Quantitative Analysis of Protein Expression to Study Lineage Specification in Mouse Preimplantation Embryos
Summary
This protocol presents a method to perform quantitative, single-cell in situ analysis of protein expression to study lineage specification in mouse preimplantation embryos. The procedures necessary for collection of blastocysts, whole-mount immunofluorescent detection of proteins, imaging of samples on a confocal microscope, and nuclear segmentation and image analysis are described.
Abstract
This protocol presents a method to perform quantitative, single-cell in situ analyses of protein expression to study lineage specification in mouse preimplantation embryos. The procedures necessary for embryo collection, immunofluorescence, imaging on a confocal microscope, and image segmentation and analysis are described. This method allows quantitation of the expression of multiple nuclear markers and the spatial (XYZ) coordinates of all cells in the embryo. It takes advantage of MINS, an image segmentation software tool specifically developed for the analysis of confocal images of preimplantation embryos and embryonic stem cell (ESC) colonies. MINS carries out unsupervised nuclear segmentation across the X, Y and Z dimensions, and produces information on cell position in three-dimensional space, as well as nuclear fluorescence levels for all channels with minimal user input. While this protocol has been optimized for the analysis of images of preimplantation stage mouse embryos, it can easily be adapted to the analysis of any other samples exhibiting a good signal-to-noise ratio and where high nuclear density poses a hurdle to image segmentation (e.g., expression analysis of embryonic stem cell (ESC) colonies, differentiating cells in culture, embryos of other species or stages, etc.).
Introduction
The mouse preimplantation embryo is a paradigm to study the emergence and maintenance of pluripotency in vivo, as well as a model for the study of cell fate specification and de novo epithelialization in mammals. The preimplantation stages of mammalian development are dedicated to the establishment of the three cell lineages that make up the blastocyst, namely the pluripotent epiblast – which gives rise to most somatic tissues and germ cells – and two extraembryonic lineages, the trophectoderm (TE) and the primitive endoderm (PrE) (Figure 1A) 1,2. This protocol describes the procedures to (1) harvest and fix preimplantation stage mouse embryos, (2) perform immunofluorescence to label proteins of interest, (3) carry out whole-mount imaging using a confocal microscope with z-sectioning capabilities and (4) perform nuclear segmentation of confocal images and subsequent quantitative image analyses. This pipeline allows the unbiased measurement of protein levels for the assignment of cell identities to characterize subpopulations of cells in situ. This protocol can be carried out in as little as 3 – 4 days for a single litter (generally up to 10 mouse embryos), from embryo collection to the data analysis (Figure 1B). The simultaneous analysis of several litters would increase the imaging and data analysis time burden, thus extending the overall length of the protocol.
The preimplantation stage mouse embryo is an experimentally tractable system which, given its small size and stereotypical morphology 3, is well suited for in toto imaging of cellular processes with single-cell resolution. To carry out an unbiased, systems-level analysis of a statistically relevant number of embryos, an automated, quantitative analysis pipeline is desirable. However, due to the high nuclear density of the inner cell mass (ICM) of the blastocyst (Figure 1A, 2D), conventional image segmentation platforms fail to provide sufficient accuracy to establish an automated or semi-automated workflow. On the other hand, manual segmentation, while accurate, does not allow the processing of large cohorts of cells and embryos, nor is it suitable for a reproducible, unbiased determination of cell identities – which is especially critical when studying developmental stages where patterns of marker expression have not fully resolved (e.g., do not exhibit a binary distribution across a population). We have recently developed and validated an image segmentation method tailored for mouse preimplantation stage embryos and for mouse embryonic stem cells (ESCs) that achieves high accuracy, while requiring minimal user input 4-8.
The analysis pipeline presented here revolves around the MATLAB-based image segmentation tool Modular Interactive Nuclear Segmentation (MINS) 4. MINS performs unsupervised nuclear segmentation on large batches of confocal Z-stacks after the user has established a minimal number of image properties, using a graphical user interface (GUI) (Table 1) 4. This pipeline has proven efficient for the generation of high throughput data on protein expression and cell localization in both wild type, experimentally treated and genetically modified embryos and ESCs 5-7. In the present protocol, we describe the application of MINS to the segmentation of preimplantation-stage embryo images. For examples of MINS performance on ESCs please refer to 4,7. The automated nuclear segmentation step significantly reduces the time burden of the cell identification process, whereas the spatial and fluorescence intensity measurements allow an unbiased determination of cell identities and the generation of three-dimensional maps of gene expression domains and cell position in the embryo (Figure 1C). Moreover, the scalability of this workflow makes it applicable to the analysis of individual litters through large cohorts of experimentally treated embryos, or embryos of different genetic backgrounds 5,6. MINS is freely available at http://katlab-tools.org (the software requires a MATLAB license).
No approach developed to date allows the generation of such in-depth data on protein expression and cell localization in mouse preimplantation embryos. All attempts thus far at quantifying these types of data have been restricted to the manual determination and quantitation of cell numbers for different populations in the embryo (either entirely manually, or software-assisted) 9-19. This approach (incorporating MINS software) has been tailored for and tested on mouse preimplantation embryos and ESCs; nevertheless its performance on other systems with high nuclear density, although yet untested, is expected to be equivalent.
Protocol
Ethics statement: All animal work, including husbandry, breeding and sacrifice was approved by Memorial Sloan Kettering Cancer Center's Institutional Animal Care and Use Committee (IACUC), protocol #03-12-017.
1. Embryo Collection
Note: All animal work must have been approved by institutional and local authorities and conform to local and institutional rules.
- Mate a virgin female mouse with a fertile stud male of the desired genotypes.
Note: If setting up natural matings, selecting females in the estrus phase of the estral cycle increases the chances of copulation on the desired date. If inducing superovulation, please refer to the protocols described in 20. - Check the presence of a vaginal plug in the morning using a blunt probe. Ideally, do this before noon (12:00 pm), as copulation plugs are lost throughout the day. Consider noon of detection of the vaginal plug embryonic day (E)0.5.
- On the desired day and time of embryonic development, warm up M2 or flushing holding medium (FHM) to RT or, preferably, 37 ºC. Note: Either M2 or FHM can be used for flushing and handling embryos. When not in use, store these media at 4 ºC. Estimate the use of ~2 ml of medium for each uterus.
- Thaw frozen 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS) or prepare some fresh solution. Estimate 500 µl of 4% PFA solution per well of a 4-well plate. Fix one litter on each well of a 4-well plate. Note: Alternatively, a 96-well plate can be used to fix and store embryos. If using a 96-well plate, use 100 µl of PFA solution per well.
- Pull a glass Pasteur pipette using an open flame to draw a capillary end at the tip.
- Sacrifice female by cervical dislocation or CO2 inhalation, or as required by local and institutional regulations. It is often desirable to confirm euthanasia of the animal by cervical dislocation.
- Spray belly of the animal with 70% ethanol to minimize hair shedding and perform an abdominal incision, first through the skin and subsequently through the body wall (peritoneum) to expose the viscera.
Note: The following steps describe the procedure to collect blastocysts from the uterus of a pregnant female mouse at any stage between E3.25 and E4.5 (Figure 1A). For protocols on collection of preimplantation embryos earlier than E3.25, refer to 20. - Locate the uterus and remove from the animal. Holding the cervical end of the uterus with a pair of forceps, cut through the cervix and pull up gently to stretch both uterine horns.
- Trim fat from the uterus, taking care not to pierce the uterine wall. This can be done easily at the time of removal, while still attached to the body of the animal, as the uterus can be stretched out. Alternatively, it can be done later, under a dissection microscope, in RT PBS. For a more detailed protocol on dissection of the uterus, see Protocols 5 & 8 20
- Cut above the oviducts, below the ovaries, to release the entire uterus, place on a petri dish and cover with PBS.
Note: This step also allows the washing of excess blood or other debris from the uterus, which facilitates subsequent blastocyst harvesting. - Separate both uterine horns by cutting through the proximal end, on both sides of the cervix and discard the cervix. Separate each oviduct from the uterus by cutting below the isthmus that connects them (Figure 1A). Transfer both horns to a drop of manipulation medium (either M2 or FHM) for embryo collection and manipulation.
- Under a dissection microscope, flush blastocysts out of the uterus and into the medium by forcing 0.5 – 1 ml of M2 or FHM through each uterine horn from the cervical opening. Use a 1 ml syringe with a hypodermal needle. Note: The needle can be blunted using a filing stone to avoid tearing the uteri, although this is not critical. A detailed diagram for uterine flushing can be found in21.
- Using the dissection microscope, locate the blastocysts, which will be scattered throughout the drop of medium. Allow up to 1 – 2 min for blastocysts to sink to the bottom of the dish after flushing. Using the mouth-controlled, glass Pasteur pipette with a capillary end, collect all blastocysts and transfer to a fresh drop of media.
- Rinse blastocysts by moving them through 2 – 3 drops of fresh M2 or FHM. Move embryos in groups of 5 – 10 blastocysts at a time.
Note: Moving larger groups at any one time will speed up the process, but it will also increase the chances of accidentally losing many embryos. If untrained in the use of mouth-controlled pipettes, practicing embryo manipulation and transfer should be considered before beginning the experiment. - Remove the Zona Pellucida (ZP) from unhatched blastocysts (generally <E4.0) by briefly washing them in acidic Tyrode's solution. As soon as the ZP is no longer visible, transfer blastocysts back to manipulation media. Only keep embryos in acid Tyrode's for as long as strictly necessary, in order to prevent damage to the cells.
- Take care when handling denuded blastocysts, as they adhere very easily to glass and plastic surfaces. Manipulation medium contain 4 mg/ml of bovine serum albumin (BSA) to prevent attachment, but acid Tyrode's or PBS do not, therefore, to reduce the risk of damage or loss of the embryos do not pick up more than 2 – 5 denuded blastocysts at a time when manipulating them in BSA-free or serum-free PBS or acid Tyrode's.
Note: Alternatively, coat the glass pipette with 1% BSA before use to reduce adherence of the embryos to the glass surface.
- Take care when handling denuded blastocysts, as they adhere very easily to glass and plastic surfaces. Manipulation medium contain 4 mg/ml of bovine serum albumin (BSA) to prevent attachment, but acid Tyrode's or PBS do not, therefore, to reduce the risk of damage or loss of the embryos do not pick up more than 2 – 5 denuded blastocysts at a time when manipulating them in BSA-free or serum-free PBS or acid Tyrode's.
- Coat the bottom of each well of a 4-well tissue culture dish with a thin layer of 1% agar, 0.9% NaCl to prevent embryos from adhering to the plastic. Alternatively, add 4 mg/ml of BSA to the PBS (PBS-BSA).
- Fill one well of the coated 4-well tissue culture dish with 500 µl of RT PBS (or PBS-BSA) and another well with 500 µl of 4% PFA in PBS.
- Rinse blastocysts in RT PBS and fix in 4% PFA in PBS for 10 min at RT. Transfer to PBS (or PBS-BSA) after fixation.
- If embryos are not going to be processed immediately, cover the PBS containing the embryos with a layer of mineral oil to prevent evaporation (leading to desiccation of the sample) and store the plate at 4 ºC.
Note: Different fixation methods and times can be used depending on the experiment, the epitopes of interest and the antibodies to be used. Whichever the method of choice, be consistent throughout equivalent experiments to facilitate subsequent comparison of the staining results.
Note: Pause point: Embryos can be stored at 4 ºC in PBS for up to several weeks before proceeding to the next step. Ensure PBS is sterile and/or add 100 U/ml of Penicillin + 100 µg/ml Streptomycin (Pen-Strep) to prevent bacterial contamination (especially when using PBS-BSA) if anticipating prolonged storage.
2. Immunofluorescence
- Perform immunofluorescence using any protocol that has previously been tested and proven to be robust for whole-mount immunofluorescence. Note: We recommend those described in 22 and 11. In the present article the former will be described. Whichever the method of choice, be consistent throughout equivalent experiments to facilitate comparison of the staining results.
- Use a flexible, polyvinyl, clear, U-bottom 96-well plate to carry out the sequential steps of the immunofluorescence protocol. Fill each well with 50 – 100 µl of solution and move embryos from one solution to another using a L-shaped mouth pipette. Note This approach minimizes the use of reagents and facilitates tracking steps as well as the parallel staining of several experimental groups.
- Optional : Coat the bottom of the wells with a thin layer of 1% agar, 0.9% NaCl to prevent the adhesion of embryos to the plastic. Do not add BSA to immunofluorescence solutions.
- Prepare PBX (0.1% Triton X-100 in PBS). If anticipating the performance of several immunofluorescence experiments, prepare 50 ml of PBX and store at 4 ºC between experiments.
- Prepare permeabilization solution (0.5% Triton X-100, 100mM glycine in PBS). Make 1 ml (or whichever amount necessary) of fresh permeabilization solution for each experiment.
- Prepare blocking solution (2% horse serum (HS) or 20% fetal bovine serum (FBS) with Pen-Strep in PBS). Prepare 10 ml and store at 4 ºC between experiments.
- No differences have been observed between either type of serum, however, be consistent in the choice of blocking solution used for equivalent experiments.
- Rinse fixed embryos for 5 min at RT in PBX (~100 µl).
- Permeabilize embryos for 5 min at RT in permeabilization solution (~100 µl).
- Rinse embryos for 5 min at RT in PBX (~100 µl).
- Block embryos for 30 min to 1 hr at RT in ~100 µl of blocking solution. Cover the solution with a layer of mineral oil to prevent evaporation or keep in a humidified chamber.
Note: Embryos can be blocked for up to O/N periods at 4 ºC without noticeable improvement or detriment when compared to 30 min blocking steps. - Incubate embryos O/N at 4 ºC in primary antibody/antibodies diluted in blocking solution (~100 µl). Cover the solution with a layer of mineral oil to prevent evaporation or keep in a humidified chamber.
- Determine optimal dilution for each primary antibody beforehand. See discussion and Materials section for tested dilutions of a number of antibodies.
- After O/N incubation, rinse embryos 3x for 5 min each at RT in PBX (~100 µl).
- Block embryos for 30 min to 1 hr at RT in ~100 µl of blocking solution. Cover the solution with a layer of mineral oil to prevent evaporation or keep in a humidified chamber.
- Incubate embryos for 1 to 2 hr at 4 ºC in secondary antibody/antibodies diluted at 4 µg/ml in ~100 µl of blocking solution. Cover the solution with a layer of mineral oil to prevent evaporation or keep in a humidified chamber.
- Rinse embryos 2x for 5 min each at RT in PBX.
- Move embryos to preferred DNA stain. If using Hoechst 33342, dilute to 5 µg/ml in PBS. Cover the solution with a layer of mineral oil to prevent evaporation or keep in a humidified chamber.
Note: Pause point: If embryos are not being imaged immediately, they can be kept for up to several weeks in nuclear staining solution. In this case, ensure PBS is sterile and/or add Pen-Strep or Sodium Azide to prevent bacterial growth and contamination.
3. Confocal Imaging
Note: Individual confocal microscope configurations will require specific acquisition parameters to be adjusted for the system and software in place. However, the following section provides a set of general rules to follow that should be applicable to any given installation.
- Using a fine mouth pipette, make microdrops of PBS or nuclear stain solution on the glass surface of a 35mm glass-bottom dish and cover with mineral oil. As an alternative, use a regular coverslip with silicone separators to avoid damaging the embryos and cover with a glass microscope slide. Note: When using a regular coverslip, embryos cannot be subsequently re-positioned or recovered for genotyping, therefore we strongly recommend the use of glass-bottom dishes.
- Place embryos in the microdrops and arrange in a consistent manner, preferably with the ICM-cavity axis parallel to the glass surface. Set up dish on the microscope holder. Note: Images obtained on laser scanning confocal microscopes suffer from a Z axis-associated loss of fluorescence intensity. Therefore, consistency in the arrangement is important for comparing intensities between equivalent regions on different embryos.
- Set up reference parameters using wild type embryos that have not been subject to any sort of treatment that may alter gene expression.
- Acquire images using a high magnification objective (i.e., 40X or higher) in order to obtain data that will facilitate accurate segmentation using the MINS tool 4.
Note: Using digital zoom on a 20X objective will not yield images of adequate resolution for segmentation using MINS. One should also note the working distance (WD) of the objective, as it should be sufficient to allow acquisition of a Z-stack that spans an entire blastocyst, thus a long WD (~130 – 150 µm) objective is usually necessary. The images shown in Figures 2 – 4 were acquired using a 40X/NA 1.30 oil immersion objective with a working distance of 210 µm. - Use the lowest laser output that provides a strong signal-to-noise ratio without bleaching the fluorophores.
- Adjust the gain and offset to obtain the widest dynamic range without overexposing the sample. Exposing the images to cover most of, or span the entire, gray scale range will facilitate the detection of small differences in intensity between images.
- Use a small (1 µm) Z-step, since Z axis resolution is critical for accurate segmentation with MINS.
Note: XY dimensions do not affect the nuclear segmentation process, only the image file size. Generally, 512 x 512 pixels is a sufficient XY resolution for publication-quality images without compromising hard-drive space. - Critical step: Keep imaging parameters consistent within and across imaging sessions of the same experiment so as to be able to compare samples.
- Acquire images using a high magnification objective (i.e., 40X or higher) in order to obtain data that will facilitate accurate segmentation using the MINS tool 4.
- Imaging batches of embryos:
- Image coherent groups (litters, experiments) in a single session whenever possible.
- Keep parameters consistent within and across imaging sessions for replicates of the same experiment.
- Image embryos throughout (whole Z axis) to capture every cell.
Note: If desired, embryo genotyping may be done after imaging as described in 23. The need for nested PCR has to be determined empirically and will depend on the locus to be analyzed and the primers used.
- Make sure to be able to match embryos to images retrospectively.
4. Image Analysis and Data Pre-processing
- Download and install MINS 4 from http://katlab-tools.org (MATLAB license required).
- Perform image segmentation:
- Follow the graphic user interface (GUI) of MINS to load a confocal image or stack from the hard drive, detect the nuclei and segment the image. Load the entire Z-stack of raw data generated by the microscope (.lsm, .lif, .oif, etc.). See Table 2 for details on each step and instructions found on http://katlab-tools.org and4. Once completed, view the outcome of each step by clicking the corresponding 'View' button. A yellow tag above the button indicates operation in process, a green tag indicates operation completed.
Note: MINS currently does not support .czi files, .tiff sequences or multi-position files – each Z-stack needs to be an independent file. - After satisfactory segmentation, save the pipeline created so it can be used directly for other embryos or litters.
- Follow the graphic user interface (GUI) of MINS to load a confocal image or stack from the hard drive, detect the nuclei and segment the image. Load the entire Z-stack of raw data generated by the microscope (.lsm, .lif, .oif, etc.). See Table 2 for details on each step and instructions found on http://katlab-tools.org and4. Once completed, view the outcome of each step by clicking the corresponding 'View' button. A yellow tag above the button indicates operation in process, a green tag indicates operation completed.
- Batch-Mode segmentation:
Note: Single files can be segmented individually. However, given that embryos within a litter or experiment are generally of comparable stage, MINS can apply the segmentation pipeline created for a single image to a group of them without supervision.- From the Batch-Mode Run menu, click 'Add files' and load all files to be processed at once. Note: Files from different sources can be added to the segmentation queue.
- Click 'Start Batch-Mode Run' and allow time for the software to process the files. Note: Segmentation output will be saved in the same directory where the original files where located.
- Segmentation assessment and manual correction
Note: MINS segments images with very high accuracy (>85%), however, the quality of the staining and imaging, as well as the stage of the embryo, may affect MINS performance. Generally, the higher the nuclear density within the embryo, the more likely segmentation errors will take place (Figures 2D, 3). Thus, segmentation accuracy should be evaluated for each sample and manually corrected if needed. Two types of errors typically occur: over-segmentation and under-segmentation.- Resolving over-segmentation:
- Identify false positives – apoptotic vesicles (Figure 3A a, b, d, asterisks) or other elements that are not intact nuclei, but which may have been identified as such by MINS.
Note: Generally such false positives have a smaller size than real nuclei and can, in the majority of cases, be easily identified and sorted on the spreadsheet. However, visual examination is strongly recommended, as oftentimes nuclei may be only partially segmented, resulting in a smaller, but valid segmented volume (Figure 3A b, arrowhead). - Delete the corresponding records for false positives from the *statistics.csv file. Preserve the original *statistics.csv file for future reference and edit only a copy of it.
- If a nucleus has been over-segmented and presented as 2 or more nuclei (Figure 3A c, arrow and arrowheads), either merge the records by averaging their intensity level or, if similar, keep one of the records for that cell and discard the rest.
- Identify false positives – apoptotic vesicles (Figure 3A a, b, d, asterisks) or other elements that are not intact nuclei, but which may have been identified as such by MINS.
- Resolving under-segmentation:
- Identify events where MINS has failed to detect the border between two or more nuclei and consequently segmented them as a single nucleus (Figure 3A d, arrow and 3B). Note: This poses a problem for an accurate cell count, but more importantly, for quantifying protein levels, as the levels measured will be the average of the cells that have been erroneously merged, and which may belong to different lineages.
- Identify events where MINS has failed to detect a nucleus altogether.
- In these instances (4.4.2.1 and 4.4.2.2), measure the average gray levels for each channel in the under- or un-segmented cell(s) using an alternative tool, such as ImageJ (http://imagej.nih.gov/ij/), and replace the erroneous record with the correct levels. Preserve the original *statistics.csv file for future reference and edit only a copy of it. To do this using ImageJ, follow these steps:
- Open the relevant multi-channel stack on ImageJ and import the corresponding *overlaid.tiff file generated by MINS as a virtual stack ('File' > 'Import' > 'TIFF virtual stack…').
- Find a medial section of the under- or un-segmented nucleus.
- Using the freehand selection tool outline the perimeter of the under- or un-segmented nucleus on the DNA stain channel.
- Press Crl+M (Cmd+M on a Macintosh) or go to the Analyze menu and select 'Measure'. This action will record the Mean gray value for the area outlined and will display it on a new window. Go to 'Analyze' > 'Set measurements…' and make sure 'Mean gray value' is selected. Select any other parameters to be measured.
- Using the same area outlined in step 4.4.2.3.3, repeat the measurement for each of the fluorescence channels of interest. The results will be appended to the previous one on the measurements 'Results' window.
- Use the measurements obtained to replace the erroneous records in the *statistics.csv file. If the nucleus had not been segmented, insert a new row, assign it a new Cell ID and introduce the values obtained in ImageJ under the corresponding column for each channel. This cell will not have spatial coordinates (X, Y and Z values). If the nucleus had been undersegmented, duplicate its row, assign different Cell IDs to each new cell and introduce the values obtained in ImageJ under the corresponding column. In this case, both cells will share spatial coordinates.
Note: If spatial distribution is to be analyzed, we recommend excluding the XYZ coordinates of these cells, as they will overlap. To this end, when creating new records, assign Cell IDs to them that are easily distinguished from the original ones (for instance, non-integer numbers).
- Score dividing cells. Note: MINS detects mitotic as well as interphase nuclei, but cannot distinguish between them.
- If mitotic nuclei are to be considered separately in the analysis, manually score these and add this information to the data file. To record mitotic cells, add a new variable (column) to the *statistics.csv data file and assign different values to mitotic and interphase nuclei.
- Resolving over-segmentation:
- Data pre-processing.
Note: Once the spreadsheets have been corrected for over- and under-segmentation they are ready to be analyzed. The analysis process and data presentation will depend on the specific experiment. However, below are some general data transformations we recommend carrying out prior to analysis:- Transform the fluorescence intensity values into their logarithm or square root to reduce the scatter of the data. This also helps visualization, as the raw data tend to concentrate near the axes of the plot (see Figure 4B, C).
- Correct the Z-associated attenuation of fluorescence:
Note: Fluorescence intensity in laser scanning confocal microscopy images decreases the deeper on the Z axis a slice is located (Figure 4E, F).- If one of the epitopes detected in the samples is a ubiquitous and homogeneously expressed housekeeping nuclear marker, normalize the fluorescence intensities of the other epitopes by dividing their values in each cell by the corresponding value of the housekeeping marker.
- In the absence of such a reference marker, compensate Z-associated loss of fluorescence in the following manner:
- Plot the values of each marker (Y axis of the graph) over the Z position (X axis of the graph) to visualize the decrease in intensity over Z – plot together the values for all control, wild type embryos (Figure 4F).
- Fit a linear model (regression curve) to the plot obtained in the previous step (intensity values over Z) for each marker (Figure 4F).
- Correct all values for each marker using the following formula:
log(original value) + Z * slope of the model (Figure 4D, G).
Note: The specific steps to perform this correction will depend on the analysis software used (R, MATLAB, etc.). If using R, run the following formula to fit a linear model to the data:
>lm(log(channel)~Z, data = dataframe)
where, channel is the fluorescence channel to be corrected, Z is the Z coordinate and dataframe is the table containing all the values for the desired embryo(s) (the *statistics.csv file generated by MINS or a modification of it).
Note: The output of the above formula will have the following format:
(Intercept) Z
4.76373 -0.01947
where, the value Z is the slope of the regression line for that dataset, which absolute value should be used to modify the original value with the formula given in 4.5.2.2.3 (see Figure 4G for an example).
Representative Results
To facilitate data interpretation and presentation, care should be taken not to damage the embryos during collection and manipulation, so that all cells and their relative position can be analyzed. Figure 2A – D shows examples of intact blastocysts at different stages with an expanded cavity. Should damage occur, extra care should be taken when analyzing and interpreting results.
The quality and reliability of the data generated using this protocol is dependent on the quality of the fixation and on the signal-to-noise ratio of the antibodies used to detect the proteins of interest. Always use fresh fixative and test new antibodies, with appropriate positive and negative controls, before beginning a set of experiments. Figure 2A shows examples of good antibodies for a number of nuclear proteins. Figure 2B shows an example of a good antibody for a cytoplasmic protein (DAB2). Figure 2C shows an example of a bad staining, with low signal-to-noise ratio, where the sample was fixed for only 10 min. In this case, the antibody used for GATA4 (Santa Cruz, sc-1237) requires O/N fixation to provide a strong signal (see 24 for instances of embryos fixed O/N and stained with this antibody). Increasing the signal level during post-processing reveals a very noisy image. By contrast the anti-GATA4 used for Figure 2A (sc-9053) provides high signal-to-noise ratio after only 10-min fixation. Details for these and other robust antibodies are provided in the Materials section.
The limiting factors for a good MINS segmentation are a) the magnification of the objective used for imaging, b) the Z-resolution and c) the quality of the nuclear staining. Figure 2D shows examples of embryos imaged with an oil immersion 40X objective with a NA of 1.30 and a 0.21 mm WD. Middle panels show magnifications of the ICMs where individual nuclei and the border between them can be distinguished. If not using DNA staining (i.e., Hoechst, DAPI, TO-PRO3, YO-YO1, etc.) fluorescence values for quantification, acquisition parameters can be individually adjusted to obtain the best signal. Bottom panels show how the more advanced the embryo, the higher the nuclear density and thus the higher the chance of segmentation errors (arrowhead and arrow). Figure 3 shows examples of errors that MINS can commit, such as detecting apoptotic nuclei as live cells (asterisks in panels in Figure 3Aa, Ab, Ad), over-segmentation (arrow and arrowheads in Figure 3Ac) or under-segmentation (arrow in Figure 3Ad). Figure 3B shows a sequence of Z-slices of an under-segmentation event, where two cells have been identified as one. Note how MINS segmentation takes the Z-axis into account to segment a volume that comprises all slices where a given cell is present.
Finally, the specifics of the data analysis will depend on the question under study, therefore, guidelines for the analysis process cannot be given here. However, we have found certain initial data transformations to be useful. Figure 4B – D shows plots of the fluorescence values for the markers NANOG and GATA6 in the ICM of the embryo shown in Figure 4A. Figure 4B shows the raw fluorescence values obtained from MINS (after manual correction of under- and over-segmentation). Figure 4C shows the same data after a logarithmic transformation, which separates the values from the plot axes (a square root transformation is also possible and yields a similar plot). Figure 4D shows the same data after automatically correcting the values for the Z-associated attenuation in fluorescence, as explained in step 4.5.2.2 of the protocol. Examples of this Z-associated fluorescence decay are given in the image in Figure 4E and the plots in 4F. Figure 4G shows the effect of the data transformation explained in 4.5.2.2. Colors representing either epiblast (red, NANOG+, GATA6-) or PrE (blue, NANOG-, GATA6+) identity have been added to the plot as a function of the NANOG and GATA6 values. In this particular example, cells where the ratio of log[GATA6]/log[NANOG] >1.25 were considered PrE, cells where the ratio of log[GATA6]/log[NANOG] < 1 were considered EPI, and cells with an intermediate ratio were considered uncommitted (none in this case). All data transformations and plots have been done in Rstudio, however, the choice of software is not critical and will depend on the end user.
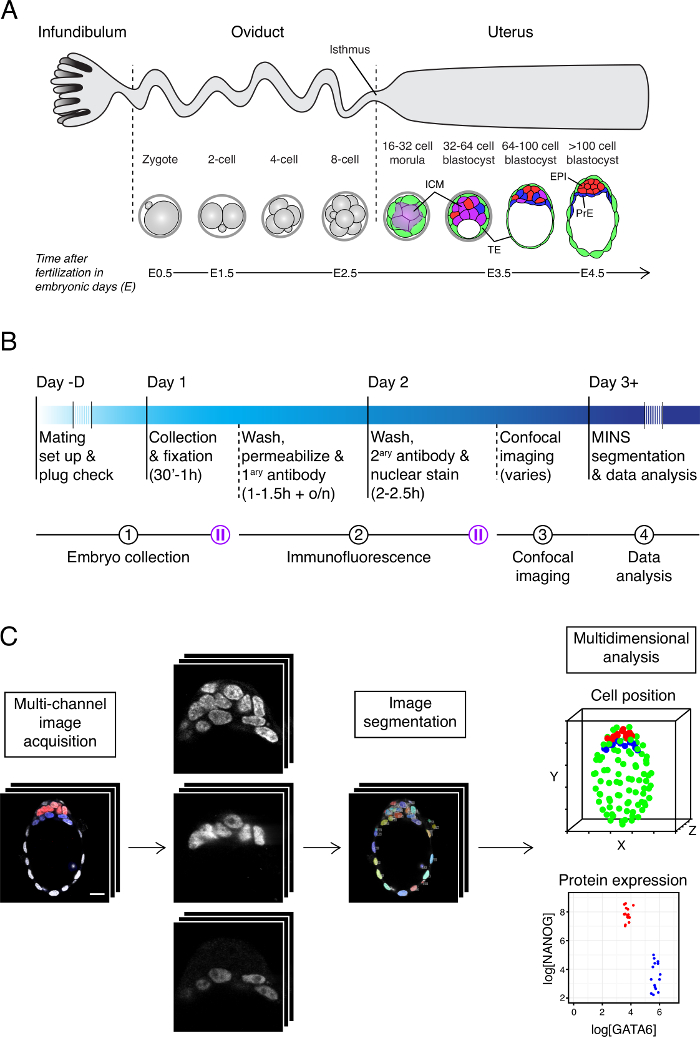
Figure 1. Embryo Location, Timeline and Analysis Pipeline. (A) Schematic of one (of two) mouse uterine horn and the stages of preimplantation development, aligned with the region of the horn where they should be found. (B) Experimental timeline with timing for each step. Steps of the protocol and pause points (blue) are indicated. (C) Summary of image acquisition and analysis pipeline using MINS. Please click here to view a larger version of this figure.
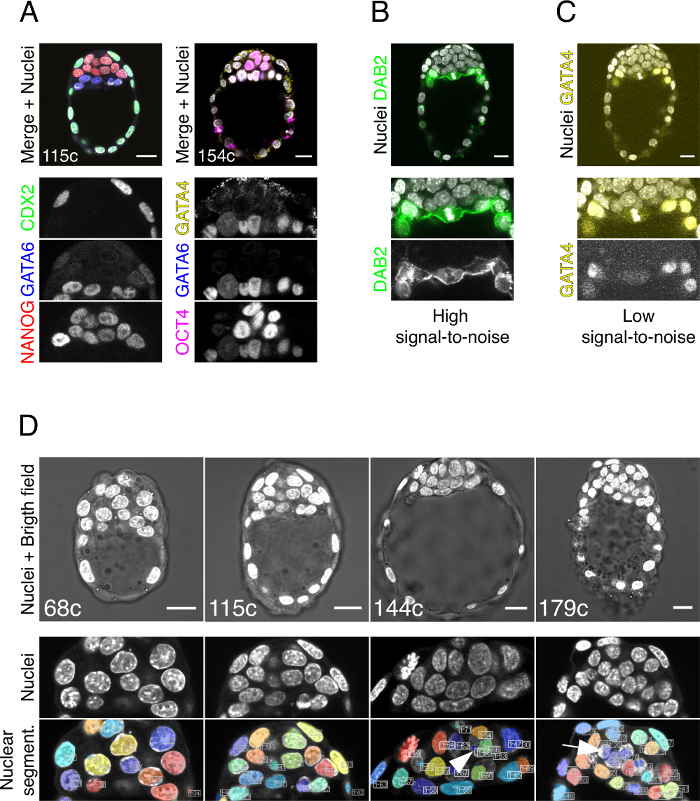
Figure 2. Examples of Immunofluorescence and Nuclear Density. (A) Examples of good immunofluorescence results with tested antibodies for nuclear transcription factors. Anti-CDX2 was detected with an AlexaFluor 488 donkey anti-mouse secondary antibody, anti-GATA6 with an AlexaFluor 568 donkey anti-goat, anti-NANOG with an AlexaFluor 647 donkey anti-rabbit, anti-GATA4 (Santa Cruz, sc-9053) with an AlexaFluor 488 donkey anti-rabbit and anti-OCT4 with an AlexaFluor 647 donkey anti-mouse. All primary antibodies are listed in the Materials Table. Cytoplasmic GATA4 nuclear staining is non-specific, and appears also in Gata4 null embryos (not shown). (B) Example of good immunofluorescence for a cytoplasmic protein, DAB2. It has been detected using an AlexaFluor 488 donkey anti-mouse. (C) Example of bad immunofluorescence, with a low signal-to-noise ratio for a nuclear factor. GATA4 has been detected with an anti-GATA4 raised in goat (Santa Cruz, sc-1237; whereas sc-9053 (rabbit anti-GATA4) was used in (A)) and an AlexaFluor 568 donkey anti-goat. (D) Examples of intact embryos with good nuclear staining showing how nuclear density increases with embryo age and how this increases errors in MINS segmentation (arrowhead – oversegmentation – and arrow – undersegmentation). Scale = 20 µm. Please click here to view a larger version of this figure.
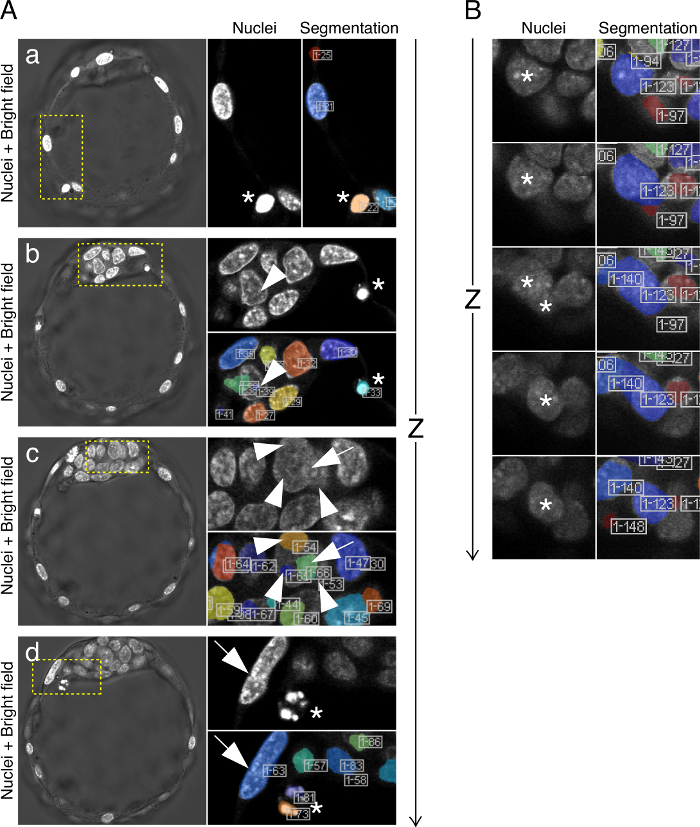
Figure 3. Examples of MINS Errors. (A) Sequence of individual Z-sections from one embryo showing examples of MINS segmentation errors. Apoptotic cells which are nevertheless identified as nuclei are marked with an asterisk (*) in panels a, b and d. In panel b, ID #39 (arrowhead), albeit very small, does identify the corresponding nucleus and can thus be left uncorrected. In panel c, ID #66 (arrow) spans two different nuclei, which have in turn been identified separately (arrowheads, IDs #65 #54 and #53). In panel d, two cells (ID #63) have been identified as a single one (see arrow marking the thin border) and this record should therefore be duplicated for cell counting purposes. (B) MINS detects cells along the Z-axis. The images show a sequence of Z-slices of two cells that have been erroneously scored as a single one (#123). Note how the blue area delimiting the nuclei is continuous along Z. Please click here to view a larger version of this figure.
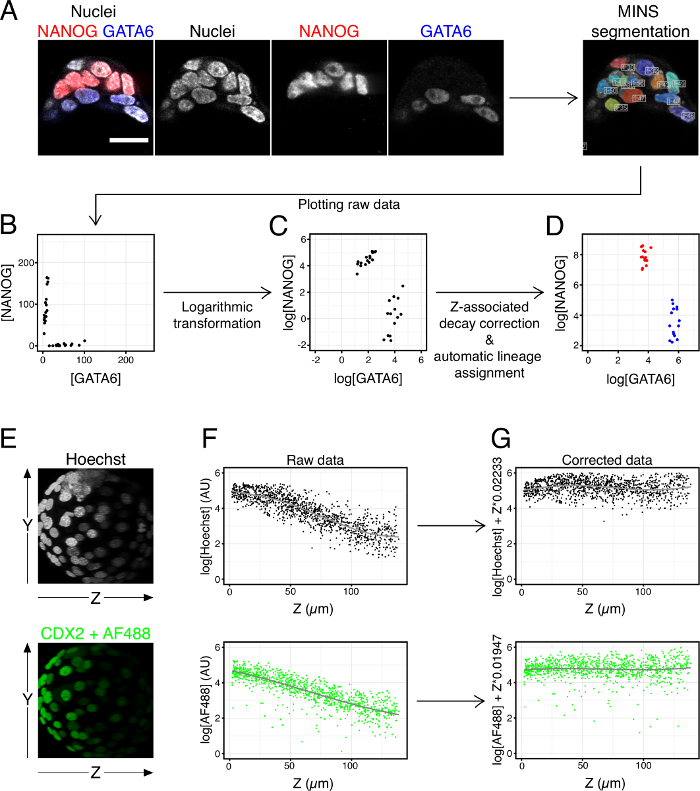
Figure 4. Example of Segmentation and Data Transformations. (A) Images of ICM cells of a blastocyst stained for NANOG and GATA6 and snapshot of nuclear segmentation of the same embryo using MINS on the nuclear staining channel (Hoechst 33342). (B-D) NANOG and GATA6 fluorescence values in ICM cells measured by MINS plotted as raw data (B), after performing logarithmic transformation (C) and after correcting values for the fluorescence decay along the Z axis and automatically assigning cell identities based on the corrected values (D). (E – G) Examples of data correction for fluorescence decay along the Z axis. (E) Images of a blastocyst labeled with Hoechst and anti-CDX2+AlexaFluor 488 (AF488), showing fluorescence decrease along the Z axis. (F) Scatter plots of fluorescence values along the Z axis for Hoechst and AF488 for a pool of embryos including the one shown in (E). Gray line represents the regression curve for each set of values. (G) Same data as in F after compensating fluorescence decay for each cell using the following factor: Z coordinate * the slope of the linear model (see step 4.5.2.2 in the protocol). Panels E – G were originally published by the authors in the Node (http://thenode.biologists.com/99problems/discussion/) Each dot represents one cell. Scale = 20µm. Please click here to view a larger version of this figure.
| User Input |
| Brightness threshold* (facilitates nuclear detection in dim images) |
| Relative X/Y-Z resolution |
| Channel to segment |
| Nuclear diameter |
| Image noise level |
| MINS Output |
| Segmentation Z-stack with nuclei IDs |
| Nuclear volume |
| XYZ coordinates for each nucleus |
| Average and summation of fluorescence values for each fluorescence channel and nucleus |
| Advantages |
| Accurate segmentation (>85%) of nuclei throughout the stack — recognizes all occurrences of a nucleus in adjacent z-sections |
| Single cell resolution |
| Unsupervised segmentation of batches of embryos |
| Segmentation pipelines can either be saved and re-used or adjusted for each particular embryo |
Table 1. MINS Features
| Load Image | At the prompt, introduce the Z relative resolution for the image. It can be obtained from the file metadata using the default reader or applying the following formula: X (or Y) resolution/Z resolution (all in µm). |
| Enter the sequence number (1 – 5) for the channel to be segmented. Using the DNA stain channel for segmentation will detect all cells in the image, however, other channels may be segmented separately too. | |
| Enter the frame to begin segmentation at (1 by default). Only applies to files with multiple time frames. | |
| Enhance Image (optional) | The white and black point of the image can be adjusted to facilitate detection. Generally lowering the white point and re-scaling to 0 – 255 (for 8-bit images) or 0 – 4096 (for 12-bit images) will make the image brighter and facilitate detection of dim nuclei. |
| Detect Nuclei | Select the estimated nuclear diameter (generally between 30 and 40 px in blastocysts). |
| For noisy images the noise level can be adjusted. Otherwise, the default value will be applied. | |
| Segment Nuclei | Use default parameters. |
| Classify Nuclei | MINS can distinguish trophectoderm (TE) from inner cell mass (ICM) cells in preimplantation embryos based on position. This can also be done manually or based on fluorescence levels if a TE marker is used. |
| Export Results | Select the files to be generated: |
| – segmentation overlaid over the channel used (.tiff sequence file), | |
| – segmentation only (.tiff sequence file), | |
| – spreadsheet with IDs for all cells detected, XYZ coordinates and fluorescence levels (average and sum) for all channels for each cell (.csv file). | |
| All files exported by default. Files are saved in the same directory where images are located. |
Table 2. MINS Segmentation Pipeline
Discussion
The present protocol describes a method to perform a quantitative analysis of whole-mount immunofluorescence on preimplantation stage mouse embryos. A robust immunofluorescence protocol 22 is followed by high-resolution, whole-mount confocal imaging and by image segmentation using a tailored piece of software 4. While the choice of immunofluorescence protocol is not critical, we find the one presented here 22 to be fast, reliable and to provide robust signal for many of the antibodies we have tested. Other protocols may be followed, provided their application is consistent across equivalent experiments. While many factors affect the outcome of each individual experiment, and direct comparison between experiments is not necessarily possible, we have found that striving for consistency across repetitions and performing sufficient technical replicates greatly reduces variability. Therefore, once optimized, fixation and immunolabeling reagents and protocols, as well as imaging conditions should be kept constant between equivalent experiments.
The main problems that may arise during the application of this protocol fall into two groups: 1) poor quality staining, with dim signal, and 2) suboptimal segmentation, with many errors (i.e., over- and under-segmentation). Low quality immunofluorescence is generally due to improper fixation of the samples or to the antibodies not being suitable for whole-mount immunofluorescence. Ensure PFA is fresh (either freshly prepared or thawed to RT from -20 ºC no longer than a week prior). Given the small size of preimplantation embryos, 10-min fixation in PFA should suffice. However, we have found certain antibodies to yield a stronger signal with longer fixation times (see Materials). If an antibody that has been previously tested provides weak signal, try different fixation protocols. Not all antibodies perform robustly on whole-mount immunofluorescence and the choice of primary antibody is critical for the outcome of the experiment. New antibodies should be tested on embryos and the optimal concentration determined empirically. In the Materials chart we provide details of a number antibodies we have tested and use routinely. Refer to the published literature or to the manufacturer's information when considering new antibodies. Note that not all antibodies suitable for Western blot work in whole-mount immunofluorescence, and the concentrations necessary for immunofluorescence may be up to one order of magnitude higher. Commercial secondary antibodies generally work well out of the box, however, be consistent in the use of primary-secondary antibody combinations throughout equivalent experiments that are going to be compared.
The quality of image segmentation depends on both the quality of the nuclear staining and on the nuclear density in the ICM of the embryo. Always strive for bright, sharp nuclei and use a high-resolution objective (40X or more). If the nuclear stain signal is low, use a fresh aliquot or a new batch. As long as the nuclear stain is not used for quantification, the acquisition parameters can be adjusted for each embryo in order to record sharp, bright nuclei. Likewise, if a channel other than the nuclear staining is used for segmentation, the signal-to-noise ratio of that particular channel will determine the quality of the segmentation. Late stage blastocysts (E4.0 onwards) present a very high nuclear density within the ICM. Therefore, it is at these stages that high quality staining is most critical. Nevertheless, segmentation errors will take place most often at these stages. Introduce the estimated nuclear diameter and the image noise level on the MINS pipeline, explore the outcome of each step using the 'View' button and adjust the parameters until a satisfactory result is obtained. Use the parameters that produce the best segmentation and manually correct errors during data processing. Note that late stage embryos (~E4.5) have a high cell count and manual data correction will be time consuming.
The limitations of this protocol are determined by the confocal microscope system used for image acquisition. As discussed, use a high-magnification objective, with a working distance that allows Z-axis imaging of the entire embryo (preimplantation mouse embryos can reach up to 150 – 160 µm in diameter). Similarly, the number of epitopes that can be detected will be determined by the number of channels the microscope can acquire at once. Using this protocol, three different epitopes besides the DNA (4 channels in total) can be labeled simultaneously on each sample. Ensure the instrument is calibrated for each fluorescence channel, the gating is optimized and that the laser output is consistent.
The methods described here allow the analysis of cell position along the XYZ axes as well as the quantification of protein expression levels in situ for every single cell in mouse blastocysts. In our experience, alternative methods using any other available software cannot provide the level of resolution that the pipeline applied in this protocol can (see 4 for a direct comparison with other tools). The high nuclear density found in the ICM of the blastocyst causes other tools to generate so many mistakes that the extent of manual correction required makes their use impractical. Alternatively, manual cell counting and fluorescence measurements have been previously used for subsets of cells or defined regions of the embryo 10,11,18,19,24. However, manual counting and measurement of all fluorescence channels and cell position, across all XYZ axes, for the tens of cells present in each embryo would be so time consuming that it would not allow for the high-throughput analysis for which our protocol was devised. Moreover, manual assessment of fluorescence levels is prone to bias, while a pipeline such as the one described here allows an objective measurement of fluorescence intensities for the subsequent determination of cell identity. In order to assign cell identities, the investigator should establish a standard criterion to be applied across all samples for a particular experimental set up. Given the different factors – biological and technical – that can affect the outcome of immunolabeling, this criterion should be determined using appropriate control experiments and previously published data wherever possible. We envision a near future where an algorithm may be designed for automatic determination of cell identity regardless of the experiment. However, such a tool will only come from more extensive application and improvement of the current method, most likely coupled with machine learning algorithms.
Finally, given the simplicity of this pipeline and the stereotypical morphology of mouse preimplantation embryos, this protocol is easily scalable for the analysis of large numbers of embryos, thus allowing high-throughput analyses of null phenotypes 5,6 or any other experimental manipulation 7. Finally, while the current protocol has been designed and optimized for the analysis of preimplantation mouse embryos and ESC colonies, we see no reason a priori why it could not be applied to other systems with similar characteristics – namely high nuclear density. We encourage others to experiment and adapt this protocol to other scenarios and provide feedback for its future improvement.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank Stefano Di Talia, Alberto Puliafito, Venkatraman Seshan and Panagiotis Xenopoulos, for input on data handling, analysis and representation, Berenika Plusa for assistance in the design of the immunofluorescence protocol and antibody testing and members of the Hadjantonakis lab for comments on the manuscript and on the development of this protocol. Work in our lab is supported by the National Institutes of Health R01-HD052115 and R01-DK084391, and by NYSTEM N13G-236.
Materials
| Embryo collection | |||
| Blunt probe for plug checking | Roboz | RS-9580 | |
| Forceps | Roboz | RS-4978 | |
| Surgery scissors | Roboz | RS-5910 | |
| Glass Pasteur pipettes | Fisher | 13-678-20C | |
| Pre-assembled aspirator tube & mouthpiece | Sigma | A5177 | |
| Longer rubber tubing | Fisher | 14-178-2AA | |
| M2 | Millipore | MR-015-D | |
| FHM | Millipore | MR-024-D | |
| Acid Tyrode's solution | Millipore | MR-004-D | |
| Penicillin/Streptomycin | Gibco | 15140 | |
| Bovine Serum Albumin | Sigma | A9647 | |
| 4-well plates | Nunc/Thermo-Fisher | 12-566-300 | |
| Name | Company | Catalog Number | Comments |
| Immunofluorescence | |||
| 96-well U-bottom plates | Fisher | 14-245-73 | |
| Triton X-100 | Sigma | T8787 | |
| Glycine | Sigma | G7403 | |
| Horse serum | Sigma | H0146 | |
| Primary antibodies | Concentration | ||
| CDX2 | Biogenex | AM392-5M | 1:200 |
| GATA6 | R&D | AF1700 | 1:100 |
| GATA4 | Santa Cruz | sc-9053 | 1:100, 10 min fixation |
| GATA4 | Santa Cruz | sc-1237 | 1:100, overnight fixation |
| SOX17 | R&D | AF1924 | 1:100 |
| Nanog | ReproCELL | RCAB0002P-F | 1:500 |
| OCT4 | Santa Cruz | sc-5279 | 1:100, 10 min to overnight fixation |
| DAB2 | BD | BD-610464 | 1:200 |
| Secondary antibodies | Life Technologies | Various | 1:500 |
| Hoechst 33342 | Life Technologies | H3570 | |
| Name | Company | Catalog Number | Comments |
| Imaging | |||
| 35 mm glass-bottom dishes | MatTek | P35G-1.5-14-C | |
| Name | Company | Catalog Number | Comments |
| Segmentation | |||
| Computer running 64-bit Windows OS | n/a | Verify minimal system requirements at http://katlab-tools.org and in Lou et al., (2014) Stem Cell Reports | |
| MATLAB (software) | Mathworks | n/a | |
| MINS (software) | Free | n/a | http://katlab-tools.org |
Referenzen
- Saiz, N., Plusa, B. Early cell fate decisions in the mouse embryo. Reproduction. 145 (3), R65-R80 (2013).
- Schrode, N., Xenopoulos, P., Piliszek, A., Frankenberg, S., Plusa, B., Hadjantonakis, A. -. K. Anatomy of a blastocyst: cell behaviors driving cell fate choice and morphogenesis in the early mouse embryo. Genesis. 51 (4), 219-233 (2013).
- Saiz, N., Plusa, B., Hadjantonakis, A. -. K. Single cells get together: High-resolution approaches to study the dynamics of early mouse development. Seminars in cell & developmental biology. , (2015).
- Lou, X., Kang, M., Xenopoulos, P., Muñoz-Descalzo, S., Hadjantonakis, A. -. K. A Rapid and Efficient 2D/3D Nuclear Segmentation Method for Analysis of Early Mouse Embryo and Stem Cell Image Data. Stem Cell Reports. 2 (3), 382-397 (2014).
- Le Bin, G. C., Muñoz-Descalzo, S., et al. Oct4 is required for lineage priming in the developing inner cell mass of the mouse blastocyst. Development. 141 (5), 1001-1010 (2014).
- Schrode, N., Saiz, N., Di Talia, S., Hadjantonakis, A. -. K. GATA6 Levels Modulate Primitive Endoderm Cell Fate Choice and Timing in the Mouse Blastocyst. Developmental Cell. 29 (4), 454-467 (2014).
- Xenopoulos, P., Kang, M., Puliafito, A., Di Talia, S., Hadjantonakis, A. -. K. Heterogeneities in Nanog Expression Drive Stable Commitment to Pluripotency in the Mouse Blastocyst. Cell Reports. 10 (9), 1508-1520 (2015).
- Saiz, N., Kang, M., et al. Quantitative analyses for elucidating mechanisms of cell fate commitment in the mouse blastocyst. Proceedings of SPIE. 9334, (2015).
- Fleming, T. P. A quantitative analysis of cell allocation to trophectoderm and inner cell mass in the mouse blastocyst. Developmental biology. 119 (2), 520-531 (1987).
- Dietrich, J. -. E., Hiiragi, T. Stochastic patterning in the mouse pre-implantation embryo. Development. 134 (23), 4219-4231 (2007).
- Plusa, B., Piliszek, A., Frankenberg, S., Artus, J., Hadjantonakis, A. -. K. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development. 135 (18), 3081-3091 (2008).
- Gerbe, F., Cox, B., Rossant, J., Chazaud, C. Dynamic expression of Lrp2 pathway members reveals progressive epithelial differentiation of primitive endoderm in mouse blastocyst. Developmental biology. 313 (2), 594-602 (2008).
- Nichols, J., Silva, J., Roode, M., Smith, A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 136 (19), 3215-3222 (2009).
- Yamanaka, Y., Lanner, F., Rossant, J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development. 137 (5), 715-724 (2010).
- Morris, S. A., Teo, R. T. Y., Li, H., Robson, P., Glover, D. M., Zernicka-Goetz, M. Origin and formation of the first two distinct cell types of the inner cell mass in the mouse embryo. Proceedings of the National Academy of Sciences of the United States of America. 107 (14), 6364-6369 (2010).
- Artus, J., Panthier, J. -. J., Hadjantonakis, A. -. K. A role for PDGF signaling in expansion of the extra-embryonic endoderm lineage of the mouse blastocyst. Development. 137 (20), 3361-3372 (2010).
- Artus, J., Piliszek, A., Hadjantonakis, A. -. K. The primitive endoderm lineage of the mouse blastocyst: Sequential transcription factor activation and regulation of differentiation by Sox17. Developmental biology. 350 (2), 393-404 (2011).
- Kang, M., Piliszek, A., Artus, J., Hadjantonakis, A. -. K. FGF4 is required for lineage restriction and salt-and-pepper distribution of primitive endoderm factors but not their initial expression in the mouse. Development. 140 (2), 267-279 (2013).
- Bessonnard, S., De Mot, L., et al. Gata6, Nanog and Erk signaling control cell fate in the inner cell mass through a tristable regulatory network. Development. 141 (19), 3637-3648 (2014).
- Behringer, R. R., Gertsenstein, M., Vintersten Nagy, K., Nagy, A. . Manipulating the mouse embryo: a laboratory manual. , (2014).
- Czechanski, A., Byers, C., et al. Derivation and characterization of mouse embryonic stem cells from permissive and nonpermissive strains. Nature Protocols. 9 (3), 559-574 (2014).
- Frankenberg, S., Shaw, G., Freyer, C., Pask, A. J., Renfree, M. B. Early cell lineage specification in a marsupial: a case for diverse mechanisms among mammals. Development. 140, 965-975 (2013).
- Artus, J., Vandormael-Pournin, S., Frödin, M., Nacerddine, K., Babinet, C., Cohen-Tannoudji, M. Impaired mitotic progression and preimplantation lethality in mice lacking OMCG1, a new evolutionarily conserved nuclear protein. Molecular and cellular biology. 25 (14), 6289-6302 (2005).
- Saiz, N., Grabarek, J. B., Sabherwal, N., Papalopulu, N., Plusa, B. Atypical protein kinase C couples cell sorting with primitive endoderm maturation in the mouse blastocyst. Development. 140 (21), 4311-4322 (2013).

