Mimicking the Function of Signaling Proteins: Toward Artificial Signal Transduction Therapy
Summary
We present guidelines for developing synthetic ‘chemical transducers’ that can induce communication between naturally unrelated proteins. In addition, detailed protocols are presented for synthesizing and testing a specific ‘transducer’ that enables a growth factor to activate a detoxifying enzyme and consequently, to regulate the cleavage of an anticancer prodrug.
Abstract
Signal transduction pathways, which control the response of cells to various environmental signals, are mediated by the function of signaling proteins that interact with each other and activate one other with high specificity. Synthetic agents that mimic the function of these proteins might therefore be used to generate unnatural signal transduction steps and consequently, alter the cell’s function. We present guidelines for designing ‘chemical transducers’ that can induce artificial communication between native proteins. In addition, we present detailed protocols for synthesizing and testing a specific ‘transducer’, which can induce communication between two unrelated proteins: platelet-derived growth-factor (PDGF) and glutathione-S-transferase (GST). The way by which this unnatural PDGF-GST communication could be used to control the cleavage of an anticancer prodrug is also presented, indicating the potential for using such systems in ‘artificial signal transduction therapy’. This work is intended to facilitate developing additional ‘transducers’ of this class, which may be used to mediate intracellular protein-protein communication and consequently, to induce artificial cell signaling pathways.
Introduction
Signal transduction pathways play a significant role in virtually every cellular process and allow the cell to rapidly respond to environmental signals.1 These pathways are often triggered by the binding of a signaling molecule to an extracellular receptor, which results in activation of intracellular enzymes. Amplification and propagation of this signal within the cell is mediated by the function of signaling proteins that form a network of protein-protein interactions in which enzymes are reversibly activated with high specificity. Because dysregulation of these networks frequently leads to cancer development, there has been much interest in establishing 'signal transduction therapy of cancer',2 whereby drugs are designed to disrupt malignant signaling pathways. We have recently proposed an alternative approach to signal transduction therapy that relies on the ability of drugs to generate unnatural signal transduction pathways.3 In particular, we believe that by designing synthetic agents that mimic the function of signaling proteins, it would be possible to modulate the cell's function indirectly. For example, these artificial networks may enable protein biomarkers to activate enzymes that cleave prodrugs. Alternatively, these signaling protein mimetics might be able to activate unnatural cell signaling pathways, resulting in therapeutic effects.
To demonstrate the feasibility of this approach, we have recently created a synthetic 'chemical transducer'4 that enables platelet-derived growth factor (PDGF) to trigger the cleavage of an anticancer prodrug by activating glutathione-s-transferase (GST), which is not its natural binding partner. The structure of this 'transducer' consists of an anti-PDGF DNA aptamer that is modified with a bivalent inhibitor for GST. Hence, this synthetic agent belongs to a family of molecules with binding sites to different proteins,5-7 such as chemical inducers of dimerization (CIDs)8-10 and also to the group of protein-binders based on oligonucleotide-synthetic molecule conjugates.11-21
The general principles underlying the design of such systems is described herein and detailed protocols for synthesizing and testing the function of this 'transducer' with conventional enzymatic assays are provided. This work is intended to facilitate developing additional 'transducers' of this class, which may be used to mediate intracellular protein-protein communication and consequently, to induce artificial cell signaling pathways.
Figure 1 schematically describes the operating principles of synthetic 'chemical transducers' that can mediate unnatural protein-protein communications. In this illustration, a 'chemical transducer', which integrates synthetic binders for proteins I and II (binders I and II), enables protein II to trigger the catalytic activity of protein I, which is not its natural binding partner. In the absence of protein II, the transducer binds the catalytic site of the enzyme (protein I) and inhibits its activity (Figure 1, state ii). The binding of the 'transducer' to protein II, however, promotes interactions between binder I and the surface of protein II (Figure 1, state iii), which reduces its affinity toward protein I. As a result, the effective concentration of the 'free' transducer in the solution is reduced, which leads to dissociation of the transducer-protein I complex and to reactivation of protein I (Figure 1, state iv). Taken together, these steps highlight three fundamental principles underlying the design of efficient 'transducers': (1) a 'transducer' should have a specific binder for each of the protein targets, (2) the interaction between binder II and protein II should be stronger than the interaction between binder I and protein I, and (3) binder I must be able to interact with the surface of protein II. This last principle does not necessarily require that binder I alone would have a high affinity and selectivity toward protein II. Instead, it is based on our recent studies which showed that bringing a synthetic molecule in proximity to a protein is likely to promote interactions between this molecule and the surface of the protein.19,22,23
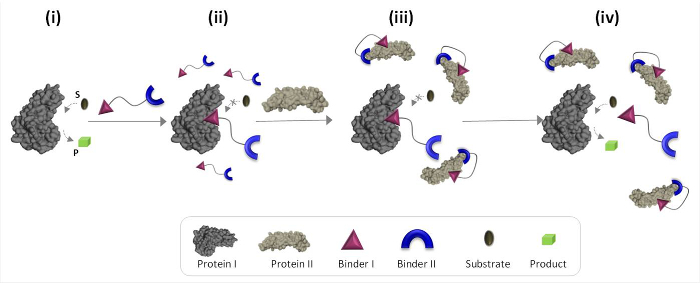
Figure 1: Operating principles of 'chemical transducers'. When the 'chemical transducer' is added to an active protein I (state i), it binds to its active site through binder I and inhibits its activity (state ii). In the presence of protein II, however, the unbound 'chemical transducer' interacts with protein II through binder II, which promotes interactions between binder I and the surface of protein II. This induced binder I-protein II interaction reduces the effective concentration of binder I, which leads to dissociation of the 'transducer'-protein I complex and to protein I reactivation (state iv). Please click here to view a larger version of this figure.
Protocol
1. Synthesis of the 'Chemical Transducer'
- Preliminary Preparations
- Prepare 2 M triethylammonium acetate (TEAA) buffer by mixing 278 ml of triethylamine with 114 ml of acetic acid and 400 ml of ultrapure water. Adjust the pH to 7 and add water to a final volume of 1 L. Keep it in a dark bottle.
Note: This solution is stable for years. - Prepare a 5 mM ascorbic acid solution by dissolving 18 mg of ascorbic acid in 20 ml of ultrapure water. Use a fresh solution; the solution is stable for one day.
- Prepare a 10 mM Cu(II)/Tris(benzyltriazolylmethyl)amine (TBTA) solution by dissolving 25 mg of copper(II) sulfate pentahydrate in 10 ml of ultrapure water and 58 mg of TBTA in 11 ml dimethyl sulfoxide (DMSO). Mix the two solutions. Keep it at room temperature and protect it from light.
- Prepare 2 M triethylammonium acetate (TEAA) buffer by mixing 278 ml of triethylamine with 114 ml of acetic acid and 400 ml of ultrapure water. Adjust the pH to 7 and add water to a final volume of 1 L. Keep it in a dark bottle.
- Conjugation Procedure
- Dissolve 100 nmol of the modified oligonucleotide (ODN-1) in 80 µl of fresh ultrapure water. Add 20 µl of 2 M TEAA, pH = 7. Add 80 µl of a freshly made solution of ascorbic acid (5 mM in water).
- Dissolve 1.5 µmol (574.5 µg) of azido-modified ethacrynic acid in 180 µl of DMSO and add it to the solution. Degas the solution using Argon for 60 sec and quickly add 40 µl from the Cu(II)/TBTA solution (10 mM in 55% (v/v) DMSO/water).
- Purge again with Argon, close tightly, and stir overnight.
- Monitor the progress of the reaction and purify the conjugate by RP-HPLC (mobile phase: A) 5% Acetonitrile, 5% TEAA, 90% ultrapure water; B) 65% Acetonitrile, 5% TEAA, 30% ultrapure water).24
2. Controlling GST Activity by PDGF
- Preliminary Preparations
- Prepare 50 ml of the Assay buffer by mixing 33.9 ml of phosphate buffered saline (PBSx1) with 16.1 ml of ultrapure water to achieve an 8 mM final phosphate concentration and add 23.8 mg of MgCl2 to achieve a 5 mM final concentration.
- Prepare a stock solution of GST M1-1 by dissolving the protein in a buffer containing 50 mM Tris pH 7.5, 50 mM NaCl, 1 mM dithiothreitol (DTT), and 5 mM ethylenediaminetetraacetic acid (EDTA) to a final concentration of 30 µM. Divide this solution into small aliquots and store at -80 ˚C. Dilute freshly, according to section 2.1.5.1, in the Assay buffer and keep on ice.
Note: The solution will be stable for about 5 hr, or until there is a reduction in the enzyme activity. - Prepare the substrate according to the following instructions:
- Dissolve 10 mg of reduced Glutathione (GSH) in 325 µl of ultrapure water to a final concentration of 100 mM stock solution. Dilute 21 µl from this stock solution in 979 µl of Assay buffer for a final concentration of 2.1 mM working solution.
- Dissolve 10 mg of 2,4-dinitrochlorobenzene (CDNB) in 492 µl of ethanol to a final concentration of 100 mM stock solution. Dilute 43.2 µl from the stock solution in 956.8 µl of an Assay buffer for a final concentration of 4.32 mM working solution.
- In a 96-well plate, put each substrate in a separate line (12 wells). Insert at least 60 µl into each well to allow fast and easy withdrawal of the solution. Cover the plate with an aluminum sheet for light protection.
- Prepare the following stock solutions in the Assay buffer:
- Dilute GST M1-1 by 50 to a final concentration of 0.6 µM of the dimer.
- Dilute the 'chemical transducer' to a 30 µM stock solution.
- Dilute PDGF to a final concentration of 40 µM.
- Dilute PDGF aptamer to a final concentration of 250 µM.
- Measure the GST Activity in the Presence of the 'chemical transducer' and PDGF.
- Set up an experimental procedure in the plate reader for kinetic measurement.
- Create a new experiment as a 'standard protocol'.
- Press on 'procedure' to open up the procedure settings window.
- In the pop up list on the upper side of the window, choose '384 plate' type according to the plate manufacturer.
- Press 'Read' on the left menu.
- Regarding the detection method choose 'Absorbance'.
- Regarding the read type choose 'End point'.
- Write 340 nm on the wavelength window.
- Press on the 'Full plate' bottom on the upper-right side and choose the well to be measured.
- Press 'ok' to close the 'Read' window.
- Choose 'start kinetic' on the left menu.
- Make the run time 10 min.
- Select the minimum intervals option.
- Press 'ok' to close the kinetic window.
- Drag the 'Read' line into the kinetic measurement.
- Press the 'validate' button and then the 'ok' button.
- Save the experiment.
- Press the 'play' button. A dialog box will appear — press the 'ok' button only when the measurement should be started.
- In order to perform the triplicates experiment, prepare four samples each containing 3.25 µl of the 'chemical transducer' and 3.25 µl of GST M1-1. Add to each sample 0, 1.2, 2.4, or 4.9 µl of PDGF and 123.5, 122.3, 121.1, or 118.6 µl of the Assay buffer, respectively.
- Incubate the solution at room temperature for 10 min.
- In a 384-transparent well plate, insert 40 µl of sample to each well. Insert the samples only in the odd wells or only in the even wells in the same line to allow the use of a multi-pipettor for substrate addition.
- Using a 12-channel multi-pipettor, quickly add 10 µl from each of the substrates that were pre-prepared in the 96-well plate (section 2.1.4). Mix gently and quickly to avoid bubbles. Insert the plate into the reader and start the kinetic measurement. Since GST kinetics is quite fast, try to minimize the time between substrate addition and the beginning of the kinetic measurement.
- Set up an experimental procedure in the plate reader for kinetic measurement.
- GST Activation/Inhibition Cycles Mediated by the 'chemical transducer'.
- In order to perform the triplicates experiment, prepare 5 samples each containing 84.5 µl of Assay buffer, 3.25 µl of 'chemical transducer', and 3.25 µl of GST M1-1. Incubate at room temperature for 3 min.
- Add 3.65 µl of Assay buffer to sample 1 and 3.65 µl of PDGF to samples 2-5. Incubate at room temperature for 3 min.
- Add 3.12 µl of Assay buffer to samples 1-2 and 3.12 µl of PDGF aptamer to samples 3-5. Incubate at room temperature for 3 min.
- Add 24.4 µl of Assay buffer to samples 1-3 and 24.4 µl of PDGF to samples 4-5. Incubate at room temperature for 3 min.
- Add 7.8 µl of Assay buffer to samples 1-4 and 7.8 µl of PDGF aptamer to sample 5. Incubate at room temperature for 5 min.
- In a 384-transparent well plate, insert 40 µl of sample into each well. Insert samples only into the odd or only into the even wells in the same line.
- Using a 12-channel multi-pipettor, quickly add 10 µl from each substrate (pre-prepared in the 96-well plate). Mix gently and quickly to avoid bubbles. Insert the plate into the reader and start the kinetic measurement.
- Calculate the V0 [mOD/min] under each condition by subtracting the OD measured at 340 nm at t = 0.5 min from the OD measured at 340 nm at t = 1.5 min to assess the activation/inhibition recyclability.25
- Evaluate the Real-time Response of the 'chemical transducer' to Changes in the Environment.
- Real-time effect of PDGF addition
- Set up an experimental procedure in the plate reader for kinetic measurement.
- Repeat steps 2.2.1.1-2.2.1.10.
- Make the run time 3.5 min.
- Select the minimum intervals option.
- Press 'ok' to close the kinetic window.
- Drag the 'Read' line into the kinetic measurement.
- Choose Plate Out/In on the left menu.
- Choose the option 'plate out (no dialog)'.
- Choose the 'Delay' option on the left menu and enter 30 sec.
- Choose Plate Out/In on the left menu.
- Choose the option 'plate in (no dialog)'.
- Create a second kinetic measurement by repeating steps 2.2.1.4 – 2.2.1.14 but in section 2.2.1.11 set the kinetic to be 25 min instead of 10 min.
- Press the 'validate' button and then the 'ok' button.
- Save the experiment.
- Press the 'play' button. A dialog box will appear – press the 'ok' button only when the measurement should be started.
- Prepare two samples by mixing 1 µl of GST M1-1 and 1 µl of the 'chemical transducer' in 38 µl of Assay buffer. Insert the samples into two wells of a 384-transparent well plate, leaving an empty well between these two wells.
- Using a 12-channel multi-pipettor, quickly add 10 µl from each substrate, mix gently and quickly to avoid bubbles, insert the plate into the reader and start the kinetic measurement.
- When the plate opens up (after 3.5 min), quickly add 1.125 µl of PDGF to one of the wells, mix gently, and allow the plate to close up for the remaining kinetic measurements.
- Real-time effect of adding the PDGF aptamer
- Repeat steps 2.4.1.1-2.4.1.15.
- Prepare two samples by mixing 1 µl of GST M1-1, 1 µl of the 'chemical transducer', and 1.125 µl of PDGF in 36.9 µl of Assay buffer. Insert the samples into two wells of a 384-transparent well plate, leaving an empty well between these two wells.
- Using a 12-channel multi-pipettor, quickly add 10 µl from each substrate, mix gently and quickly to avoid bubbles, insert the plate into the reader, and start the kinetic measurement.
- When the plate opens up (after 1.5 min), quickly add 1.2 µl of PDGF aptamer to one of the wells, mix gently and allow the plate to close up for the remaining kinetic measurements.
- Real-time effect of PDGF addition
- Measure JS-K Prodrug Activation by GST in the Presence of the 'chemical transducer' and PDGF.
- Set up an experimental procedure in the plate reader for kinetic measurement.
- Create a new experiment as a 'standard protocol'.
- Press on 'procedure' to open up the procedure settings window.
- In the pop up list on the upper side of the window choose '384 plate' type according to the plate manufacturer.
- Press 'Read' on the left menu.
- Regarding the detection method chose 'Absorbance'.
- Regarding the read type choose 'End point'.
- Write 305 nm on the wavelength window.
- Press on the 'Full plate' button on the upper-right side and choose the well to be measured.
- Press 'ok' to close the 'Read' window.
- Choose 'start kinetic' on the left menu.
- Make the run time 10 min.
- Select the minimum intervals option.
- Press 'ok' to close the kinetic window.
- Drag the 'Read' line into the kinetic measurement.
- Press the 'validate' button and then the 'ok' button.
- Save the experiment.
- Press the 'play' button. A dialog box will appear – press the 'ok' button only when the measurement should be started.
- For NO production measurements use a nitrite/nitrate calorimetric kit. In a 96-well plate insert 50 µl of assay buffer into one row, 70 µl of Griess I reagent into a second row, and 70 µl of Griess II reagent into a third row.
- In order to perform the triplicates experiment, prepare four samples each containing 4.8 µl of the 'chemical transducer'. To sample 1, add 155.2 µl of Assay buffer; to sample 2, add 3.2 µl of GST-M1-1 and 152 µl of Assay buffer; to sample 3, add 9.6 µl of PDGF and 145.6 µl of Assay buffer; and to sample 4, add 3.2 µl of GST-M1-1, 9.6 µl of PDGF, and 142.4 µl of Assay buffer.
- Incubate the solution at room temperature for 10 min.
- In a 384-transparent well plate, insert 50 µl of sample into each well. Insert samples only in the odd or only in the even wells in the same line to allow the use of a multi-pipettor for substrate addition.
- Add 0.54 µl of JS-K (5 mM in DMSO) to each well.
- Using a 12-channel multi-pipettor, quickly add 10 µl from the GSH solution (pre-prepared in the 96-well plate), mix gently and quickly to avoid bubbles. Insert the plate into the reader and start the kinetic measurement.
- Immediately after the kinetic measurement, using a 12-channel multi-pipettor, take 50 µl from each sample into the assay buffer row in the pre-prepared 96-well plate and quickly add to it 50 µl of Griess I reagent and 50 µl of Griess II reagent. Incubate while protecting from light for 10 min at RT and measure the absorbance at 550 nm.
Note: The assay buffer and reagents volumes are dependent on the kits' protocol.
- Set up an experimental procedure in the plate reader for kinetic measurement.
Representative Results
The design, synthesis, and mechanism of action of a 'chemical transducer' that can induce artificial communication between PDGF and GST are presented in Figure 2. The structure of the 'transducer' integrates a PDGF DNA aptamer and a bis-ethacrynic amide (bEA), which is a known GST inhibitor (Figure 2a).19 These binders enable the 'transducer' to bind both PDGF and GST with different affinities, namely, with dissociation constants (kd's) of 26 nM and 144 nM, respectively.4 In addition, according to this design, the binding to PDGF should induce non-specific interactions between the bEA unit and the surface of PDGF, which would markedly reduce the potency of the bEA inhibitor.19,22,23 Figure 2b illustrates the operating mechanism underlying this system. Upon adding the 'transducer' to an active GST, the two EA units simultaneously bind both active sites of this dimeric enzyme and inhibit its activity. In the presence of PDGF, however, a PDGF-'transducer' complex is formed, which prevents the bEA unit from inhibiting GST. This consequently leads to the dissociation of the GST-'transducer' complex and to GST reactivation. GST can then be re-inhibited by adding an unmodified PDGF aptamer that displaces the 'chemical transducer' and enables it to inhibit GST again.
The ability of PDGF to control GST activity was first demonstrated by measuring GST (10 nM) activity with and without the 'chemical transducer' (500 nM) and measuring the activity of the GST-'chemical transducer' complex in the presence of different concentrations of PDGF (250, 500, and 1,000 nM) (Figure 3a). After establishing that the 'chemical transducer' induces artificial PDGF-GST communication, we next established that this artificial communication, similar to the signal transduction steps, is also reversible and rapidly adapts to changes in the environment. Reversible activation/inhibition of GST was performed by sequential additions of PDGF and unmodified PDGF aptamer to the GST-'chemical transducer' complex (Figure 3b). The response of the system to real-time changes in its environment was evaluated by measuring GST activity while adding the different inputs. A rapid increase in GST activity was observed upon the addition of PDGF (750 nM) to the GST-'transducer' complex (10 nM and 500 nM, respectively) 3.5 minutes after adding substrates (Figure 4a). Similarly, a decrease in GST activity was observed upon the addition of PDGF aptamer (5 µM) to a mixture of GST (10 nM), PDGF (750 nM), and the 'chemical transducer' (500 nM) (Figure 4b).
Finally, we demonstrated the ability of such a system to control prodrug activation in response to changes in the environment. JS-K is an anticancer prodrug activated by GST to release toxic NO (Figure 5a). The amount of NO released upon the addition of JS-K (45 µM) to the 'chemical transducer' (750 nM) and different combinations of GST (10 nM) and PDGF (2 µM) were measured (Figure 5b), thus confirming that only the presence of both GST and PDGF will result in prodrug activation.
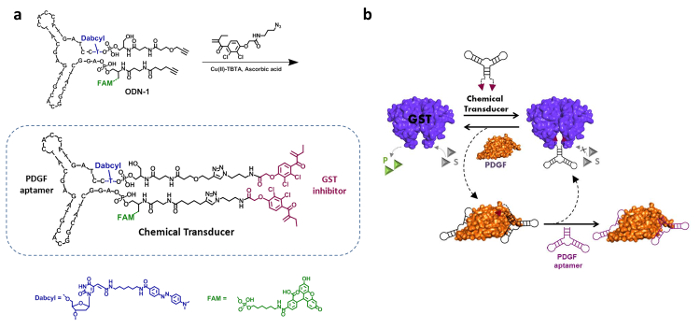
Figure 2. 'Chemical transducer' — synthesis and operating mechanism. (a) The 'chemical transducer' is composed of a PDGF aptamer, a bivalent ethacrynic amide (EA) GST inhibitor, a fluorophore (FAM), and a quencher (Dabcyl). This aptamer-inhibitor conjugate is synthesized by attaching an azide-modified ethacrynic amide (EA) derivative to a dialkyne-modified and fluorescently labeled DNA aptamer (ODN-1). Re-printed with permission from reference4. (b) The enzymatic activity of GST is inhibited by the 'chemical transducer', due to the binding of the EA groups at the enzyme's active sites. Addition of PDGF leads to the formation of the PDGF-'chemical transducer' complex, which disrupts the 'transducer'-GST interaction, therefore restoring the enzymatic activity. The following addition of an unmodified PDGF aptamer releases the 'chemical transducer' and allows it to re-inhibit the enzyme. Please click here to view a larger version of this figure.
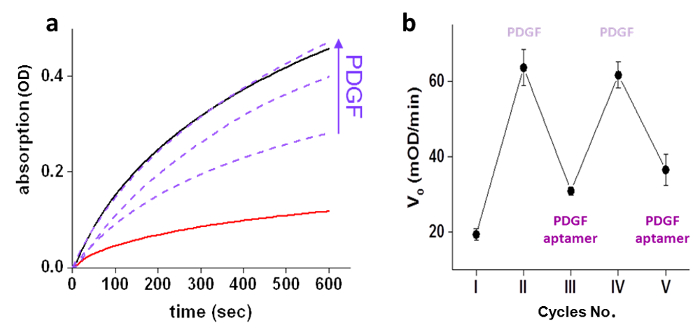
Figure 3: PDGF-controlled GST activity. (a) GST (10 nM) enzymatic activity in the presence (–) and absence (–) of the 'chemical transducer' (500 nM), and in the presence of the 'chemical transducer' (500 nM) with increasing concentrations (250, 500, or 1000 nM) of PDGF (—). (b) Inhibition-activation cycles of GST enzymatic activity manifested by changes in the initial velocity (V0) in response to sequential additions (IIV) of PDGF and unmodified PDGF aptamer to the GST-'chemical transducer' complex: (I) none, (II) PDGF (750 nM), (III) PDGF aptamer (4 µM), (IV) PDGF (5 µM), and (V) PDGF aptamer (10 µM). The graph presents the mean ± stdev of triplicates. Re-printed with permission from reference4. Please click here to view a larger version of this figure.
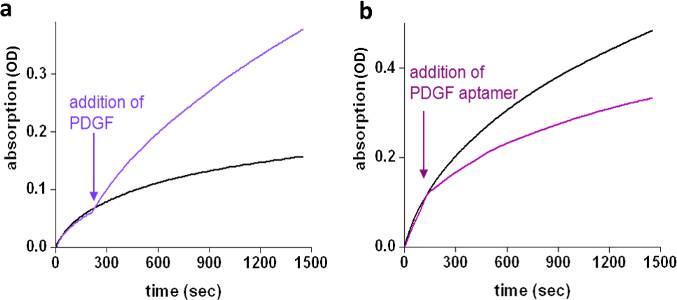
Figure 4: Real-time control of GST activity. (a) Enhancement of GST enzymatic activity detected immediately after the addition of 750 nM PDGF (-) to a solution containing GST (10 nM) and 'chemical transducer' (500 nM) (-) at t = 3.5 min. (b) Addition of a unmodified PDGF aptamer (5 µM) (-) to a solution containing GST (10 nM), PDGF (750 nM), and the 'chemical transducer'(500 nM) (-) at t = 1.5 min leads to an immediate decrease in the enzymatic reaction rate. Re-printed with permission from reference4. Please click here to view a larger version of this figure.
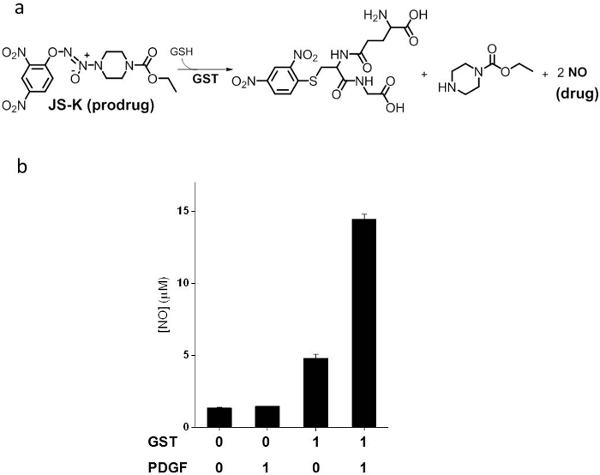
Figure 5: Controlled prodrug activation. (a) GST activation of JS-K prodrug to release toxic NO. (b) NO release in the presence of the 'chemical transducer' and different combinations of GST (10 nM) and PDGF (2 µM). The graph presents the mean ± stdev of triplicates. Changed with permission from reference4. Please click here to view a larger version of this figure.
Discussion
We presented a method for designing and testing of a ‘chemical transducer’ that can induce artificial communication between two naturally unrelated proteins, GST and PDGF, without modifying the native proteins. The unnatural GST-PDGF communications could be detected in real time by using enzymatic assays that follow the changes in the activity of GST in the presence of the ‘chemical transducer’ and increasing the concentrations of PDGF. In addition to detecting the activation of GST by PDGF, these assays were used to follow the deactivation of GST by a competing aptamer, as well as the PDGF-mediated cleavage of an anticancer prodrug. Other switchable protein binders that can control the activity of proteins by responding to oligonucleotide inputs have been described.26 The protocols describe herein, however, demonstrates a means to follow the function of a new class of protein binders that respond to the presence of natural proteins. In this way, the regulation of an enzyme by a protein biomarkers can be detected in real time, which opens the way for using such systems to selectively activate prodrugs and other substrates of GST.
It is important to keep in mind that the kinetics of GST is very rapid and hence, to obtain reliable results from this enzymatic assay, it is essential to minimize the time between the substrate addition and the beginning of the measurement. In addition, because small changes in enzyme or substrate concentrations may have significant effects on the kinetic measurement, it is recommended to perform all experiments in triplicate and to perform positive and negative control experiments in order to avoid experimental errors and misinterpretation of the results.
One limitation of using enzymatic assays to follow unnatural protein-protein communication is the need to have an enzyme as one of the protein partners. In addition, the current assays follow changes in absorbance and hence, they are less suitable for detecting the activation of the enzyme in living cells, which would require using fluorogenic substrates. However, considering that this field is in its infancy, we believe that similar design principles and experimental protocols could also be used for developing future ‘chemical transducers’, which may be used to alter the response of cells to environmental signals or to provide them with new properties. If such changes will also result in therapeutic effects, these developments may lead to artificial signal transduction therapy, which relies on drugs that can mediate unnatural protein-protein communication.3
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
This research was supported by the Minerva Foundation, the HFSP Organization, and a European Research Council Grant (Starting Grant 338265).
Materials
| 1-chloro-2,4-dinitrobenzene | Sigma-Aldrich | 237329 | |
| Acetic acid | Bio Lab | 01070521 | |
| Acetnitrile | J.T.Baker | 9017-03 | |
| Ascorbic acid | Sigma-Aldrich | A4544 | |
| Copper(II) Sulfate pentahydrate | Merck-Millipore | 102790 | |
| Dimethyl sulfoxide | Merck-Millipore | 802912 | |
| Dulbecco's Phosphate Buffered Saline | Biological Industries | 02-023-5A | |
| Ethacrynic acid | Tokyo Chemical Industry Co. Ltd | E0526 | |
| Glutathione-s-transferase M1-1 | Israel Structural Proteomics Center (Weizmann Institute of Science, Rehovot, Israel) | ||
| JS-K | Sigma-Aldrich | J4137 | |
| L-glutathione reduced | Sigma-Aldrich | G4251 | |
| Magnesium Chloride | J.T.Baker | 0162 | |
| nitrate/nitrite colorimetric assay kit | Cayman Chemical | 780001 | |
| Oligonucleotides | W. M. Keck Foundation Biotechnology at Yale University | custom order | |
| PDGF-BB | Israel Structural Proteomics Center (Weizmann Institute of Science, Rehovot, Israel) | ||
| TBTA | Sigma-Aldrich | 678937 | |
| Triethylamine | Sigma-Aldrich | T0886 | |
| Desalting column | GE Healthcare | illustra MicroSpin G-25 Columns | |
| HPLC | Waters | 2695 separation module | |
| HPLC column | Waters | XBridgeTM OST C18 column (2.5 μM, 4.6 mm × 50 mm) | |
| HPLC column | Waters | XBridgeTM OST C18 column (2.5μM, 10 mm × 50 mm) | |
| Plate reader | BioTek | synergy H4 hybrid |
Referenzen
- Hunter, T. Signaling—2000 and Beyond. Cell. 100, 113-127 (2000).
- Levitzki, A., Klein, S. Signal transduction therapy of cancer. Mol Aspects Med. 31, 287-329 (2010).
- Peri-Naor, R., Motiei, L., Margulies, D. Artificial signal transduction therapy: a futuristic approach to disease treatment. Future Med. Chem. 7, 2091-2093 (2015).
- Peri-Naor, R., Ilani, T., Motiei, L., Margulies, D. Protein-Protein Communication and Enzyme Activation Mediated by a Synthetic Chemical Transducer. J. Am. Chem. Soc. 137, 9507-9510 (2015).
- Corson, T. W., Aberle, N., Crews, C. M. Design and Applications of Bifunctional Small Molecules: Why Two Heads Are Better Than One. ACS Chem. Biol. 3, 677-692 (2008).
- Rutkowska, A., Schultz, C. Protein Tango: The Toolbox to Capture Interacting Partners. Angew. Chem. Int. Ed. 51, 8166-8176 (2012).
- Meyer, C., Köhn, M. A Molecular Tête-à-Tête Arranged by a Designed Adaptor Protein. Angew. Chem. Int. Ed. 51, 8160-8162 (2012).
- Klemm, J. D., Schreiber, S. L., Crabtree, G. R. Dimerization as a Regulatory Mechanism in Signal Transduction. Annu. Rev. Immunol. 16, 569-592 (1998).
- DeRose, R., Miyamoto, T., Inoue, T. Manipulating signaling at will: chemically-inducible dimerization (CID) techniques resolve problems in cell biology. Pflugers Arch. 465, 409-417 (2013).
- Gestwicki, J. E., Marinec, P. S. Chemical control over protein-protein interactions: beyond inhibitors. Comb. Chem. High Throughput. Screen. 10, 667-675 (2007).
- Battle, C., Chu, X., Jayawickramarajah, J. Oligonucleotide-based systems for input-controlled and non-covalently regulated protein binding. Supramol. Chem. 25, 848-862 (2013).
- Diezmann, F., Seitz, O. DNA-guided display of proteins and protein ligands for the interrogation of biology. Chem. Soc. Rev. 40, 5789-5801 (2011).
- Röglin, L., Ahmadian, M. R., Seitz, O. DNA-Controlled Reversible Switching of Peptide Conformation and Bioactivity. Angew. Chem. Int. Ed. 46, 2704-2707 (2007).
- Röglin, L., Altenbrunn, F., Seitz, O. DNA and RNA-Controlled Switching of Protein Kinase Activity. ChemBioChem. 10, 758-765 (2009).
- Harris, D. C., Chu, X., Jayawickramarajah, J. DNA-Small Molecule Chimera with Responsive Protein-Binding Ability. J. Am. Chem. Soc. 130, 14950-14951 (2008).
- Harris, D. C., Saks, B. R., Jayawickramarajah, J. Protein-Binding Molecular Switches via Host-Guest Stabilized DNA Hairpins. J. Am. Chem. Soc. 133, 7676-7679 (2011).
- Kim, Y., Cao, Z., Tan, W. Molecular assembly for high-performance bivalent nucleic acid inhibitor. Proc. Nat. Acad. Sci. U.S.A. 105, 5664-5669 (2008).
- Han, D., et al. A Logical Molecular Circuit for Programmable and Autonomous Regulation of Protein Activity Using DNA Aptamer-Protein Interactions. J. Am. Chem. Soc. 134, 20797-20804 (2012).
- Motiei, L., Pode, Z., Koganitsky, A., Margulies, D. Targeted Protein Surface Sensors as a Tool for Analyzing Small Populations of Proteins in Biological Mixtures. Angew. Chem. Int. Ed. 53, 9289-9293 (2014).
- Ranallo, S., Rossetti, M., Plaxco, K. W., Vallée-Bélisle, A., Ricci, F. A Modular, DNA-Based Beacon for Single-Step Fluorescence Detection of Antibodies and Other Proteins. Angew. Chem. Int. Ed. 54, 13214-13218 (2015).
- Franzini, R. M., et al. Identification of Structure-Activity Relationships from Screening a Structurally Compact DNA-Encoded Chemical Library. Angew. Chem. Int. Ed. 54, 3927-3931 (2015).
- Unger-Angel, L., et al. Protein recognition by bivalent, ‘turn-on’ fluorescent molecular probes. Chem. Sci. 6, 5419-5425 (2015).
- Nissinkorn, Y., et al. Sensing Protein Surfaces with Targeted Fluorescent Receptors. Chem. Eur. J. 21, 15981-15987 (2015).
- Huber, C. G., Oefner, P. J., Bonn, G. K. High-Resolution Liquid Chromatography of Oligonucleotides on Nonporous Alkylated Styrene-Divinylbenzene Copolymers. Anal. Biochem. 212, 351-358 (1993).
- Lyon, R. P., Hill, J. J., Atkins, W. M. Novel class of bivalent glutathione S-transferase inhibitors. Biochemie. 42, 10418-10428 (2003).
- Battle, C., Chu, X., Jayawickramarajah, J. Oligonucleotide-based systems for input-controlled and non-covalently regulated protein binding. Supramol. Chem. , 1-16 (2013).

