A Miniaturized Glycan Microarray Assay for Assessing Avidity and Specificity of Influenza A Virus Hemagglutinins
Summary
Using a printed glycan microarray strategy, a conventional 96-well plate assay was miniaturized for analysis of influenza A virus hemagglutinin avidity and specificity for sialic acid containing receptors.
Abstract
Influenza A virus (IAV) hemagglutinins recognize sialic acids on the cell surface as functional receptors to gain entry into cells. Wild waterfowl are the natural reservoir for IAV, but IAV can cross the species barrier to poultry, swine, horses and humans. Avian viruses recognize sialic acid attached to a penultimate galactose by a α2-3 linkage (avian-type receptors) whereas human viruses preferentially recognize sialic acid with a α2-6 linkage (human-type receptors). To monitor if avian viruses are adapting to human type receptors, several methods can be used. Glycan microarrays with diverse libraries of synthetic sialosides are increasingly used to evaluate receptor specificity. However, this technique is not used for measuring avidities. Measurement of avidity is typically achieved by evaluating the binding of serially diluted hemagglutinin or virus to glycans adsorbed to conventional polypropylene 96-well plates. In this assay, glycans with α2-3 or α2-6 sialic acids are coupled to biotin and adsorbed to streptavidin plates, or are coupled to polyacrylamide (PAA) which directly adsorb to the plastic. We have significantly miniaturized this assay by directly printing PAA-linked sialosides and their non PAA-linked counterparts on micro-well glass slides. This set-up, with 48 arrays on a single slide, enables simultaneous assays of 6 glycan binding proteins at 8 dilutions, interrogating 6 different glycans, including two non-sialylated controls. This is equivalent to 18x 96-well plates in the traditional plate assay. The glycan array format decreases consumption of compounds and biologicals and thus greatly enhances efficiency.
Introduction
Wild waterfowl are the natural reservoir for IAV, but IAV is able to cross the species barrier to poultry and mammals, including, humans. Avian IAVs recognize α2-3 linked sialic acids (avian-type receptors), whereas human viruses bind α2-6 linked sialic acids (human-type receptors). To be able to efficiently replicate and transmit between humans an avian IAV needs to bind to human-type receptors 1.
IAVs are divided based on serology that characterizes the antigenicity of their hemagglutinin (HA) and neuraminidase (NA) envelope glycoproteins. HA binds to sialic acids, whereas NA is the receptor-destroying enzyme at the other end of the viral lifecycle and cleaves sialic acid 2. All human infecting viruses, including H1N1, H2N2 and H3N2, have an avian origin 3. Over the last two of decades several avian to human crossovers have occurred, with H5N1, H7N7, and H7N9 being the most well-known; however, other subtypes have infected humans more sporadically (H6N1, H7N1, H7N2, H9N2, H10N7, H10N8) 4. Fortunately, it seems that none of these viruses have been able to fully adapt to human-type α2-6-linked sialic acid receptors 5-8. Adaptation of avian or other zoonotic viruses to accommodate replication and transmission in human hosts could have a devastating effect on human health. Therefore, prior knowledge of how these viruses are evolving to bind human-type receptors will help world-wide surveillance of emerging influenza viruses.
Determination of receptor preference was first elucidated using erythrocytes of different species and remains a favored assay among flu researchers 9-12. The demonstration that avian viruses recognize α2-3 sialic acid and human viruses α2-6 linked sialic acid was originally based on an assay using hemagglutination of erythrocytes enzymatically engineered to contain each of the linkages 13,14. Although the readout is hemagglutination, a standard assay for virologists, the underlying glycan structures are not defined, only the terminal linkage. Additionally, the limited availability of the sialyltransferases, used to re-sialylate the cells, have limited the use of this assay 15-18. Subsequently, other methods of determining receptor-binding preferences were introduced using sialylated glycan structures linked to poly-acrylamide (PAA) or poly-glutamate (PGA) structures in plate-based assays 19,20. Several variations are possible in coating either the glycans or viruses to microtiter plates, each of which results in a robust, reliable and very sensitive ELISA-type assay 21-23. Alternatively, biotin-linked glycans can replace PAA/PGA and can be conjugated to streptavidin-coated plates 2,24. Although some specific sera may be required, ELISAs are standard and several glycans linked to PAA are readily commercially and non-commercially available (Consortium for Functional Glycomics (http://www.functionalglycomics.org)).
Glycan microarray technologies have emerged as an invaluable tool to determine receptor specificity, as multiple different glycans are spotted, and binding to a wide array of different structures can be assessed within a single assay 25-29. The binding of IAV to these structures provides a better understanding of the glycan structures that IAV preferentially recognizes 30-33. Glycan microarrays require small amounts of sample volume to perform a binding assay and only uses minute amounts of glycan per spot (2 nl). However, these arrays are typically used only to evaluate specificity of glycans receptors. Analysis of multiple viruses, or hemagglutinin proteins, at multiple concentration ranges can be prohibitive due to the number of slides required. Furthermore, to date, no relative avidity assay has been developed using glycan array techniques.
To combine the low sample requirement afforded by glycan microarray techniques and the sensitivity of PAA-linked glycans in ELISA-based assays, we sought to develop a multi-well glycan array that would allow for high-throughput analysis with similar or better resolution as compared to the traditional ELISA-based assay. Simultaneously, we wanted to minimize the amount of biologicals and reporter chemicals consumed. The ultimate result is a miniaturized avidity assay, specifically developed for monitoring IAV specificity and is equally applicable to assess other glycan binding proteins. Using a glass slide separated into 48 micro-wells by a Teflon mask, 6 different glycans are spotted in 6 replicates per well. The microarray platform affords the same trends in receptor binding seen in the macro ELISA format with several advantages. These include (I) printing of compound in 6 replicates, using minimal sample, versus the coating of several rows in a plate, using 100 µl per well; (II) multiple different compounds analyzed simultaneously in a single well, including controls; (III) a massive decrease in incubation volume and; (IV) a larger dynamic range using a fluorescent readout. A single slide can be calculated to be equivalent to 18x 96-well plates.
With the following protocol, any lab capable of fabricating and analyzing spotted microarrays should be able to manufacture this miniaturized ELISA format.
Protocol
NOTE: All steps are carried out at room temperature unless otherwise indicated.
1. Array Construction
- Glycan Preparation and Plate Setup
- Prepare a stock of printing buffer. First make 500 ml of a 150 mM stock solution of dibasic sodium phosphate. Also make 50 ml of a 150 mM stock solution of monobasic sodium phosphate solution. In a flask, using a magnetic stir bar and pH meter, slowly titrate the dibasic sodium phosphate solution with the monobasic solution until a pH of 8.5 is reached. Add Tween-20 to 0.005%.
- Prepare the glycan samples. Dilute PAA conjugated glycans, (diLacNAc (Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ-PAA), 3SLNLN (Neu5Acα2-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ-PAA) and 6SLNLN (Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ-PAA (Figure 1A)) are to be diluted to 100µg/ml in printing buffer from step1.1.1. Monovalent glycans, diLacNAc (Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ-(CH2)3NH2, 3SLNLN (Neu5Acα2-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ–(CH2)3NH2) and 6SLNLN (Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ–(CH2)3NH2)) to 100µM in printing buffer from step 1.1.1. Finally, prepare the amine-functionalized dyes, to use as grid markers/landing lights. Dye marker spots are printed at 1 µM.
NOTE: Atto488-NHS dye is used in our prints. However, any amine-functionalized dye, readable by the slide scanner is appropriate for this purpose. - Transfer 10 µl of each glycan sample to 384-well microtiter plates, used for the printing robot. Store unused glycans in printing buffer solution at -20 °C in sealed tubes. Tansfer 10 µl of the dye marker to the printing plate.
- Printing
NOTE: All steps that concern touching or moving the slides must be done with gloves.- Place arrayer pins in the printing head according to the correct configuration of the layout. For this layout use one pin to print all samples and arrays. Print the glycans in blocks of six, skipping one feature (a feature is defined as single spot of a certain compound), on each side, such that spots of the same sample are not printed exactly beside each other.
NOTE: The end user must decide the configuration. - Remove slides from bags, open boxes, and remove any dust particles or small pieces of plastic that might be on their surfaces by blowing ultra-high purity Nitrogen gas on each slide. Place the desired number of slides, coated side up, on the arrayer stage.
- Program the arrayer, use the parameters of 27 spots per visit to the source plate, using 3 "pre-spots" onto poly-L-lysine coated slides, to remove excess liquid at the tip of the printing pins
- Turn on MPU (Main Power Unit) and Climate Control Unit. Open Tiling Analysis software on computer.
- Select layout to the print. Select Options → Pin Tool to be used for printing the array 1 x 1 print tool – 1 single pin. Select the Target tab → Tool Array Definition → Print Pattern and add the pitch (spot-to-spot distance), number of rows and columns to accommodate the samples to be printed, and the printing pattern of the samples (for this array an 8 x 8 printing pattern is used at 340 µm pitch for 7 total samples).
- Select Target → Slide Layout and program the block-to-block distance to allow printing of each replicate array into the 48-well (4 x 12) slide template. Select Source and add the number of source pickups (1 for each sample when using 1 pin) (for this print 7 source pickups will be used to accommodate 6 sample wells and 1 dye marker well).
NOTE: The source dialogue should turn GREEN to show that the number of source visits matches the samples to be printed in the print pattern. - Select Target → Adapter Plate and Slide Layout and adjust the slide number (5 slides are printed at a time for this print with 1 pre-spotting slide).
- Release the tray by clicking the T1 button to activate the tray and add the desired number of slides to the tray, paying close attention to the locations of the pre-spotting slide (BLUE) and "real" slides.
- Place plain glass slides on all remaining vacuum ports to block the vacuum and click OK to activate the vacuum. Check that all slides are held in place and click OK to return the tray to the home position.
- Load the printing plate into the plate holder and ensure the rack is set correctly in place. Click GO and allow the printing process to proceed.
- Print the remaining 24 spots in replicates of 6 spots per array (4 arrays/source visit).
- Load the 384-well plates into the printer and begin printing. Maintain relative humidity, while printing, throughout the print between 55 and 65%. The printer stops after printing 5 slides.
NOTE: While the microarray in this assay is printed in a specific 8 x 8 pattern, different microarray printing robots will require equipment specific programming to achieve the desired layout. It is up to the end user to determine the most appropriate pattern given their equipments’ specifications.
- Place arrayer pins in the printing head according to the correct configuration of the layout. For this layout use one pin to print all samples and arrays. Print the glycans in blocks of six, skipping one feature (a feature is defined as single spot of a certain compound), on each side, such that spots of the same sample are not printed exactly beside each other.
- Humidification/Immobilizing
NOTE: Once the slides have finished printing, they undergo a temporary immobilization step, also called humidifying. Humidifying post-print helps to homogenize the sample attachment by re-wetting the spots and ensuring complete blocking.- Put slides in a 100% humidity chamber immediately after printing for a period of 30 min. Construct the chamber by placing very wet paper towels flat in the bottom of a large glass dish, placing the slides on some sort of rack, such as a test tube rack, print side up, and sealing with plastic wrap to trap in the moisture.
- Number slides with an alcohol/solvent resistant marking pen and store in a desiccation box. Desiccate overnight.
- Numbering, Blocking, and Storage
- Prepare the blocking buffer – 50 mM ethanolamine in 50 mM borate buffer. First, dissolve boric acid, in ddH2O, to a final concentration of 50 mM in a flask containing a magnetic stir bar. Stir the solution vigorously on a stirring plate until all solid has dissolved. While constantly stirring, add the appropriate amount of ethanolamine from a 16.54 M stock solution to reach the desired 50 mM concentration. Add concentrated sodium hydroxide to adjust to a final pH of 9.2.
- Immerse the printed slides in the blocking buffer solution for 1 hr.
- Following 1 hr, rinse the slides in ddH2O to remove any excess traces of blocking buffer. Dispose blocking buffer in an appropriate waste container.
- Transfer the slides to glass staining holders and spin dry. Dry arrays by placing them in a centrifuge equipped with swinging plate holders, at a speed of 10 x g for 5 min.
- Once the slides are dry, they can be incubated immediately or stored. Storage conditions are -20 °C in a sealed plastic bag.
2. Analyzing Glycan Binding Proteins
- Prepare a humidified chamber in which the arrays to be hybridized will fit. A simple chamber consists of a Pyrex dish, layered on the bottom with dampened paper towels and a rack, upon which the slides will rest.
- Use biotinylated lectins SNA (Sambuca nigra agglutinin) and ECA (Erythrina cristagalli agglutinin) at 10 μg/ml + 2 µg/ml of Streptavidin-555 dye, in probing buffer (PBS + 0.01% Tween-20). For HA (A/Vietnam/1203/04 H5N1 and A/KY/07 H1N1, in this example), use a starting concentration of 20 μg/ml pre-complexed, as previously described 34. Briefly, make a HA:Mouse-α-Strep:Goat-α-Mouse-Alexa647 mixture in blocking buffer (PBS + 3%BSA + 0.01% Tween-20), at a 4:2:1 molar ratio.
NOTE: The array is probed with lectins to ascertain that glycans are immobilized and detectable (Figure 1B). - Transfer the glycan binding protein-antibody mix solution of 20 µl to a 384-well plate and incubate on ice for 30 min. Serially dilute lectin and HA samples 1:1, in their appropriate buffer, 8 times, by mixing 10 µl of sample with 10 µl of buffer.
- Pipette 8 µl of the sample onto the micro-well surface and incubate in the sealed humidification (100% RH) chamber for 90 min.
- Wash the slides one at a time by holding the edges and dipping them four times in a mixture of PBS and 0.05% Tween, then four times in PBS, and finally four times in deionized H2O. Using the same drying protocol described in the blocking step, spin the arrays dry in a centrifuge for 5 min at 10 x g.
3. Scanning and Data Analyses
NOTE: The glycan microarrays can be scanned at different settings, depending on the microarray scanner’s manufacturer. By varying the laser power and resolution, the instrument can produce an image with as many signals as possible within its dynamic range (Figure 1C).
- Use a fluorescent slide scanner and scan the arrays immediately after incubation with the glycan binding proteins. Take care to ensure the slide is correctly inserted into the scanning instrument.
- Open scanning software. Click start to open the program. Connect to scanner: Scanner → Connect or green button in the Scanner navigation panel menu.
- Open autoloader door of the scanner. Place slides in slots in autoloader in specific order. Close autoloader door. Click "read loader" in the Tools navigation panel to auto detect slides in the scanner.
- Set scan parameter: Scanner → Scan parameters appropriate to the experiment (e.g. 635 laser power high, detection gain 50%). Scan slides: Scanner → scan. Select path by creating a folder to save the scan images and name individual scans. Do this prior to the actual scanning. Repeat for each individual slide. Click OK to start scanning.
- Use the scanner software to set up the instrument and scan the slides. Set scanner to iteratively scan at low laser power (5 mW) with increasing gain settings of 25%, 50% and 75%.
NOTE: The settings can be tested and optimized, but keep in mind that each scan can possibly lower the fluorescence output of the dyes due to photobleaching.- Open data acquisition and analysis software such as Mapix software. Click start to open the program. Open scan image: File → open image. Open GAL file: Analyze → open grid and select the appropriate GAL file.
- Enter blocks mode: Image → blocks mode to move the grid. Use the grid markers to set the grid to the appropriate location on the slide. Adjust individual blocks as necessary to match the printed array.
- Enter spots mode: Image → spots mode to adjust location and size of individual spots within the block. Once all blocks and spots are adjusted and in place, measure signal intensities by Analyze → photometric calculations. Save the output file.
- Once the slides have finished scanning, images are analyzed for binding signals with the image-processing program installed (Figure 1D–G). For image analysis, load a GAL (Array List) file over the scanned TIFF image.
NOTE: The GAL file contains both the spot layout pattern and identities of each spot location. GAL files for arrays are created by the printer software using a tab-delimited text file that contains the identities of the samples and their location in the 384-well source plate. - Use the grid marker/landing lights printed on the microarrays to properly place the GAL grid. Individual spots may need to be moved individually, but a well-printed array will generally allow blocks within the grid to be placed with ease. Following grid placement, collect spot intensities by the software and saved as a tab-delimited text file (.txt).
- Using spreadsheet program, open the output file from the scanner. Open the appropriate macro file from the folder location. Click View → Macro → RunMacro.
NOTE: The pre-written will copy the raw output data, remove unnecessary rows and columns, sort the data by sample, calculate the average mean signal minus background values and plot the resultant values in to an spread-sheet worksheet containing the graphs for each of the six samples tested on the array.
- Using spreadsheet program, open the output file from the scanner. Open the appropriate macro file from the folder location. Click View → Macro → RunMacro.
Representative Results
Printing, Scanning & Data analyses
To ensure proper printing, it is vital to have the correct alignment of the spotted grid within the teflon mask, which delineates each array on the slide. During printing, due to the nature of the teflon coating, spots cannot be seen by the naked eye on the MPX slides. Attention is paid then to the appearance of the pre-spots on the poly-L-Lysine coated slides. Directly after printing, each slide should be checked by eye for the presence of each spotted compound that is visible due to the buffer's salts dried in the spot. Arrays are inspected for correct alignment of spots within the Teflon borders and correct number of spotted features.
Slides are scanned in an iterative process of lower to higher scan power to avoid photo bleaching and allowing the comparison of higher avidity to lower avidity binding proteins, which have higher and lower output signals, respectively. Following scanning, the fluorescent intensity of each spot in the output images are measured using an imaging program. Each array is overlayed with a grid file that matches the number of features printed on the array surface (Figure 1D–G). Using printed fluorescent dyes, the borders of the printed glycans are defined and a mask that is integrated with the identity of the printed sample lassoes each spot. The imaging mask lasso will record all aspects of the imaged spot (size, signal intensity, coordinates in the grid, etc.) and output this information to a tab-delimited text file. Using tabulation software (spreadsheet), the samples are sorted by identity and the mean signal intensity minus background is averaged over 4 of the 6 spots for each sample, leaving out the highest and lowest signal as outliers. A line graph of average mean signal minus background is plotted against the eight protein concentrations applied to the array and contains one line for each sample. The final output graph is smoothed using a non-linear regression calculation (Figure 1I).
Glycan binding proteins
The PAA-Array is used to assess receptor-binding specificity of influenza A virus hemagglutinins. An additional feature of our array, not present in the analogous plate assay, is the inclusion of non-sialylated controls in the same micro-well. To assess that printed compounds are present after printing, commonly used plant lectins with known specificity were used. ECA binds to terminal galactose linked β1-4 to N-acetyl-glucosamine (Galβ1-4GlcNAc or LacNAc) and will not bind if the terminal galactose is capped with sialic acid. ECA only detects the non-sialylated glycans on our miniaturized glycan array (Figure 2A). The lack of binding to any of the sialylated glycan samples is also indicative that these are fully capped with terminal sialic acid. For α2-6 linked sialic acid, SNA was used as a lectin control (Figure 2B). As expected, SNA only binds to α2-6 linked sialic acids, but with notable differences in affinity for PAA-linked non-PAA-linked di-LacNAc repeats. This difference reflects the sensitivity of PAA-linked glycans and allows the discrimination of proteins that may bind with low avidity. For α2-3 linked sialic acid, an H5 hemagglutinin derived from A/Vietnam/1203/04 virus that is known to recognize α2-3 linked sialic acids only was used 6. The binding profile of the H5 HA indeed shows specificity for α2-3 linked sialic acids, with a higher avidity for the PAA-linked structures (Figure 2C). Finally, the H1 hemagglutinin from a human seasonal H1N1 strain was used that, as expected, only bound to α2-6 linked sialic acid containing structures (Figure 2D).
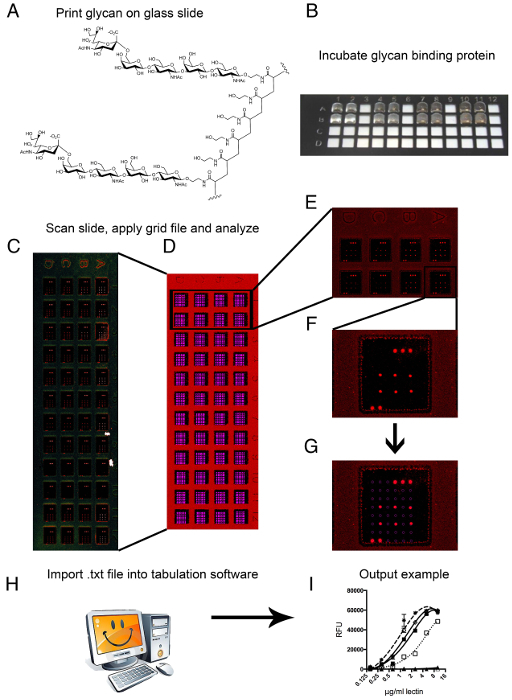
Figure 1: Printing, scanning and image analyzes. (A) PAA-conjugated 6SLNLN is shown as a representative glycan structure that is printed on the glass slides. (B) Incubating a glycan binding protein on the multi-well glycan array is shown with an 8 µl volume that creates a droplet on the array surface. (C) Following incubation of the glycan binding proteins on the array and scanning in a confocal fluorescent slide scanner, a representative image is obtained. (D) The image is overlayed with a grid and, using grid markers for proper alignment, the single spots can be analyzed. (E) A close up of a complete set of 8 arrays, in which a single glycan binding protein was analyzed using eight 1:1 dilutions. (F) A single array, in a single well is represented; the array is demarked by the grid markers in the top right (3 spots) and bottom left (2 spots), this glycan binding protein binds a single compound on the array as a replicate of six is clearly visible. (G) The grid is shown with lassos encircling specific individual spots. (H) Imaging software calculates signal values from the image file and outputs a tab-delimited data file. (I) The data can be tabulated in spreadsheet or stastical software to create a representative output graph. Please click here to view a larger version of this figure.
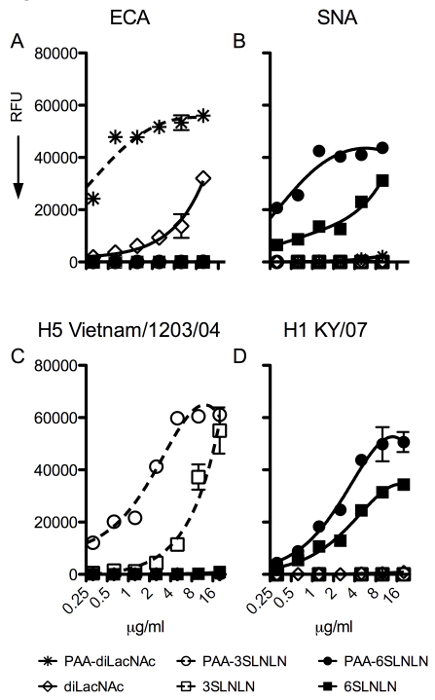
Figure 2: Output of plant lectins (ECA, SNA) and IAV hemagglutinins with different specificities (H5 from A/Vietnam/1203/04, H1 from A/Kentucky/07). (A) ECA binds terminal LacNAc or Galb1-4GlcNAc specifically and detects the presence of the non-sialylated compounds, used as controls for IAV hemagglutinins. (B) SNA recognizes only glycans bearing a terminal α2-6 sialic acid. (C) The recombinant hemagglutinin of the H5N1 (A/Vietnam/1203/04) strain binds to α2-3 sialic acids and provides an avian type receptor binding profile. (D) The recombinant hemagglutinin of a human seasonal H1N1 (A/Kentucky/07) binds to human-type receptors only. Fluorescent signal intensity was measured for each spot, and mean intensity minus mean background was calculated using spreadsheet program. For each glycan, the mean signal intensity is calculated from 6 replicates spots. The highest and lowest signals of the 6 replicates are removed and the remaining 4 replicates are used to calculate the mean signal, standard deviation (SD), and standard error measurement (SEM). The graphs represent the averaged mean signal minus background for each sample and error bars are the SEM value. Please click here to view a larger version of this figure.
Discussion
Assessing IAV receptor specificity is an important step in analyzing pandemic potential of avian viruses. Sialic acid recognition by the virus is linked to several biological properties such as binding to and release from the cell. Knowledge of which amino-acid mutations are necessary for avian viruses to achieve α2-6 binding and cross the species barrier enables pandemic preparedness. Several assays are used to determine receptor specificity; however, all have their drawbacks, including only measuring avidity and not specificity and vice versa.
Here we describe a novel miniaturized tool based on the widely used and accepted ELISA, which uses PAA-linked terminal fragments of more complex glycans. Using 4 well-defined lectins, 2 of plant and 2 of influenza A virus origin, we show that we maintain specificity and relative avidity.
When we calculate the amount of reduction of chemicals and biological used by miniaturizing the ELISA onto a glycan array chip, we reach strikingly large differences. First, the biologicals; we use 10x less assay volume and use 6 test compounds in 6 replicates; this can be calculated to be the equivalent of 36x rows in 96-well plates. The result is that we use 360x less biologicals. Critical steps include proper printing and the risk of high backgrounds using unpurified glycan binding agents or poor detecting antibodies.
The compounds used in this assay, although still available, are not easily made by chemo-enzymatic synthesis. Using microarray-printing technology, each spot is only 2 nl, compared to 100 µl per well of a microtiter plate. Printing a single slide containing 48 wells and 6 compounds, in 6 replicates, we consume approximately 1,152 nl. The same coating procedure in 96-well plates results in coating a total of 1,842 wells with 100 µl of compound in the same concentration. Therefore, printing the compound compared to the plate assay, we attain a striking reduction of 1,500x. Therefore, in total, biologicals are reduced 360-fold and glycan consumption by 1,500-fold. Using robotic liquid handlers, we envision that most steps could be automated, allowing the entire assay be used as a high-throughput screen.
Although printing microarrays and subsequent image analyses do require special machinery and tools, these instruments are not rare and are present at many universities and institutes. Printing is direct and does not require chemical alterations. Therefore, we believe that, with this minimal amount of compounds and simple printing technique, this assay could be more generally applicable. As it reduces the incubation volume, the costs of antibodies and precious biologicals, this technique will make it feasible for labs with limited resources to assess specificity of IAV or other glycan binding proteins with high fidelity while maintaining lower costs.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
This work was supported in part by the Scripps Microarray Core Facility, and a contract from the Centers for Disease Control (J.C.P.). RPdV is a recipient of a Rubicon and VENI grant from the Netherlands Organization for Scientific Research (NWO). The Consortium for Functional Glycomics (http://www.functionalglycomics.org/) funded by NIGMS grant GM62116 (J.C.P.) provided several glycans used in this study. This is publication 29113 from The Scripps Research Institute.
Materials
| NEXTERION® Slide H MPX-48 | Schott | 1091525 | Microwell slides |
| ProScanArray Plus | PerkinElmer | discontinued | confocal microarray scanner |
| Innoscan 1100AL Scanner/Mapix Software | Innopsys | – | confocal microarray scanner |
| MicroGrid II | Digilab | – | microaray printer |
| SNA | Vector Labs | B-1305 | Plant Lectin |
| ECA | Vector Labs | B-1145 | Plant Lectin |
| Anti-Strep Antibody | IBA | 2-1509-001 | Anitbody for HA binding |
| Anti-Mouse Alexa-647 | Life | A-21235 | Anitbody for HA binding |
| Tween-20 | Sigma | P2287 | detergent |
| di-basic Sodium Phosphate | Sigma | 255793 | printing buffer component |
| mono-basic Sodium phosphate | Sigma | 229903 | printing buffer component |
| poly-l-lysine solution | Sigma | P8920 | pre-spotting slide component |
| sodium hydroxide | Sigma | 221465 | pre-spotting slide component |
| ethanol | Sigma | 493546 | pre-spotting slide component |
| phosphate buffered saline | Corning | 46-013-CM | incubation/washing buffer |
| SMP4B pins | Telechem | SMP4B | printing pin |
| Compressed Nitrogen (Grade5) | Praxair | UN1066 | general dusting/drying tool |
| Boric Acid | Sigma | B6768-500G | Slide blocking reagent |
| ethanolamine | Sigma | E9508-500ML | Slide blocking reagent |
| Atto 488 | AttoTec | AD 488-91 | Gridmarker on array |
| PAA-LNLN | Consortium for Functional Glycomics | PA368 | Spotted glycans |
| PAA-3SLNLN | Consortium for Functional Glycomics | PA362 | Spotted glycans |
| PAA-6SLNLN | Consortium for Functional Glycomics | PA343 | Spotted glycans |
| LNLN | Consortium for Functional Glycomics | Te98 | Spotted glycans |
| 3SLNLN | Consortium for Functional Glycomics | Te175 | Spotted glycans |
| 6SLNLN | Consortium for Functional Glycomics | Te176 | Spotted glycans |
| 384-well microtiter plate | Matrix TechCorp | 4361 | Printing plate |
| VWR lab marker | VWR | 52877-150 | Slide Numbering |
| Wheaton slide staining dish | Sigma | Z103969-1EA | Blocking and Drying |
Referenzen
- Tumpey, T. M., et al. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science. 315 (5812), 655-659 (2007).
- Xu, R., et al. Functional balance of the hemagglutinin and neuraminidase activities accompanies the emergence of the 2009 H1N1 influenza pandemic. J Virol. 86 (17), 9221-9232 (2012).
- Bouvier, N. M., Palese, P. The biology of influenza viruses. Vaccine. 26, D49-D53 (2008).
- Freidl, G. S., et al. Influenza at the animal-human interface: a review of the literature for virological evidence of human infection with swine or avian influenza viruses other than A(H5N1). Euro Surveill. 19 (18), (2014).
- Xu, R., et al. Preferential recognition of avian-like receptors in human influenza A H7N9 viruses. Science. 342 (6163), 1230-1235 (2013).
- Paulson, J. C., de Vries, R. P. H5N1 receptor specificity as a factor in pandemic risk. Virus Res. 178 (1), 99-113 (2013).
- Tzarum, N., et al. Structure and receptor binding of the hemagglutinin from a human H6N1 influenza virus. Cell Host Microbe. 17 (3), 369-376 (2015).
- Zhang, H., et al. A Human-Infecting H10N8 Influenza Virus Retains a Strong Preference for Avian-type Receptors. Cell Host Microbe. 17 (3), 377-384 (2015).
- Carroll, S. M., Higa, H. H., Paulson, J. C. Different cell-surface receptor determinants of antigenically similar influenza virus hemagglutinins. J Biol Chem. 256 (16), 8357-8363 (1981).
- Gambaryan, A. S., et al. Specification of receptor-binding phenotypes of influenza virus isolates from different hosts using synthetic sialylglycopolymers: non-egg-adapted human H1 and H3 influenza A and influenza B viruses share a common high binding affinity for 6′-sialyl(N-acetyllactosamine). Virology. 232 (2), 345-350 (1997).
- Rogers, G. N., D’Souza, B. L. Receptor binding properties of human and animal H1 influenza virus isolates. Virology. 173 (1), 317-322 (1989).
- Rogers, G. N., et al. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 304 (5921), 76-78 (1983).
- Paulson, J. C., Rogers, G. N. Resialylated erythrocytes for assessment of the specificity of sialyloligosaccharide binding proteins. Methods Enzymol. 138, 162-168 (1987).
- Paulson, J. C., Sadler, J. E., Hill, R. L. Restoration of specific myxovirus receptors to asialoerythrocytes by incorporation of sialic acid with pure sialyltransferases. J Biol Chem. 254 (6), 2120-2124 (1979).
- Chutinimitkul, S., et al. In vitro assessment of attachment pattern and replication efficiency of H5N1 influenza A viruses with altered receptor specificity. J Virol. 84 (13), 6825-6833 (2010).
- Glaser, L., et al. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J Virol. 79 (17), 11533-11536 (2005).
- Herfst, S., et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 336 (6088), 1534-1541 (2012).
- Nobusawa, E., Ishihara, H., Morishita, T., Sato, K., Nakajima, K. Change in receptor-binding specificity of recent human influenza A viruses (H3N2): a single amino acid change in hemagglutinin altered its recognition of sialyloligosaccharides. Virology. 278 (2), 587-596 (2000).
- Gambaryan, A. S., Matrosovich, M. N. A solid-phase enzyme-linked assay for influenza virus receptor-binding activity. J Virol Methods. 39 (1-2), 111-123 (1992).
- Totani, K., et al. Chemoenzymatic synthesis and application of glycopolymers containing multivalent sialyloligosaccharides with a poly(L-glutamic acid) backbone for inhibition of infection by influenza viruses. Glycobiology. 13 (5), 315-326 (2003).
- Gambaryan, A., et al. Receptor specificity of influenza viruses from birds and mammals: new data on involvement of the inner fragments of the carbohydrate chain. Virology. 334 (2), 276-283 (2005).
- Ito, T., et al. Receptor specificity of influenza A viruses correlates with the agglutination of erythrocytes from different animal species. Virology. 227 (2), 493-499 (1997).
- Watanabe, Y., et al. Acquisition of human-type receptor binding specificity by new H5N1 influenza virus sublineages during their emergence in birds in Egypt. PLoS Pathog. 7 (5), e1002068 (2011).
- Chandrasekaran, A., et al. Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nat Biotechnol. 26 (1), 107-113 (2008).
- Blixt, O., et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci U S A. 101 (49), 17033-17038 (2004).
- Childs, R. A., et al. Receptor-binding specificity of pandemic influenza A (H1N1) 2009 virus determined by carbohydrate microarray. Nat Biotechnol. 27 (9), 797-799 (2009).
- Nycholat, C. M., et al. Recognition of Sialylated Poly-N-acetyllactosamine Chains on N- and O-Linked Glycans by Human and Avian Influenza A Virus Hemagglutinins. Angew Chem Int Ed Engl. , (2012).
- Rillahan, C. D., Paulson, J. C. Glycan microarrays for decoding the glycome. Annu Rev Biochem. 80, 797-823 (2011).
- Song, X., et al. A sialylated glycan microarray reveals novel interactions of modified sialic acids with proteins and viruses. J Biol Chem. 286 (36), 31610-31622 (2011).
- Gulati, S., et al. Human H3N2 Influenza Viruses Isolated from 1968 To 2012 Show Varying Preference for Receptor Substructures with No Apparent Consequences for Disease or Spread. PLoS One. 8 (6), (2013).
- Stevens, J., Blixt, O., Paulson, J. C., Wilson, I. A. Glycan microarray technologies: tools to survey host specificity of influenza viruses. Nat Rev Microbiol. 4 (11), 857-864 (2006).
- de Vries, R. P., et al. Evolution of the hemagglutinin protein of the new pandemic H1N1 influenza virus: maintaining optimal receptor binding by compensatory substitutions. J Virol. 87 (24), 13868-13877 (2013).
- Yang, H., et al. Structure and receptor binding preferences of recombinant human A(H3N2) virus hemagglutinins. Virology. 477, 18-31 (2015).
- de Vries, R. P., et al. The influenza A virus hemagglutinin glycosylation state affects receptor-binding specificity. Virology. 403 (1), 17-25 (2010).

