S., Frodl, R., Bernard, K. A., Funke, G. Identities of Arthrobacter spp. And Arthrobacter-Like Bacteria Encountered in Human Clinical Specimens. J Clin Microbiol. 46 (9), 2980-2986 (2008).
- Brown, D. R., Holt, J. G., Pattee, P. A. Isolation and Characterization of Arthrobacter Bacteriophages and Their Application to Phage Typing of Soil Arthrobacters. Appl Environ Microbiol. 35 (1), 185-191 (1978).
- Petrovski, S., Seviour, R. J., Tillett, D. Prevention of Gordonia and Nocardia Stabilized Foam Formation by Using Bacteriophage GTE7. Appl Environ Microbiol. 77 (21), 7864-7867 (2011).
- Le Marrec, C., Moreau, S., Loury, S., Blanco, C., Trautwetter, a. Genetic Characterization of Site-specific Integration Functions of phi AAU2 infecting “Arthrobacter aureus” C70. J Bacteriology. 178 (7), 1996-2004 (1996).
- Casida, L. E., Liu, K. C. Arthrobacter globiformis and Its Bacteriophage in Soil. J Appl Microbiol. 28 (6), 951-959 (1974).
- Einck, K. H., Pattee, P. A., Holt, J. G., Hagedorn, C., Miller, J. A., Berryhill, D. L. Isolation and Characterization of a Bacteriophage of Arthrobacter globiformis. J Virol. 12 (5), 1031-1033 (1973).
- Twest, R., Kropinski, A. M. Bacteriophage Enrichment from Soil and Water. Methods Mol Bio. 501, 15-21 (2009).
- Fullner, K. J., Hatfull, G. F. Mycobacteriophage L5 Infection of Mycobacterium bovis BCG: Implications for Phage Genetics in the Slow-growing Mycobacteria. Mol Microbiol. 26 (4), 755-766 (1997).
- Robb, S. M., Woods, D. R., Robb, F. T. Phage Growth Characteristics on Stationary Phase Achromobacter cells. J Gen Virol. 41 (2), 265-272 (1978).
- Bolger-Munro, M., Cheung, K., Fang, A., Wang, L. T4 Bacteriophage Average Burst Size Varies with Escherichia coli B23 Cell Culture Age. Journal of Experimental Microbiology and Immunology. 17, 115-119 (2013).

