Optical Control of a Neuronal Protein Using a Genetically Encoded Unnatural Amino Acid in Neurons
Summary
Here, a procedure to selectively activate a neuronal protein with a short pulse of light by genetically encoding a photo-reactive unnatural amino acid into a target neuronal protein expressed in neurons in culture or in vivo is presented.
Abstract
Photostimulation is a noninvasive way to control biological events with excellent spatial and temporal resolution. New methods are desired to photo-regulate endogenous proteins expressed in their native environment. Here, we present an approach to optically control the function of a neuronal protein directly in neurons using a genetically encoded unnatural amino acid (Uaa). By using an orthogonal tRNA/aminoacyl-tRNA synthetase pair to suppress the amber codon, a photo-reactive Uaa 4,5-dimethoxy-2-nitrobenzyl-cysteine (Cmn) is site-specifically incorporated in the pore of a neuronal protein Kir2.1, an inwardly rectifying potassium channel. The bulky Cmn physically blocks the channel pore, rendering Kir2.1 non-conducting. Light illumination instantaneously converts Cmn into a smaller natural amino acid Cys, activating Kir2.1 channel function. We express these photo-inducible inwardly rectifying potassium (PIRK) channels in rat hippocampal primary neurons, and demonstrate that light-activation of PIRK ceases the neuronal firing due to the outflux of K+ current through the activated Kir2.1 channels. Using in utero electroporation, we also express PIRK in the embryonic mouse neocortex in vivo, showing the light-activation of PIRK in neocortical neurons. Genetically encoding Uaa imposes no restrictions on target protein type or cellular location, and a family of photoreactive Uaas is available for modulating different natural amino acid residues. This technique thus has the potential to be generally applied to many neuronal proteins to achieve optical regulation of different processes in brains. The current protocol presents an accessible procedure for intricate Uaa incorporation in neurons in vitro and in vivo to achieve photo control of neuronal protein activity on the molecular level.
Introduction
Compared to conventional electric stimulation, photostimulation offers greater temporal and spatial resolution with minimum interference to the physiological system of specimens. Since the demonstration of using lasers to stimulate neurons in 19711, many creative ways have been invented to exogenously control neuronal activity with light. Optical release of photocaged agonists has long been used to study physiological response of the neuronal network to ligands2,3,4. This technique has limited specificity due to diffusion of caged agonists. Genetic specificity is achieved by ectopically expressing light-sensitive opsin channels and pumps5,6,7, and it has been successfully applied to modulate selected neuronal networks in diverse model organisms. However, it would be difficult to apply this method to optically control various other neuronal proteins, since grafting photoresponsiveness from the opsin proteins to other proteins would require intense engineering that may alter the natural characteristics of the protein under study. Chemically tethering an exogenous photosensitive ligand to a protein has demonstrated another way to control the functionality of channel proteins8,9,10. The ligand is presented or withdrawn from the binding site of the protein through the photoisomerization of the azobenzene moiety. Tethering chemistry limits the application mainly to the extracellular side of membrane proteins, excluding the intracellular side and intracellular proteins.
Photoresponsive Uaas, after being incorporated into proteins, provide a general strategy to manipulate proteins with light. In early efforts, tRNAs chemically acylated with photocaged Uaas were microinjected into Xenopus oocytes to incorporate the Uaas into membrane receptors and ion channels11, which have advanced the understanding of their structure-function relationships12,13,14. This microinjection approach is mainly limited to large oocytes. Genetic incorporation of a Uaa bypasses the technically challenging tRNA acylation and microinjection by using an orthogonal tRNA/synthetase pair, which incorporates the Uaa through endogenous protein translation in live cells15,16,17,18. Uaa incorporation into neuronal proteins has been demonstrated in primary neurons and neural stem cells19,20. More recently, photoresponsive Uaa has been genetically incorporated into a neuronal protein in the mammalian brain in vivo for the first time21. These advancements make it possible to study neuronal proteins with Uaas in their native cellular environment.
Inwardly rectifying potassium channel Kir2.1 is a strong rectifier that passes K+ currents more readily into than out of the cell, and it is essential in regulating physiological processes including cell excitability, vascular tone, heart rate, renal salt flow and insulin release22. Overexpression of Kir2.1 hyperpolarizes the membrane potential of the target neuron, which becomes less excitable23,24. To optically control Kir2.1 in its native cells, Kang et al. genetically incorporated a photo-responsive Uaa into Kir2.1 expressed in mammalian cells, neurons and embryonic mouse brains21. A brief pulse of light was able to convert the Uaa into a natural amino acid Cys, thus activating the target Kir2.1 protein. When this photo-inducible inwardly rectifying potassium (PIRK) channel protein was expressed in rat hippocampal primary neurons, it suppressed neuronal firing in response to light activation. In addition, the PIRK channel was expressed in the embryonic mouse neocortex, and the light-activated PIRK current in cortical neurons was measured. The successful implementation of the Uaa technology in vivo in the mammalian brain opens the door to optically control neuronal proteins in their native environment, which will enable optical dissection of neuronal processes and mechanisms at the molecular level.
In this protocol we describe procedures for genetic incorporation of Uaas into primary neurons in culture and in the embryonic mouse brain in vivo. The photoresponsive Uaa Cmn and Kir2.1 are used to illustrate the process. Methods to assess successful Uaa incorporation and optical control of neuronal protein activity are provided. This protocol provides a clear guide to genetically encoding Uaas in neurons and in vivo, and to optically regulating neuronal protein function via a photoresponsive Uaa. We expect this protocol to facilitate the adoption of the in vivo Uaa technology for neuroscience and optogenetic biological studies.
Protocol
All procedures in the current study were performed using Institutional Animal Care and Use Committee (IACUC) approved protocols for animal handling at The Salk Institute for Biological Studies, La Jolla, CA.
1. Uaa Incorporation in Kir2.1 and Expression of the Resultant PIRK in the Primary Neuronal Culture
- DNA Construction
- Select a target site of Kir2.1 for Uaa incorporation. Exploit prior knowledge and information about structure and function of Kir2.1, so that the chosen site with the photoresponsive Uaa incorporated enables optical modulation of Kir2.1 function21.
- Construct a recombinant DNA encoding Kir2.1 gene with the chosen site mutated to the TAG amber stop codon. Use the standard cloning techniques25 to clone the Kir2.1-TAG DNA into a mammalian expression plasmid.
- Clone tRNA/synthetase genes that are specific for the Uaa and incorporate the Uaa in response to the TAG stop codon into another mammalian expression plasmid21.
Note: For optimal Uaa incorporation, different promoters and combinations of gene cassettes (for tRNA, synthetase, and Kir2.1) can be tested in different plasmids. - Obtain high quality plasmid DNAs using miniprep or maxiprep commercial kits according to manufacturer's protocol. Around 1.9 of the 260/280 ratio is optimal for the purified DNA. When necessary, perform an agarose gel electrophoresis to check purity of the prepped DNA25. Supercoiled plasmid DNAs with high purity are best for neuronal transfection.
- Culture Rat Hippocampal Primary Neurons
- Place glass coverslips (circles, 12 mm in diameter) in 24-well plates, one coverslip on each well, and coat each well with 250 µl of 0.5 mg/ml poly-D-lysine (in 100 mM borate buffer: 1.24 g boric acid and 1.90 g sodium tetraborate in 400 ml of deionized distilled H2O or ddH2O at pH 8.5) for over 10 hr at room temperature, sealed. Prepare around 10-12 coated coverslips per pup.
- On the day of dissection, collect neonatal brains from postnatal rat pups (1-4 postnatal day) after anesthetizing them with isoflurane, and decapitating. Anesthetize the pups in an anesthetization chamber containing isoflurane (2-4%). Wait until they lose consciousness and fail to respond to tactile stimuli and to pinching of the paws.
- Using the standard technique26, dissect hippocampi from the brain in warm (~37 °C) saline (10 mM HEPES and 20 mM D-glucose added in Hank's balanced salt solution), and collect hippocampi in a conical tube with 4.5 ml warm saline.
- Add 500 µl of 2.5% trypsin to the hippocampi (0.25% final trypsin concentration), and incubate for 10 min in a water bath at 37 °C.
- Thoroughly rinse the tissue with saline (3 x 10 ml) and triturate.
- Recover the dissociated neurons in 1 ml warm growth media containing Minimum Essential Medium (MEM) supplemented with 5% Fetal Bovine Serum (FBS), 21.2 mM D-glucose, 2 mM L-glutamine, 2% B-27, and 0.1% serum extender.
Note: Add L-glutamine and B-27 on the day of dissection to prepare fresh growth media. - Count the neurons with a hemocytometer, and plate them onto coverslips in 24-well plates at 1.0-1.5 x 105 cell/well density with 500 µl growth media after filtered through a 40 µm nylon mesh.
- Incubate the neuronal culture at 35 °C in a 5% CO2: 95% air humidified incubator for 2-3 weeks. For the best result, avoid disturbing the culture as much as possible during incubation.
- Calcium phosphate (Ca-P) Transfection of Primary Neuronal Culture
- Make stock solutions and store at 4 °C: 0.5 M BES (N,N-bis[2-hydroxy-ethyl]-2-aminoethanesulfonic acid) buffer (10x); 150 mM Na2HPO4 (100x); 2.8 M NaCl (10x); sterile ddH2O; sterile 1 N NaOH.
- On the day of transfection, make fresh 2.5 M CaCl2 solution in ddH2O and sterile filter with 0.22 µm filter. Make fresh 2x BES-buffered Saline (BBS) buffer containing 50 mM BES, 1.5 mM Na2HPO4, and 280 mM NaCl at pH 7.00 (adjust pH with NaOH) and sterile filter with 0.22 µm filter.
Note: Calculate the amount of CaCl2 solution and BBS buffer depending on the number of coverslips to be transfected. Also look at 1.3.4. - Replace the culture growth media with 500 µl fresh pre-warmed transfection growth media. (Transfection media can be made with MEM plus 21.2 mM D-glucose).
Note: Do not discard the old media. - Calculate the amount of ddH2O and prepare to add in order to make the transfection solution (33 µl per each coverslip) containing 1.65 µl CaCl2, 0.7 µg DNA, and 16.5 µl 2x BBS.
- Prepare the transfection solution immediately before adding it to the culture. To prepare, first combine CaCl2 and ddH2O while slowly agitating the tube. Continue agitating and slowly add DNA into the solution. Lastly, add 2x BBS buffer drop-wise while agitating.
- Immediately add 30 µl of the transfection solution to each coverslip of neuronal culture.
- Rock the culture dish a few times to mix the solution and incubate at 35 °C in a 5% CO2: 95% air humidified incubator for 45 min-1 hr. After neurons are incubated with the transfection solution, very fine Ca-P precipitates would form a layer covering neurons.
- Replace the transfection media with 500 µl pre-warmed washing buffer (135 mM NaCl, 20 mM HEPES, 4 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 1 mM Na2HPO4, 10 mM glucose at pH 7.3, sterile filtered with 0.22 µm filter) and incubate at 35 °C in a 5% CO2: 95% air humidified incubator for 15-20 min. Ca-P precipitates would disappear after the wash step.
- Replace the washing buffer with 500 µl fresh growth media. Again, replace the growth media with the saved original media.
- Add the Uaa Cmn (pre-mixed in 50 µl warm growth media) to the culture to reach 1 mM final concentration.
- Incubate the transfected culture at 35 °C in a 5% CO2: 95% air humidified incubator for 12-48 hr before assays.
- Whole Cell Recording with Light Activation
- Prepare extra/intracellular solutions. The intracellular solution contains 135 mM potassium gluconate, 10 mM NaCl, 2 mM MgCl2, 10 mM HEPES, 1 mM EGTA, 2.56 mM K2ATP, and 0.3 mM Li2GTP at pH 7.4. The extracellular recording solution contains 150 mM NaCl, 3 mM KCl, 5 mM MgCl2, 0.5 mM CaCl2, 5 mM glucose, and 10 mM HEPES at pH 7.4.
- Set up the electrophysiology rig for whole-cell patch clamping: microscope fitted with 4X and 20X objectives, differential interference contrast (DIC), and mCitrine filter (excitation: 495/10 nm, emission: 525/25 nm); manipulator; patch-clamp amplifier; digitizer; data acquisition and analysis software.
- Install a light-emitting diode (LED) with emission of 385 nm by the microscope at the rig to deliver light to the focal point from 1 cm away at a 45° angle. Check the power of the LED with a light power meter.
- Pull patch pipettes from glass electrodes using a commercial micropipette puller to have 3-6 MΩ pipette resistance. Follow manufacturer's instruction to set up the micropipette puller.
- To test pipette resistance, first fill the pipette with the intracellular solution and position it on the electrode holder. Dip the pipette in a 35 mm culture dish filled with the extracellular solution, placed on the microscope platform. Immerse a ground electrode into the dish to complete a circuit.
- Turn on the amplifier/digitizer and start a data acquisition software. Monitor pipette resistance with a membrane test protocol.
- Take out a coverslip of the neuron culture from the incubator and rinse once in the extracellular solution. Using vacuum grease, hold down the coverslip in the middle of a 35 mm culture dish filled with the fresh extracellular solution.
- Place the coverslip/dish on the electrophysiology microscope platform.
- Using the standard patch clamping techniques, patch a neuron with mCitrine fluorescence27. Record neuronal activity using current clamp (I-clamp) method. First, adjust the resting potential to around -72 mV by injecting a small current. Then, inject a step current (10-200 pA) to induce continuous firing (5-15 Hz) of action potentials.
- Manually, or using the data acquisition software, flash a pulse (a single pulse of 100 msec-1 sec duration) of the LED light to the neuron while recording, and see if action potentials are affected.
- Add 0.5 mM BaCl2 to the bath and verify if action potentials are recovered.
2. Uaa Incorporation in Kir2.1 and Expression of the Resultant PIRK in the Mouse Embryonic Brain In Vivo
- DNA Construction
- Design the plasmid DNA similarly as in step 1.1. For in vivo expression, use a strong promoter such as CAG (chicken beta-actin promoter with CMV enhancer).
- Purify DNA with an endotoxin-free maxiprep commercial kit according to the manufacturer's protocol. Perform phenol-chloroform extraction followed by ethanol precipitation25 to acquire extremely high quality DNA (condensed to 2-5 µg/µl).
- In Utero Electroporation to the Mouse Embryonic Neocortex
Note: The basic technique for in utero electroporation has been described previously28.- Anesthetize a timed pregnant mouse at the embryonic day 14.5 (E14.5) with sodium pentobarbital (intraperitoneal injection, 50 µg per gram body weight) or isoflurane (inhalation, 2-4%). Monitor anesthetic depth by checking loss of consciousness and no response to tactile stimuli and to pinching of the paws.
- Make a small incision at the abdominal midline. Gently expose the uterine horns with forceps and fingertips.
- Inject about 1 µl DNA solution (2-5 µg/µl of each plasmid, depending on the construct) into the lateral ventricle of each littermate with a glass pipette inserted through the uterine wall. In most cases, about 60-80% of total embryos are injected.
- Electroporate the embryos with an electroporator (33-35 V, 50 msec duration, 950 msec interval, 4-8 pulses).
- Return the uterine horns to the abdominal cavity gently with forceps and fingertips. Suture the muscle wall and then the skin with surgical suture to allow the embryos to continue development.
- Uaa Microinjection
- Make a small incision at the abdominal midline again at E16.5, and gently expose the uterine horns with forceps and fingertips.
- Inject about 2-5 µl Cmn-Ala (500 mM) to the electroporated side or both sides of the lateral ventricle with a glass pipette inserted through the uterine wall. To increase Cmn bioavailability, use the dipeptide Cmn-Ala (Cmn-alanine) to deliver Cmn in vivo21.
- Again, return the uterine horns to the abdominal cavity gently with forceps and fingertips. Suture the muscle wall and then the skin with surgical suture to allow the embryos to continue development.
- Obtain Acute Brain Slices
- Make 1 L artificial cerebrospinal fluid (ACSF) containing 119 mM NaCl, 2.5 mM KCl, 1.3 mM MgCl2, 2.5 mM CaCl2, 1 mM NaH2PO4, 26.2 mM NaHCO3, and 11 mM glucose at pH 7.3.
- Take 200 ml ACSF and fast-freeze at -80 °C for 20-30 min. Bubble the rest of ACSF with 5% CO2: 95% O2 gas at room temperature.
- Prepare and sterilize surgery tools to harvest brains from the embryos. Also prepare a bucket of ice.
- Make ~100 ml 4% low melting point agarose solution in a flask by heating in a microwave. Cool down solution for about 5 min before it starts to solidify.
- Euthanize the mouse 12-24 hr after Cmn-Ala injection by CO2 overdose. Make large incisions at the abdominal area, and dissect out the electroporated/microinjected embryos from the uterus with fine scissors and forceps.
- Harvest brains from the embryos with fine scissors and forceps.
- Place each brain on a 10 cm culture dish placed on the ice bucket. Divide two hemispheres using a sharp blade, and place each hemisphere on the dish with the midsagittal plane touching the bottom of the dish. Quickly pour over the agarose solution over brains (around 500 µl per hemisphere).
- Using a sharp blade, square cut the agarose around a brain to make a brain-embedded agarose block.
- Using tissue adhesive, glue down the midsagittal plane surface of the agarose block on a mount for vibratome. Fill the vibratome chamber with pre-chilled ACSF and cut 200 µm sagittal brain slices.
- Incubate acute slices in ACSF supplemented with 3 mM myo-inositol, 0.4 mM ascorbic acid, and 2 mM sodium pyruvate at 33 °C for 42 min while bubbling with 5% CO2: 95% O2 gas.
- After 42 min, turn off the heater and continue to incubate the slices at room temp with bubbling. Start whole-cell patch recording at least 15 min after turning off the heater.
- Whole Cell Recording with Light Activation
- Prepare the intracellular solution containing 130 mM potassium gluconate, 4 mM MgCl2, 5 mM HEPES, 1.1 mM EGTA, 3.4 mM Na2ATP, 10 mM sodium creatine phosphate, and 0.1 mM Na3GTP at pH 7.3 adjusted with KOH.
- Pull patch pipettes from glass electrodes using a commercial micropipette puller to have 3-6 MΩ pipette resistance.
- Set up the slice electrophysiology rig for whole-cell patch clamping and superfuse the recording chamber with ACSF at 2 ml/min rate with a perfusion pump. Adjust the chamber temperature to around 33 °C.
Note: The slice electrophysiology rig is equipped with a perfusion chamber, a water immersion objective and a temperature controller. Also, GFP filter (excitation: 480/30 nm, emission: 535/40 nm) and mCherry filter (excitation: 580/20 nm emission: 675/130 nm) are required.- Install an LED with emission of 385 nm by the microscope at the rig to deliver light to the focal point from 1 cm away at a 45° angle. Check the power of the LED with a light power meter.
- Carefully pick up a brain slice with a glass pipette and place in the perfusion chamber. Hold down the slice with a harp.
- Patch a neuron with mCherry/GFP fluorescence from the neocortical region27. Record PIRK activity using voltage clamp (V-clamp) method. First, hold the membrane potential at -60 mV. Then, record currents at fixed negative membrane potentials (-100 mV) or voltage ramps (-100 mV to +40 mV). Specifically, monitor Kir2.1 specific inward currents at -100 mV.
- Manually, or using the data acquisition software, flash a pulse (a single pulse of 100 msec-1 sec duration) of LED light to the neuron while recording, and see if PIRK proteins are activated. Once PIRK is activated, inward currents at -100 mV would increase significantly.
- Add 0.5 mM BaCl2 to the bath and verify if PIRK is inactivated again.
Representative Results
To genetically incorporate a Uaa into a protein in neurons, the first important step is to design appropriate gene constructs to deliver and express genes efficiently in neurons. There are three genetic components for Uaa incorporation: (1) the target gene with the TAG amber stop codon introduced at the chosen site for Uaa incorporation (2) an orthogonal tRNA to recognize the mutated TAG stop codon, and (3) an orthogonal aminoacyl-tRNA synthetase to charge the Uaa onto the orthogonal tRNA. Each component needs to be driven by the appropriate promoter. These three gene cassettes can be included in one plasmid or separated in several plasmids. To express PIRK in neurons, the photo-reactive Uaa Cmn is incorporated into Kir2.1 using the orthogonal tRNALeuCUA/CmnRS (Cmn-tRNA synthetase) pair evolved to be specific for Cmn21,29. Residue Cys169 of Kir2.1 gene is mutated to the TAG amber stop codon (Kir2.1-TAG), and the fluorescent protein mCitrine is fused to the C-terminus of Kir2.1 to visualize PIRK expression in neuronal culture (Figure 1).
Obtaining good-quality neuronal culture is a prerequisite for successful PIRK experiments. Postnatal day 1-4 Sprague Dawley rat pups are generally used for neuronal culture preparation, yet rat embryos are also frequently used. When plated onto coverslips in 24-well plates at 1.0-1.5 x 105 cell/well density, one should see well-separated healthy neurons with not too much microglia or other debris (Figure 2A). Healthy neurons have extensive processes, and cell bodies (soma) are plump (Figure 2B). Distinctively visible suborganelle structure is usually a sign of unhealthy culture. Neuronal culture can be maintained for 2-3 weeks at 35 °C in the 5% CO2: 95% air humidified incubator.
Calcium phosphate (Ca-P) transfection is a very gentle transfection method appropriate for sensitive neuronal culture. However, it requires extreme precision and caution to achieve high transfection efficiency in neurons. Stock solutions need to be stored at 4 °C, and mixed to make 2x BBS buffer on the day of transfection. Precise pH adjustment is key to reproducibly get successful results. The 2.5 M CaCl2 solution should also be made fresh on the day of transfection. All buffers need to be filtered. Moreover, fresh DNA plasmids with high purity and quality improve the results. For best results, one can obtain fresh DNA mini-prepped from not overgrown Escherichia coli culture (~12 hr at 37 °C in the shaking incubator). After all the buffers are ready, bring the solutions in the tissue culture hood and prepare for transfection. Using a vortex at a low speed, agitate the transfection solution while mixing. Immediately add the mixed solution to the neuronal culture, and bring the culture dish back to the incubator. Stop the transfection after 45 min-1 hr, and add the original growth media back to the culture for continuous incubation. The transfected neurons can be visualized from 6 hr after transfection is terminated (Figure 3). In the presence of Cmn in the culture media, PIRK is expressed and PIRK-expressing neurons are green-fluorescent due to the mCitrine protein fused to the C-terminal of PIRK (Figure 3D). PIRK-expressing neurons have normal basal physiology and they are capable of firing action potentials in response to injected currents (Figure 4). However, this series of action potentials is suppressed immediately with a brief light pulse shone to the cell (Figure 4A). When 0.5 mM BaCl2, a Kir2.1 specific inhibitor, is added to the bath, neuronal firing is then resumed, indicating that the previous suppression was due to the activation of Kir2.1 by the light (Figure 4B). Untransfected control neurons do not respond to the light or Ba2+ (Figure 4C, D).
To express PIRK in vivo, highly pure and concentrated DNA plasmids (2-5 µg/µl) should be prepared. Specific plasmids used in these experiments are shown in Figure 5A. In addition to the orthogonal tRNALeuCUA/CmnRS pair and the target Kir2.1-TAG gene, a gene coding green fluorescent protein with a TAG mutation (GFP-TAG) is also co-electroporated as a fluorescent reporter. Detection of GFP fluorescence would indicate the successful delivery of all three plasmids, since TAG suppression in GFP would require both the tRNALeuCUA and the CmnRS; Cmn incorporation in GFP-TAG would suggest Cmn incorporation in Kir2.1-TAGas well, because both genes are present in the same cell. The three gene constructs are all injected into the lateral ventricle of mouse embryos on E14.5 (Figure 5B, left panel). After electroporation, return the embryos to the abdominal cavity to allow the embryos to continue development. Two days later, inject about 2-5 µl Cmn-Ala (500 mM) to the electroporated side or both sides of the lateral ventricle, in the similar manner as DNA injection (Figure 5B, middle panel). Again, return the embryos to the abdominal cavity for continuous development. The embryos can be harvested on E17.5 for imaging or electrophysiological assays. As expected, only with Cmn-Ala injection, green fluorescent cells are observed in the mouse neocortex. Cells with both red and green fluorescence should have Cmn incorporated into Kir2.1-TAG to make PIRK channels (Figure 6A). Whole-cell recording from the embryonic neocortical slice is done similarly as it is done on neuronal culture. The green and red fluorescent neurons have no inward current at negative holding potential, but a brief pulse of light rapidly activates the inward current (Figure 6B). The current is completely blocked by adding Ba2+, confirming that it is generated by PIRK.
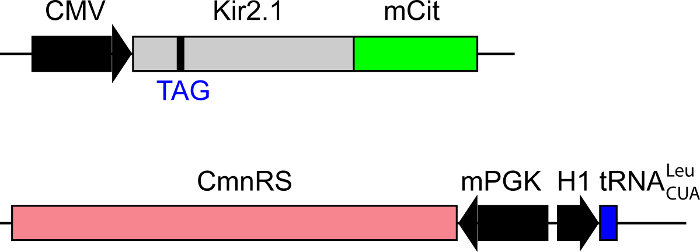
Figure 1: PIRK expression plasmid set for neuron culture. Scheme showing exemplary plasmid design for PIRK expression in neuronal culture. Top plasmid encodes Kir2.1 gene (grey) with a single amino acid mutation in C169 site under cytomegalovirus (CMV) promoter. C169 is mutated to the TAG amber stop codon, where the Uaa Cmn would be incorporated. Kir2.1 gene is followed by mCitrine gene (green). mCitrine is fused to the C-terminal of the Kir2.1 gene to visualize PIRK expressing cells. Bottom plasmid encodes CmnRS (pink), the Cmn specific synthetase, driven by the mouse phosphoglycerate kinase-1 (mPGK) promoter. tRNALeuCUA (blue), the orthogonal tRNA21, is also expressed in the same plasmid driven by the H1 promoter, but in a reverse direction.
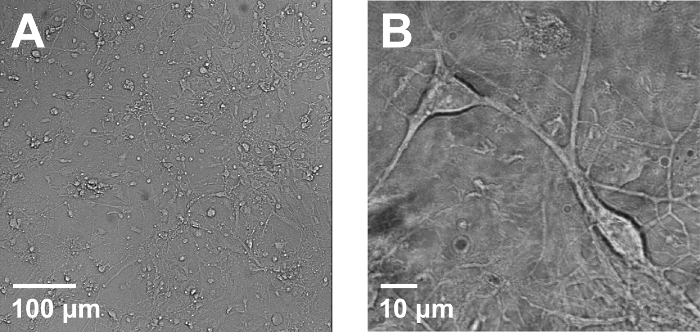
Figure 2: Rat hippocampal primary neuronal culture. Exemplary pictures showing healthy rat hippocampal neuronal cultures. (A) A DIC image of neuronal culture on 10 days in vitro (DIV). Neurons mature as they branch out dendrites and axons. Microglia cells (small and round cells without processes) coexist but do not overwhelm the culture. (B) A magnified DIC image of healthy cultured neurons. A healthy neuron exhibits plump cell body (soma) and pronounced dendrites/axons. Please click here to view a larger version of this figure.
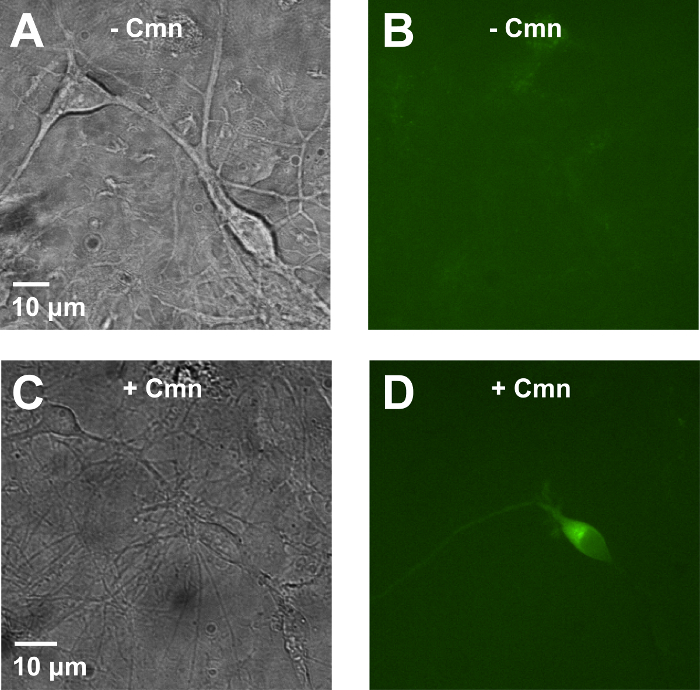
Figure 3: PIRK expression in cultured primary neurons. DIC and fluorescence images of rat hippocampal neurons cultured in vitro and transfected with gene constructs depicted in Figure 1 for PIRK expression. When Cmn is not added in the culture after transfection (A and B), no green fluorescence is detected from the neurons. In the presence of Cmn (1 mM) in the growth media, transfected neurons are capable of expressing full-length PIRK-mCitrine proteins, thus showing green fluorescence (C and D). Please click here to view a larger version of this figure.
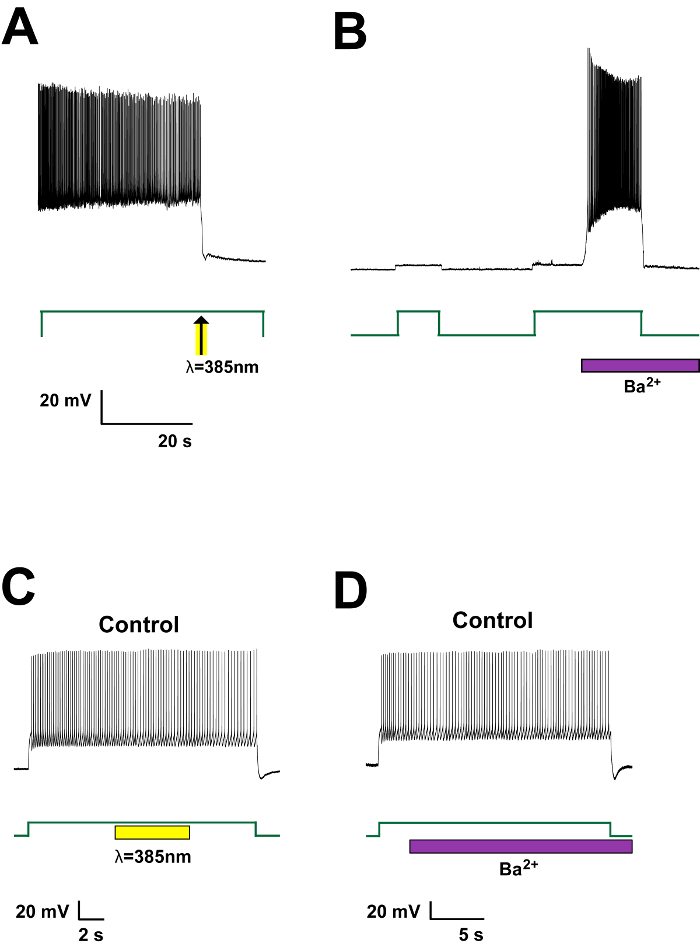
Figure 4: Light activation of PIRK suppresses firing from rat hippocampal neurons. (A) A single light pulse suppresses activity of the hippocampal neuron expressing PIRK. Representative voltage traces are recorded continuously in current-clamp. Action potential firing is evoked by 20 pA current injection (I-step). Light exposure (385 nm, 40 mW/cm2, 1 sec; indicated with arrow) rapidly and completely suppresses neuronal firing. Firing is restored with extracellular 500 mM BaCl2, which selectively inhibits Kir2.1 channels. (B) Ultraviolet (UV) illumination (385 nm, 40 mW/cm2, 6 sec; indicated with yellow box) does not alter excitability of control, untransfected neurons. Action potential firing is evoked by 50 pA current injection (I-step). (C and D) Neither UV illumination nor BaCl2 (500 mM) affects excitability of control neurons. Action potential is evoked by 50 pA current injection (I-step).
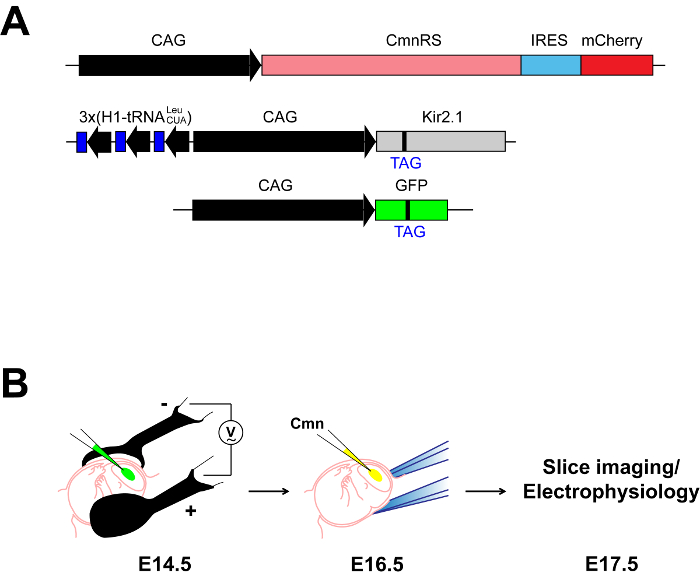
Figure 5: Procedures for PIRK expression in the mouse neocortex in vivo. (A) PIRK expression plasmid set. Top plasmid: the plasmid for CmnRS driven by the CAG promoter and mCherry via internal ribosome entry site (IRES). mCherry fluorescence would verify successful expression of CmnRS. Middle plasmid: the plasmid for Kir2.1 gene coupled with three copies of tRNALeuCUA, the orthogonal tRNA for Cmn incorporation21. Bottom plasmid: the plasmid for GFP_Y182TAG. The gene for GFP is engineered with an amber stop codon at the permissive Tyr182 site (GFP_Y182TAG) to visualize successful incorporation of Cmn to TAG sites21. (B) Cartoon showing an experimental procedure for PIRK expression in vivo. Gene constructs for PIRK expression are injected into the mouse neocortex (E14.5) and electroporated in utero (left panel). Two days later, Cmn-Ala is injected to the ventricle in the electroporated side or both sides of cerebral hemispheres (middle panel). Slice imaging and electrophysiological assay can be performed on E17.5-E18.5 (right panel).
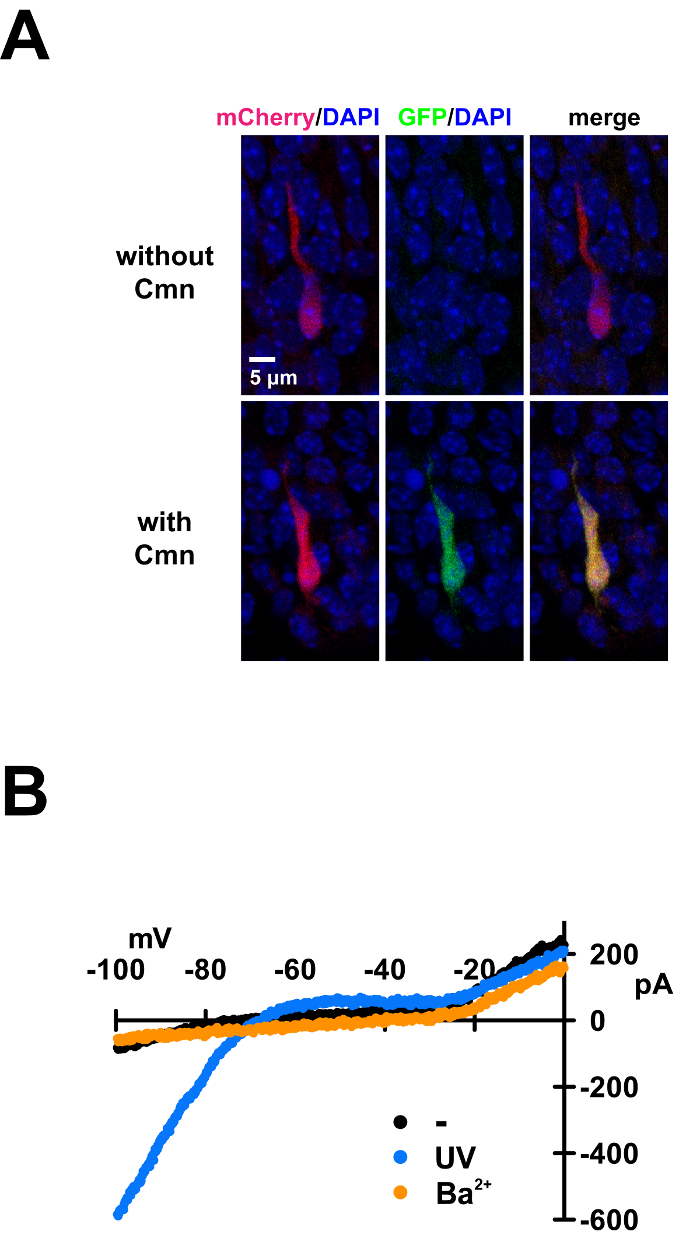
Figure 6: PIRK expression in the mouse neocortex and light activation. (A) Fluorescence images of mouse embryonic cortical neurons showing the incorporation of Cmn into GFP and Kir2.1 proteins in vivo. The three gene constructs in Figure 5A are electroporated in utero. mCherry fluorescence demonstrates successful expression of the Cmn specific synthetase gene, CmnRS. GFP fluorescence is detected only with Cmn-Ala injection (bottom), indicating Cmn incorporation in GFPTAG and likely Cmn incorporation in Kir2.1TAG. Thus, green and red fluorescence indicates successful expression of all three plasmids and Cmn incorporation. (B) I-V plot of currents recorded from mice neocortical neurons showing light-dependent activation of PIRK. Two days after gene constructs in Figure 5A were electroporated, Cmn-Ala was injected in utero; 12-48 hr after Cmn-Ala injection, neocortical acute slices were prepared from the embryos. PIRK-expressing neurons in the slices, detected by both red and green fluorescence, are recorded before (black) and after (blue) light exposure (385 nm, 8 mW/cm2, 10 sec for saturated exposure). BaCl2 (500 mM) is added to verify PIRK-specific currents after photoactivation (orange).
Discussion
To achieve effective photo-modulation, the important initial step is to decide where to incorporate the photoresponsive Uaa in the target protein. Structural and functional information of the target protein is very helpful to guide the selection of candidate sites. At the same time, the purpose of light regulation would determine which site is most suitable. After choosing candidate sites, we recommend to test the sites in mammalian cell lines such as the Human Embryonic Kidney (HEK) cells for easier culture and manipulation, before proceeding to primary neurons and in vivo. To identify a site for Kir2.1 photoactivation, multiple sites along the pore of Kir2.1 have been initially tested in HEK cells. C169 of Kir2.1 was found optimal, because the Kir2.1 pore size where C169 resides is big enough to accommodate Cmn side chains but small enough for Cmn side chains to block ionic passage. When Cmn was incorporated at C169 in Kir2.1 expressed in HEK cells, the resultant mutant Kir2.1 was inactive but became activated upon a short pule of light, confirming successful PIRK development21.
Efficient expression of the orthogonal tRNA and synthetase is critical to obtain high Uaa incorporation efficiency in cells and in vivo. The orthogonal synthetase gene is efficiently expressed using polymerase II promoter and poly-A signals often used in mammalian cells and neurons. For expression of the orthogonal tRNA, a special type-3 polymerase III promoter, such as the H1 promoter used here, together with a 3'-flanking sequence are needed as described previously18,20. For in vivo studies, we found that three copies of the tRNA expression cassette increased Cmn incorporation efficiency compared to one copy (Figure 5A). In another study in mammalian cells, increasing the number of tRNA expression cassette also increased Uaa incorporation efficiency30. Therefore, we suggest optimizing the number of tRNA expression cassettes for different Uaas, cells, and target proteins when Uaa incorporation efficiency needs improvement. Bioavailability of Uaas in cells and in vivo is another critical factor for Uaa incorporation. Previous studies have shown that esterification of Uaas increases their uptake in mammalian cells31, and that preparation of Uaas in the dipeptide form increases Uaa uptake into cells in Caenorhabditis elegans32. We found that directly feeding Cmn to neuronal culture media is sufficient for Cmn incorporation in Kir2.1 expressed in cultured primary neurons, but inadequate for incorporation in the mouse brain in vivo21. Therefore, the dipeptide Cmn-Ala was synthesized and injected into the mouse brain. Oligopeptide transporter PEPT2 is highly expressed in rodent brains33, which may facilitate transportation of the dipeptide into neurons. The internalized dipeptide would be hydrolyzed by cellular peptidases to yield the Uaa Cmn for incorporation. Indeed, with this optimization we achieved efficient Cmn incorporation into neuronal proteins at multiple regions of the embryonic mouse brain, including neocortex, thalamus and hypothalamus21.
Successful PIRK expression in neurons depends strongly on the preparation of healthy neuronal culture. Every step in the protocol should be performed in a precise and meticulous manner. After culture preparation, it is best to maintain the culture in the incubator without disturbance. Opening and closing the incubator door, as well as moving the culture dish in and out of the incubator, can add up to undermining the quality of culture. Lower incubator temperature (35 °C instead of 37 °C) helps reduce excitotoxicity. On a similar note, Ca-P transfection should be done with extra caution. Preparation of 2x BBS buffer is a critical step. It is a good idea to calibrate the optimal pH for 2x BBS buffer between pH 6.90-7.15, since new DNA constructs or preps could change transfection conditions. The buffer pH could be adjusted with precision to the 0.01 digits if it helps to achieve consistent results. Prior to transfection, the growth media of neuronal culture is replaced with fresh one. It is crucial to save the original growth media, and add back to the culture after transfection to restore the original condition. Maintaining neuron culture in fresh media after transfection would interfere with neurons from recovering, since it lacks key molecules for cell proliferation such as growth factors released from neurons.
For light activation of PIRK in cultured cells and tissue slices, proper light source and delivery method need to be determined. Cmn absorbs long and medium wave UV lights (280-400 nm). However, extended exposure to UV light, especially to shorter wavelength UV light, would be harmful to cells. At the same time, far-red light could heat up the specimen perturbing the cells. Therefore, an LED with a single-wavelength emission is best for PIRK activation. For Cmn photolysis, an LED with emission of 385 nm (~40 mW; Prizmatix) was selected. The LED is externally installed near the microscope, and an optical fiber is used to deliver light precisely to the neuron in focus. For reproducible data acquisition, the optical fiber is set up 1 cm away at a 45° angle from the neuron in focus. Light power at the sample was measured 40 mW/cm2. It is also recommended to periodically check the LED performance using an optical power meter.
We demonstrate that in utero electroporation is an effective technique to genetically incorporate Uaas in vivo. In utero electroporation of plasmid DNAs into the mouse embryonic neocortex is performed as described previously34, with minor modifications. To express PIRK proteins in neonatal mouse brains, two surgical procedures are required (Figure 5B). The first step is to inject gene constructs for PIRK expression followed by electroporation. Two days later, the second brain injection is performed to deliver Cmn-Ala. It would require practice to proficiently perform surgeries without stressing the animals too much. After each surgery, the animals should be looked after and checked on a regular basis throughout the recovery. Sufficient and timely availability of Cmn in the brain is essential for proper PIRK expression. 2-5 µl of Cmn-Ala (500 mM) is typically injected to an embryonic brain. Sometimes, it helps to inject Cmn-Ala in the ventricle on both the electroporated and the opposite hemisphere, since Cmn-Ala from the opposite side would diffuse to the electroporated side for extended Cmn supply. Considering general utility of in utero electroporation, this technique could be applied not only to neocortical neurons but also to neurons in other brain regions such as striatum, diencephalon, and cerebellum, when electroporation/injection site is adjusted for each region. Embryonic/neonatal brains are much softer than adult brains, so it would be difficult to get acute slices for electrophysiology experiments. Agarose embedding helps stabilize the embryonic brain structure for vibratome cutting. Although low melting point agarose minimizes temperature shock to brain tissues, caution is needed when pouring melted agarose over cold brains. The effect of temperature shock to the tissue from agarose embedding was unnoticeable during the whole cell recording.
Genetically encoding photoresponsive Uaas in cultured neurons and in vivo, exemplified by PIRK here, will afford an optogenetic technique to control various neuronal protein activities with light in their native environment. The current protocol presents a step-by-step procedure to perform PIRK expression and activation experiments in neuronal culture in vitro and in rodent brains in vivo, representing the first successful development of the Uaa system for use in mammalian brains. It has potential to benefit research of many natural proteins, as well as their implication in diseases. For example, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors and N-methyl-D-aspartate (NMDA) receptors share similar transmembrane structures with Kir2.1 channels. It would therefore be feasible to apply PIRK methodology to these ligand-gated ion channels. Moreover, PIRK technique could also be utilized to address the pathophysiology of Kir2.1 related genetic diseases, such as Andersen's syndrome and cardiac short QT syndrome35,36. In addition to "block-and-release" Cys via Cmn, multiple amino acids such as tyrosine, serine, lysine, glutamate, aspartate, and glycine have been caged with different photoreleasable groups37. Thus, similar strategies can be used to photo-regulate these amino acids in neurons in vitro and in vivo. Furthermore, reversible optical control can be achieved with azobenzene-containing Uaas, and such Uaas have now been genetically encoded in E. coli and mammalian cells38,39.
In summary, this protocol presents a general method to control the activity of neuronal proteins in their native settings with light. In comparison to other optogenetic methods involving opsin-family and other light sensitive proteins or domains, this method uses genetically encoded light-sensitive Uaas and changes only a single residue. Therefore, it should have minimal interference to the function, trafficking and localization of the target protein under study. High site-selectivity can also be achieved with these genetically encoded light-responsive Uaas to provide greater flexibility and specificity for detailed molecular investigation. This protocol also provides the first comprehensive, easy-to-follow procedure how to successfully incorporate Uaas into proteins in neurons in vitro and in vivo. Optical control of general proteins via genetically encoded Uaas in neurons in vitro and in vivo has a great potential to benefit both basic and translational science, and this protocol would help promote its application.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
We thank Dr. S. Szobota for helpful discussion on neuron culture and Ca-P transfection. L.W. acknowledges support from The Salk Innovation Grant, the California Institute for Regenerative Medicine (RN1-00577-1), and the National Institutes of Health (1DP2OD004744).
Materials
| Cover Glasses, Circles, 12 mm, Thickness 0.13-0.17 mm | Carolina Biologicals | 633029 |
| Corning BioCoat Poly-D-Lysine | Corning Discovery Labware | 354210 |
| D-(+)-Glucose solution | Sigma-Aldrich | G8769 |
| Iris Spatula-curved | Fine Science Tool | 10092-12 |
| Dissecting Knife – Fine Angled Tip | Fine Science Tool | 10056-12 |
| GlutaMAX-I Supplement | Life Technologies | 35050-061 |
| MITO+ Serum Extender | BD Biosciences | 355006 |
| Falcon 40 µm Cell Strainer | Corning Life Sciences | 352340 |
| BES (N,N-Bis(2-hydroxyethyl)-2-aminoethanesulfonic acid, N,N-Bis(2-hydroxyethyl)taurine) | Sigma-Aldrich | B6420 |
| LED LIGHT SOURCE – Black LED 385 | Prizmatix Ltd. | |
| Agarose, Low Melting Point, Analytical Grade | Promega | V2111 |
Referenzen
- Fork, R. L. Laser Stimulation of Nerve Cells in Aplysia. Science. 171 (3974), 907-908 (1971).
- Callaway, E. M., Katz, L. C. Photostimulation using caged glutamate reveals functional circuitry in living brain slices. Proc. Natl. Acad. Sci. USA. 90 (16), 7661-7665 (1993).
- Cambridge, S. B., et al. Doxycycline-dependent photoactivated gene expression in eukaryotic systems. Nat. Methods. 6 (7), 527-531 (2009).
- Yoshimura, Y., Dantzker, J. L., Callaway, E. M. Excitatory cortical neurons form fine-scale functional networks. Nature. 433 (7028), 868-873 (2005).
- Bernstein, J. G., Boyden, E. S. Optogenetic tools for analyzing the neural circuits of behavior. Trends Cogn. Sci. 15 (12), 592-600 (2011).
- Fenno, L., Yizhar, O., Deisseroth, K. The development and application of optogenetics. Annu. Rev. Neurosci. 34 (1), 389-412 (2011).
- Yizhar, O., Fenno, L. E., Davidson, T. J., Mogri, M., Deisseroth, K. Optogenetics in neural systems. Neuron. 71 (1), 9-34 (2011).
- Banghart, M., Borges, K., Isacoff, E., Trauner, D., Kramer, R. H. Light-activated ion channels for remote control of neuronal firing. Nat. Neurosci. 7 (12), 1381-1386 (2004).
- Volgraf, M., et al. Allosteric control of an ionotropic glutamate receptor with an optical switch. Nat. Chem. Biol. 2 (1), 47-52 (2006).
- Szobota, S., Isacoff, E. Y. Optical control of neuronal activity. Annu. Rev. Biophys. 39 (1), 329-348 (2010).
- England, P. M., Lester, H. A., Davidson, N., Dougherty, D. A. Site-specific, photochemical proteolysis applied to ion channels in vivo. Proc. Natl. Acad. Sci. USA. 94 (20), 11025-11030 (1997).
- England, P. M., Lester, H. A., Dougherty, D. A. Mapping disulfide connectivity using backbone ester hydrolysis. Biochemie. 38 (43), 14409-14415 (1999).
- Philipson, K. D., Gallivan, J. P., Brandt, G. S., Dougherty, D. A., Lester, H. A. Incorporation of caged cysteine and caged tyrosine into a transmembrane segment of the nicotinic ACh receptor. Am. J. Physiol. Cell. Physiol. 281 (1), C195-C206 (2001).
- Tong, Y., et al. Tyrosine decaging leads to substantial membrane trafficking during modulation of an inward rectifier potassium channel. J. Gen. Physiol. 117 (2), 103-118 (2001).
- Liu, C. C., Schultz, P. G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 79 (1), 413-444 (2010).
- Wang, L., Brock, A., Herberich, B., Schultz, P. G. Expanding the genetic code of Escherichia coli. Science. 292 (5516), 498-500 (2001).
- Wang, L., Xie, J., Schultz, P. G. Expanding the genetic code. Annu. Rev. Biophys. Biomol. Struct. 35 (1), 225-249 (2006).
- Wang, Q., Parrish, A. R., Wang, L. Expanding the genetic code for biological studies. Chem. Biol. 16 (3), 323-336 (2009).
- Shen, B., et al. Genetically encoding unnatural amino acids in neural stem cells and optically reporting voltage-sensitive domain changes in differentiated neurons. Stem Cells. 29 (8), 1231-1240 (2011).
- Wang, W., et al. Genetically encoding unnatural amino acids for cellular and neuronal studies. Nat. Neurosci. 10 (8), 1063-1072 (2007).
- Kang, J. Y., et al. In vivo expression of a light-activatable potassium channel using unnatural amino acids. Neuron. 80 (2), 358-370 (2013).
- Bichet, D., Haass, F. A., Jan, L. Y. Merging functional studies with structures of inward-rectifier K(+) channels. Nat. Rev. Neurosci. 4 (12), 957-967 (2003).
- Burrone, J., O’Byrne, M., Murthy, V. N. Multiple forms of synaptic plasticity triggered by selective suppression of activity in individual neurons. Nature. 420 (6914), 414-418 (2002).
- Johns, D. C., Marx, R., Mains, R. E., O’Rourke, B., Marban, E. Inducible genetic suppression of neuronal excitability. J. Neurosci. 19 (5), 1691-1697 (1999).
- Sambrook, J., Russell, D. W. . Molecular Cloning: A laboratory manual. , (2001).
- Beaudoin, G. M., et al. Culturing pyramidal neurons from the early postnatal mouse hippocampus and cortex. Nat. Protoc. 7 (9), 1741-1754 (2012).
- Molleman, A. . Patch Clamping: An Introductory Guide To Patch Clamp Electrophysiology. , (2003).
- Walantus, W., Castaneda, D., Elias, L., Kriegstein, A. In utero intraventricular injection and electroporation of E15 mouse embryos. J. Vis. Exp. (6), e239 (2007).
- Lemke, E. A., Summerer, D., Geierstanger, B. H., Brittain, S. M., Schultz, P. G. Control of protein phosphorylation with a genetically encoded photocaged amino acid. Nat. Chem. Biol. 3 (12), 769-772 (2007).
- Coin, I., et al. Genetically encoded chemical probes in cells reveal the binding path of urocortin-I to CRF class B GPCR. Cell. 155 (6), 1258-1269 (2013).
- Takimoto, J. K., Xiang, Z., Kang, J. Y., Wang, L. Esterification of an unnatural amino acid structurally deviating from canonical amino acids promotes its uptake and incorporation into proteins in mammalian cells. Chembiochem. 11 (16), 2268-2272 (2010).
- Parrish, A. R., et al. Expanding the Genetic Code of Caenorhabditis elegans Using Bacterial Aminoacyl-tRNA Synthetase/tRNA Pairs. ACS Chem. Biol. 7 (7), 1292-1302 (2012).
- Lu, H., Klaassen, C. Tissue distribution and thyroid hormone regulation of Pept1 and Pept2 mRNA in rodents. Peptides. 27 (4), 850-857 (2006).
- Tabata, H., Nakajima, K. Efficient in utero gene transfer system to the developing mouse brain using electroporation: visualization of neuronal migration in the developing cortex. Neurowissenschaften. 103 (4), 865-872 (2001).
- Plaster, N. M., et al. Mutations in Kir2.1 cause the developmental and episodic electrical phenotypes of Andersen’s syndrome. Cell. 105 (4), 511-519 (2001).
- Priori, S. G., et al. A novel form of short QT syndrome (SQT3) is caused by a mutation in the KCNJ2 gene. Circ. Res. 96 (7), 800-807 (2005).
- Beene, D. L., Dougherty, D. A., Lester, H. A. Unnatural amino acid mutagenesis in mapping ion channel function. Curr Opin Neurobiol. 13 (3), 264-270 (2003).
- Hoppmann, C., et al. Genetically encoding photoswitchable click amino acids in Escherichia coli and mammalian cells. Angew Chem Int Ed Engl. 53 (15), 3932-3936 (2014).
- Hoppmann, C., et al. In Situ Formation of an Azo Bridge on Proteins Controllable by Visible Light. J Am Chem Soc. 137 (35), 11218-11221 (2015).

