The Use of Induced Somatic Sector Analysis (ISSA) for Studying Genes and Promoters Involved in Wood Formation and Secondary Stem Development
Summary
Here we present a protocol that facilitates the medium to high throughput functional characterization of gene and promoter constructs in tree secondary stem tissue within comparatively short time frames. It is efficient, easy to use and widely applicable to a range of tree species.
Abstract
Secondary stem growth in trees and associated wood formation are significant both from biological and commercial perspectives. However, relatively little is known about the molecular control that governs their development. This is in part due to physical, resource and time limitations often associated with the study of secondary growth processes. A number of in vitro techniques have been used involving either plant part or whole plant system in both woody and non-woody plant species. However, questions about their applicability for the study of secondary stem growth processes, the recalcitrance of certain species and labor intensity are often prohibitive for medium to high throughput applications. Also, when looking at secondary stem development and wood formation the specific traits under investigation might only become measurable late in a tree's lifecycle after several years of growth. In addressing these challenges alternative in vivo protocols have been developed, named Induced Somatic Sector Analysis, which involve the creation of transgenic somatic tissue sectors directly in the plant's secondary stem. The aim of this protocol is to provide an efficient, easy and relatively fast means to create transgenic secondary plant tissue for gene and promoter functional characterization that can be utilized in a range of tree species. Results presented here show that transgenic secondary stem sectors can be created in all live tissues and cell types in secondary stems of a variety of tree species and that wood morphological traits as well as promoter expression patterns in secondary stems can be readily assessed facilitating medium to high throughput functional characterization.
Introduction
Tree stems comprise a significant amount of the planets biomass and are of immense biological, cultural and commercial importance. Secondary stems create habitat by providing resources and shelter for many other life forms. They deliver many other services to the ecosystems they inhabit and act as a renewable resource for the production of timber, pulp and paper and other wood and non-wood products. Secondary stem development and more specifically wood formation is governed by complex molecular system that regulate the development of specific cell types, the biochemical composition of their cell walls and how they are arranged to form tissues and organs. Dissecting the molecular basis of secondary stem development and wood formation is confounded by many factors including the variability of wood and stem properties within and between stems, long generation times, out-crossing mating systems, high heterozygosity, high genetic load, seasonal dormancy, long mature trait establishment periods and the sheer physical size of mature trees. As a result, understanding of secondary stem development relative to the detailed knowledge of most other aspects of the molecular control of plant development, is still in its infancy.
A number of in vitro techniques have been used to study and understand secondary stem development, in particular wood and secondary cell wall formation. These protocols involve the use of whole plant or plant part systems, where either transgenic plants are created or specific secondary cells or tissue are transformed for the study of specific aspects of wood and/or secondary stem development1. Transgenic plants can be recovered post genetic transformation from a wide variety of plant tissues and cell types, however, progress is slow, particularly when analyzing wood fiber traits due to long regeneration and stem maturation times (in the order of years), high technical and labor demands, low throughput of candidate genes as well as difficulties in propagating some woody plant species. Similar techniques have been developed in non-woody model system such as Arabidopsis that successfully overcome some of these limitations, but not all secondary stem cell types a present in these stems and traits related to seasonality or longevity cannot be studied in such species2. Alternatively, plant part systems, such as Pinus radiata callus cultures3 reduce the associated timeframes. These methods however are restricted to the study of an individual cell type and suffer similar constraints as noted for in vitro experiments. Similarly, apical stem cultures4 involving whole stem explants have shown promise but as yet have not been applied for the study of specific genes or promoters of interest. More recently, an alternative protocol involving hairy root cultures has been developed for eucalypts and has been successfully applied5, however, this method still requires in vitro cultivation, involves secondary roots rather than stems and to date it is limited to a single tree species.
Induced somatic sector analysis (ISSA), as described here, was developed to overcome some of these problems providing a medium to high throughput functional screening tool for genes and promoters with suspected roles in wood formation and secondary stem tissue development. ISSA is an in vivo transformation and screening system which was developed to reduce the time taken to produce transgenic cells and tissue in an intact secondary stem while overcoming labor, technical and throughput limitations of routinely used in vitro methods. The protocols described here allow for the simultaneous creation of hundreds of independently transformed tissue sectors and cells in secondary stems within a short period of time, in the tree species and tissue of interest without genetic and/or environmental variation within relatively short time frames and low labor cost. ISSA in vivo techniques were first described for secondary stem6 and bud7 tissues and have since been refined in secondary stem tissue through studies of genes and/or promoters involved in cambial differentiation and include: tubulin (TUB)8, fasciclin-like arabinogalactan (FLA)9, cellulose synthase (CESA)10, secondary cell wall-associated nac domain (SND2)11, ARBORKNOX (ARK1)12 and really interesting new gene (RING) H2 protein13. These studies were conducted in secondary stems of poplar and eucalypt plants and provided insights into cell morphology, cell wall chemistry and gene expression.
The protocols described here are intended to bring together the experience and knowledge gained through the development and application of ISSA from a range of published and unpublished studies over the last decade. They focus on the in vivo transformation of secondary stem tissues6 and concentrate on studies involving Populus alba 'pyramidalis' clone, Eucalyptus globulus as well as 11 Eucalyptus globulus x camaldulensis clones. This document takes researchers through the protocol from the cultivation of plants and bacteria, transformation of stem tissues, growth and harvest of tissue, identification of transgenic cells and tissues, preparation for phenotypic assessments and methods for the collection and analysis of data. While techniques have successfully been applied to measure cell wall monosaccharide composition also9,11, due to space limitations, this document concentrates on techniques used for measuring cell and tissue morphology and understanding gene expression patterns in secondary stems only. Accordingly, the protocol as outlined is suited to those looking to gain further insights into the role and/or expression of genes linked to secondary stems using a low cost, technically easy, and medium to high throughput method.
Protocol
1. Preparation of Plant Material
- Prior to experimentation, raise new seedlings of the preferred tree species from seed or cutting and grow tree/s until the diameter of the stem in the area intended for experimentation is approximately 1 cm in diameter.
Note: The time needed may vary due to plant growth rates therefore allow between three to nine months for this step.
2. Binary Vector Creation
- Conduct work from this section to section 7 in a laboratory or greenhouse where Agrobacterium tumefaciens can be handled and that appropriate personal protective equipment prescribed for the laboratory is used.
- Prepare a binary vector containing either a gene of interest knockdown or overexpression cassette and a β-glucuronidase (GUS) reporter gene cassette or a promoter of interest fused to a GUS gene depending on the type of study.
Note: There are a number of binary vector backbones that can be sourced commercially or through research networks as well as numerous techniques available to insert genes and promoters of interest into binary vectors. In the context of this protocol is up to the experimenter to make the decision on how to create a binary vector. - Transform binary vector into a disarmed strain of A. tumefaciens using electroporation or heat shock14 and store appropriately until needed for experimentation.
- Repeat step 2.2 for any positive and/or negative controls.
Note: Negative controls should include a binary vector containing no gene of interest for a gene of interest study and a promoterless GUS for a promoter of interest study. Positive control for a promoter of interest study should include a Cauliflower Mosaic Virus 35s promoter (CaMV35s) fused to a GUS reporter.
3. Preparation of A. tumefaciens for Inoculation
- At least a week prior to experimentation, take a small amount (approximately 1 µl) of A. tumefaciens prepared during step 2.2 and 2.3 and spread thinly on separate LB medium with agar14 plates containing appropriate antibiotics for bacterial selection. Grow at 28 °C in an incubator to form small colonies and then store at 4 °C until needed.
- 48 hours prior to experimentation, transfer one colony of A. tumefaciens from each plate into separate 50 ml screw top tubes containing 5 ml of pre-warmed (28 °C) LB media14 containing appropriate antibiotics for bacterial selection and agitate at 200 rpm on a shaker incubator for approximately 48 hr at 28 °C until mixture is very cloudy.
- Between 4-6 hr prior to inoculation of plant stems (section 4) add 1 ml of the LB/A. tumefaciens mixture into a fresh 50 ml screw top tube containing 19 ml (1:20 dilution) of fresh warm (28 °C) LB media with appropriate antibiotics for bacterial selection. If optical density (OD) at 600 nm (OD600, as measured by spectroscopy) is above 0.1, dilute further with warm LB until this is achieved.
- Place diluted LB/A. tumefaciens mixture back on the shaker incubator and shake under the same conditions (200 rpm, 28 °C) until OD600 is between 0.4-0.6 and remove.
- Centrifuge LB/A. tumefaciens mixture for 15 min at 1,000 x g and 4 °C.
- Decant liquid and immediately resuspend A. tumefaciens pellet into 1 ml of cooled MS media15, transfer to 2 ml microtube and store on ice until required for inoculation (section 4). This solution is referred to as Inoculation Media.
4. Inoculation of Plant Stem with A. tumefaciens
- Start experimentation during late spring or early summer using plants created in section 1 that have an active and fast growing cambium. A good diagnostic for an active and fast growing cambium is that the phloem can be easily peeled from the xylem.
- Find a clear straight section of stem near the base of the plant and remove any leaves and branches.
- Using a sharp scalpel (preferably No. 11) or razor blade, create a 1 cm2 'cambial window' in a clear section of stem by introducing two vertical parallel incision through the phloem of 20 mm in length and one 5 mm apart then a horizontal cut that connects the two vertical cuts at their basal end.
- Peel the phloem strip created by the incision upwards exposing the developing xylem tissue and add sufficient Inoculation Media from step 3.6 to wet the surface of the exposed developing xylem using a pipette (typically 5-10 µl). Immediately reinsert the phloem strip.
- Bind the phloem strip to the stem tightly with Parafilm.
- Repeat steps 4.2 to 4.5 for any additional cambial windows for the gene(s) or promoter(s) of interest creating any new cambial windows least 1 cm above or below other cambial windows and at a 90° offset.
- Complete steps 4.2 to 4.5 for the positive and negative control vectors (step 2.4) ensuring that each vector is added to the same stem of each plant used in the experiment.
- Monitor stem diameter growth periodically and harvest for GUS assay (section 5) when radial growth of at least 5 mm has been observed in stems.
Note: The amount of radial growth needed is dependent on the amount of tissue required for downstream analysis.
5. Harvesting for GUS Histological Assay
- Excise the 'cambial window' from the stem removing any tissue not part of new growth within the cambial window.
- For studies relating to mature xylem and phloem cell/tissue morphology, peel the phloem from the xylem.
- For studies where the cambial zone is required to remain intact or promoter expression patterns are to be assessed, slice the cambial window transversely using a razor blade or other sharp blade into discs of between 0.5 and 1 mm in thickness.
- Place processed cambial window tissues in 14 ml round bottom tubes and rinse twice in 0.1 M phosphate buffer at pH 714. Ensure that the tissue is completely submerged, remains in solution for at least five minutes and all excess solution is removed at the end of the second rinse.
- Add 5 ml of GUS reagent (0.5 mM X-gluc (5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid), 10 mM EDTA, 0.5% Triton X-100 v/v, 0.5 mM potassium ferricyanide(III), 0.05 mM potassium ferrocyanide(II), made up to final volume with 0.1 mM phosphate buffer pH 7; see Hawkins et al.16) to each tube. If tissue is not completely submerged, add additional GUS reagent.
- Incubate tubes for 10 min at 55 °C in the dark.
- Incubate tubes for a further 12-16 hr on a shaker incubator at 37 °C in the dark using gentle agitation (between 30 and 60 rpm) to allow for mixing.
- Upon removal from shaker incubator randomly check the pH of GUS reagent in a small subset of tubes using litmus paper with a pH range 0-7.
- If any of the tubes have a pH below 6 then label them. Continue to check the remainder of the tubes to confirm pH and label if pH is 6 or below.
Note: Samples with a pH below 6 may not be suitable for analysis. These samples have to be excluded from further analysis.
- If any of the tubes have a pH below 6 then label them. Continue to check the remainder of the tubes to confirm pH and label if pH is 6 or below.
- Decant GUS reagent and replace with sufficient 70% ethanol to cover tissue.
- Store at 4 °C until needed.
6. Identification of Transgenic Tissue
- Using forceps, take out all the cambial window tissue from a tube and place in small tray or petri dish to allow for microscopic visualization.
- Using a dissecting microscope between 1X and 4X magnification, identify and tally the number of cells or tissues that have a bright blue staining. Blue stained cells or tissue are referred to as a sector from this point onwards. Tally them in the following ways.
- For samples where the phloem has been removed (step 5.1.1), count the number of sectors in cambial tissue found on the surface of the developing xylem (cambial sector, Figure 1a).
- For samples that have been cut into discs (step 5.1.2), count the number sectors found in the different cell or tissue types (Figure 1b, 1c, 1d). Sectors can be found in the following tissue types: the periderm (Periderm sector, Figure 1e), phloem (Phloem sector, Figure 1f), cambial tissue (Cambial sector, Figure 1g, 1h, 1I), wound parenchyma (Wound Parenchyma Sector, Figure 1j) and in tyloses (Tylose sector, Figure 1k).
- During microscopic assessment ensure that both sides of a disc have been viewed and that sectors that occur across two discs are matched so that numbers are not overestimated.
- Place only the tissue containing sectors back into the tube containing 70% ethanol and store until needed.
- Repeat step 6.1 to 6.3 for any additional cambial windows including the positive and/or negative control.
- Calculate the average transformation efficiency for each sector type for each gene or promoter of interest and the controls separately by dividing the total number of sectors by the total number of 1 cm2 cambial windows to derive an Average Number of Transformation Events per cm2 of Cambium Inoculated (ATScm-2).
- Proceed to step 7.2 and 8 for analysis of promoter expression patterns and step 7.1, 7.2 or 7.3 for techniques to assess cell and tissue morphology in cambial tissue.
7. Analysis of Cell and Tissue Morphology in Cambial Tissue
Note: Below are a selection of techniques that have been used successfully for analysis of ISSA derived samples.
- Analysis of xylem cell area, lumen area, cell wall thickness and cell wall area using Scanning Electron Microscopy (SEM).
Note: This protocol requires minimal sample preparation and allows for relatively high throughput of sector analysis relating to the morphological traits outlined.- From this step onwards please ensure lab coat, eye protection and gloves are used whenever woody tissue is being handled and treated.
- Identify a cambial sector for analysis and make an incision approximately 0.5 mm to either side of it using a single edge razor blade to create a disc (Figure 2a).
- Trim off excess xylem tissue leaving approximately 1 mm of xylem tissue to the tangential side of the sector (Figure 2b).
- Carefully cut transversely through the center of the sector using a fresh double edge razor blade (Figure 2c).
- Make two additional shallow radial incisions on the transverse plane either side of sector to demarcate the transgenic tissue (Figure 2d). As GUS staining will not penetrate deep into the tissue follow along the radial files of cells to either side of stained tissue found at the cambial surface.
- Attach prepared cambial sector face up to stage of an SEM pin stub mount using SEM conductive tape (Figure 2e) and keep in desiccator until needed for SEM visualization.
- Repeat steps 7.1.2 to 7.1.6 for any additional sectors including those from the negative control.
- Using a SEM in a low vacuum mode (energy 5 kV, spot 3.0 nm), visualize and take photos of cells/tissue within the sector as well as the cells/tissue directly adjacent the sector (Figure 2f). The amount of magnification is dependent on the features of interest. For xylem fibers, 2,000X is suitable (Figure 2g).
- Once a cambial sector has been visualized and photographs taken at an appropriate magnification, measure xylem cell/tissue morphological traits of interest using image measuring software.
- For statistical analysis, undertake multiple pairwise analyses using paired t-tests to compare the difference between morphological measured in the transgenic sector and the adjacent non-transgenic control cells/tissue (measured within 0.5 mm from sector edge) for each sector to determine a p-value.
- Repeat step 7.1.10 for the negative control.
- In cases where the positive control shows a low p-value (α <0.05), calculate the difference between the transgenic and adjacent non-transgenic cells/tissue for a morphological trait. Use this 'difference value' in an unpaired t-test to compare the effect of the gene of interest with the negative control to determine a p-value.
- Histochemical analysis of cambial cell/tissue morphology or promoter expression patterns in stem cells/tissues using light microscopy.
Note: While more time consuming, this method allows for more analysis options including aspects of cambial dynamics, e.g., cambial width as well as promoter expression patterns. In addition, if unable to access a SEM, this protocol can be used as an alternative for step 7.1.- Excise sector of interest using razor blade and some surrounding tissue as small blocks (no greater than 1 mm3) and place directly in 1-3 ml of 100% ethanol in a sample vial with screw cap for at least 2 days at 4 °C on a shaker. Prepare the small block in a way that will allow for the surface of interest to be accessed by a microtome.
- Repeat for any additional sectors including those from the negative control.
- Remove liquid and replace with 25% ethanol: 75% LR white mixture for 2 days maintaining conditions.
- Repeat step 7.2.3 with 50% ethanol: 50% LR white, 25% ethanol: 75% LR white and then finally twice with 100% LR white.
- Place small block in Embedding Mold with the surface of interest aligned to short end and carefully cover with fresh LR white and polymerize as per manufacturer instructions.
- Cut 5 µm sections from the surface of interest on a rotary microtome and mount on a glass slide. Use Safranin O (0.01%) staining to visualize cell wall and/or other features.
- Add mounting media, place a cover slip and allow to set overnight.
- View under a light microscope between 100X and 600X magnification and capture image.
- Capture morphological traits from images using image measuring software.
- For statistical analysis of quantitative morphological traits, follow on from step 7.1.10.
- For qualitative analysis of morphological traits, compare and describe patterns observed for the gene or promoter of interest and the negative and/or positive control.
- Analysis of Microfibril Angle (MFA) in Macerated Fibers Using Light Microscopy.
- Excise transgenic xylem tissue directly from a sector as well as from non-transgenic tissue adjacent to it (within 0.5 mm, Figure 2h) and place in separate 1.5 ml tubes. Repeat for any additional sectors including those from the negative control.
- Complete steps 7.3.3 to 7.3.5 in a fume hood.
- Add 250 µl of hydrogen peroxide and 250 µl of glacial acetic acid to each tube.
- Place tube in heat block at 90 °C for 2 hr in fume hood.
- Remove tissue from tubes and carefully rinse with distilled water at least twice.
- Using a water soluble mounting media, mount tissue on a glass slide and tease apart with sharp forceps prior to affixing a cover slip.
- View under a light microscope and capture images of individual fibers at magnification greater than 400X.
- To determine microfibril angle of fibers, measure the angle between the pit apertures and/or cell wall striations and the long axis of the fiber (Figure 2i) using image measuring software.
- For statistical analysis, follow on from step 7.1.10.
8. Analysis of Promoter Expression Patterns
- Analysis of Promoter Expression Patterns in Secondary Stem Tissues.
- Tally the frequency of each sector type for the promoter of interest as well as the positive and negative controls as noted in step 6.2.2.
- Compare the frequency of the different sector types for the promoter of interest with the positive control using Chi-square tests to establish p-values. Further analysis of cell/tissue specificity can be undertaken using step 7.2 as required.
- If more than one promoter of interest is being investigated then repeat this analysis making comparison between all promoters of interest and the positive control.
- If sectors or blue staining are observed in the negative control then add this to the analysis or abandon depending on extent or if endogenous staining is suspected (see Discussion).
- Analysis of Promoter Expression Patterns During Cambial Derivative Development and Differentiation.
- Using a dissecting microscope, visualize all the cambial sectors (Figure 1g, 1h, 1i) identified in step 6.2 and determine the presence/absence of blue staining in three tissues types derived from the cambium; the phloem (P), developing xylem (X1) or developed xylem tissue (X2) (see Figure 3a).
- As per step 8.1.2, compare the frequency of presence/absence of GUS staining in the P, X1 and X2 regions of promoter of interest with the positive control using Chi-square tests to establish p-values. Further analysis of cell/tissue specificity can be undertaken using step 7.2 as required.
- See steps 8.1.2.1 and 8.1.2.2 for additional considerations.
Representative Results
Using this protocol all live secondary stem cell and tissue types have been shown to be susceptible to A. tumefaciens transformations and have been defined into sector types based on the cell type initially transformed and it subsequent developmental growth pattern. Sector types include periderm, phloem, cambial, wound parenchyma and tylose (Figure 1b, 1c, 1d) and can be found in consistent locations describe in the remainder of this paragraph. A periderm sector comprises of a group of transformed cells found exclusively in the periderm and never extending into the phloem (Figure 1e). A phloem sector can be found at various locations between the initiating layer and the periderm, however, never extending into these surrounding tissues (Figure 1f). A cambial sector is comprised of transformed xylem/cambium/phloem tissues derived from transformation of cambial initial and can occur in three distinct patterns: i) 'full' cambial sector, transformed xylem/cambial/phloem cells extending from the boundary of the wound parenchyma tissue through the xylem and cambial zone to the phloem tissue containing both ray and fusiform cells or ray elements only (Figure 1g), ii) 'lost initial' cambial sector, a variant of the full cambial sector where an initial has been lost from the initiating layer leaving mirrored xylem and phloem sectors and an untransformed initiating layer (Figure 1h), and iii) 'xylem only' cambial sector, transformed xylem tissue extending from the boundary of the wound parenchyma tissue to variable lengths into the newly formed xylogenic tissue but never reaching the initiating layer (Figure 1i). A wound parenchyma sector contains transformed wound parenchyma cells found as either an individual cell or groups of cells, often in radial files, located between the pre-existing wood and newly formed xylem tissue (Figure 1j). A tylose Sector comprised of transformed vessel tyloses found within the pre-existing wood (Figure 1k).
Representative transformation efficiency data for both gene and promoter of interest studies as well as their positive controls is presented in Table 1 and Table 2. Data has been drawn from two large trials conducted across the same time period (Dec-Apr) in the same genotypes of eucalypt and poplar across two successive years involving a number of wood formation genes and promoters of interest. Focusing on positive controls from the promoter of interest studies (Table 2), the most susceptible tissue type to A. tumefaciens T-DNA transfer is cambial tissues with approximately 60% and 40% of the sectors identified in eucalypts and poplar respectively being cambial sectors. In eucalypts, the next most abundant sector type is wound parenchyma at 35% while the other sector types all had ATScm-2 values below 5%. In poplars, the next most abundant sector type is wound parenchyma at 20% followed by phloem at 15% while the remainder of the sector types were found at a frequency below 5%. A higher likelihood of finding a sector in a cambial window as well as greater numbers of cambial sectors identified are typical where the phloem peel protocol is used (Table 1, step 5.2.1) due to the ability for staining and the possibility to visualize the entire area of the cambium.
During the development of these protocols a number of variables were explored as part of protocol optimization trials. These included bacterial concentration in Inoculation Media, media type used in Inoculation Media, addition of Acetosyringone to Inoculation Media, tissue age and A. tumefaciens strain as well as time of inoculation and genotype. Cambial sector ATScm-2 data from these trials is presented in Table 3 and Table 4. The majority of variables investigated, when altered, lead to changes in cambial ATScm-2 in both species and included in particular increasing the concentration of bacteria in the Inoculation Media, use of MS in the Inoculation Media and choice of A. tumefaciens strain. In addition, time of inoculation and tree genotype showed significant differences in eucalypts with inoculations in early summer showing higher ATScm-2 while eucalypt clones SG21 and SG44 showed higher cambial ATScm-2, 41.6 and 40.4, respectively compared to others across all months combined.
Average cambial sector size varies and is dependent on the amount of its tangential and radial growth in a cambial window. In one subsample of 53 sectors in poplar, grown for approximately 4 months, the tangential width of sectors at the cambium varied between 0.09 and 0.58 mm with an average of 0.27 mm while radial growth across cambial windows ranged from 0.7 to 2.2 mm with an average of 1.35 mm. When combined, these provided between 0.063 mm2 and 1.276 mm2 of transgenic tissue for analysis. In another subsample of 188 sectors in the same species, where between 1.5-4 mm of radial growth across the cambial window was observed over approximately 5 months, the average sector mass was 72 µg (Table 5).
The amount of transgenic tissue created has been shown to provide sufficient cells and/or tissue to undertake morphological measurements as well as for the study of promoter activity. For example, the influence of three Eucalyptus grandis FLAs on a number of xylem fiber morphological traits9 was explored in eucalypt clones and revealed possible roles for EgrFLA2 in MFA determination and EgrFLA1 in cell size determination (Table 6). In addition, patterns of expression of Eucalyptus grandis CESA gene promoter in the developing xylem (X1), developed xylem (X2) and phloem (P) tissues10 identified significantly different expression patterns between EgrCESA1, 2, 3, and EgrCESA4, 5, 7 in eucalypt stems. In this analysis, EgrCESA1, 2, 3 were shown to be primarily expressed in developing xylem tissue while EgrCESA4, 5, 7 were shown to be expressed in both developing xylem and phloem tissue (Figure 3b, Table 7). All EgrCESA promoters showed significantly different expression patterns to the positive control (CAMV35S promoter) (Table 7).
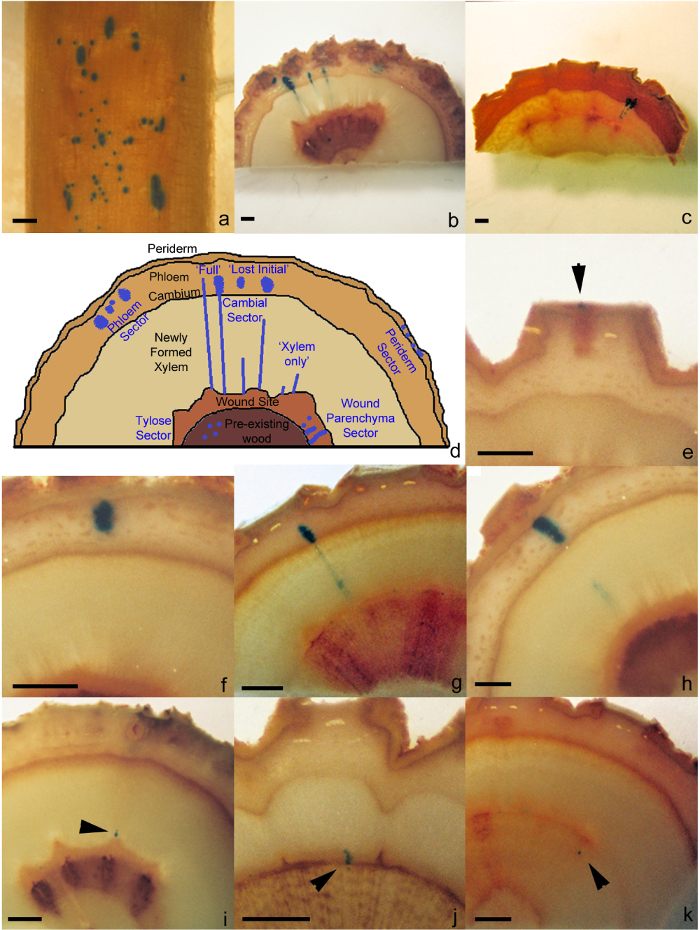
Figure 1: Transgenic sectors types. (a) Cambial sectors as viewed on the surface of the exposed xylem after processing using the phloem peel protocol (step 5.2.1). Discs showing a range of sector types on transverse surface of in poplar (b) and eucalypt (c) stems after processing using the transverse cut protocol (step 5.2.2). (d) Schematic diagram of the range of sector types found in plant stems. Examples of periderm sector (e), phloem sector (f), 'full' cambial sector (g), 'lost initial' cambial sector (h), 'xylem only' cambial sector (i), wound parenchyma sector (j) and tylose sector (k) in poplar. Black arrows indicate where smaller sized sector are located. All scale bars = 1 mm. Please click here to view a larger version of this figure.
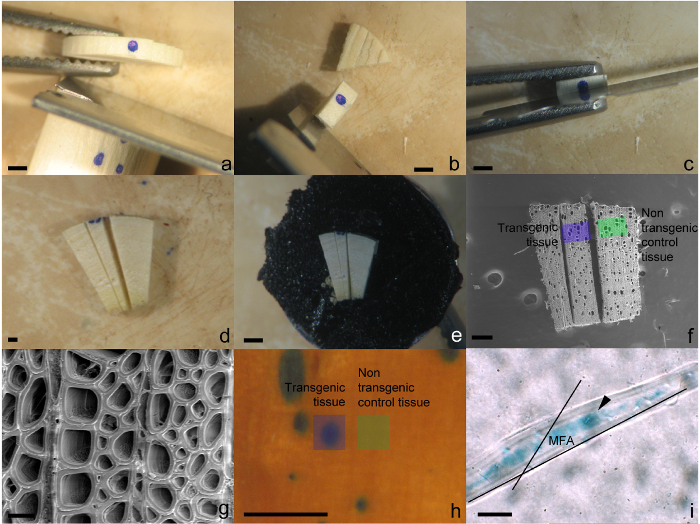
Figure 2: Preparation of cambial sectors for morphological analysis using SEM and light microscopy. (a) Excision of disc containing sector. (b) Disc trimmed to remove excess tissue. Cutting through transverse surface of the transgenic sector (c). (d) Introduction of two radial cuts to assist in locating the transgenic tissue under SEM. (e) Tissue mounted on an SEM pin using conductive tape. (f) Depiction of the region of transgenic and non-transgenic tissues that should be imaged for analysis. (g) Typical image of xylem fiber and ray cells captured at 2,000X magnification. (h) Area where transgenic and non-transgenic fibers should be excised from for MFA analysis. (i) Macerated fiber viewed under light microscopy depicting the angle of the pits and/or cell wall striations and to the long axis of the cell. Black arrow indicates pit aperture. Scale bars = a-f, h = 1 mm, g, i = 20 µm. Please click here to view a larger version of this figure.
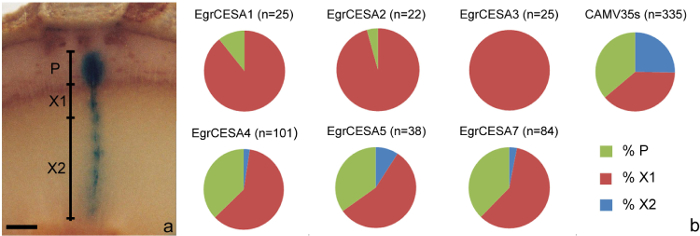
Figure 3: Analysis and results of gene promoter expression patterns of cambial derivatives. (a) Depiction of the P (phloem), X1 (developing xylem) and X2 (developed xylem) regions. (b) Results indicating proportion of P, X1 and X2 staining in cambial sectors from a trial involving a range of CESA promoters from Eucalyptus grandis transformed into stems of eucalypt hybrids. Data sourced from Creux et al.10. n = number of sectors assessed. Scale bar = 0.5 mm. Please click here to view a larger version of this figure.
| Tree species | Vector type | Number of genes of interest used | Total number of windows created | Total number of windows where a sector was identified (%) | Cambial ATScm-2 cambial (in windows where a sector was identified) |
| Eucalyptus globulus x camaldulensis hybrids (total of three clones) | Positive Control (Genes of interest) | 1 (GUS only) | 96 | 97% | 45.1 (1.5) |
| Genes of Interest | 20 | 630 | 92% | 46.1 (2.2) | |
| Populus alba 'pyramidalis' | Positive Control (Genes of interest) | 1 (GUS only) | 110 | 74% | 10.8 (1.5) |
| Genes of Interest | 20 | 700 | 81% | 14.6 (0.8) |
Table 1: Typical cambial ATScm-2 for the positive control and genes of interest. Figures have been sourced from studies undertaken over two consecutive years in the same eucalypt and poplar clones using a range of genes and the phloem peel harvesting protocol (step 5.2.1). Standard error values in brackets.
| Tree species | Vector type | Number of promoter/genes | Total number of windows created | Total number of windows where a sector was identified (%) | ATScm-2 all sectors | ATScm-2 Periderm | ATScm-2 Phloem | ATScm-2 cambial | ATScm-2 Wound Parenchyma | ATScm-2 Tylose |
| Eucalyptus globulus x camal- dulensis hybrids (total of three clones) |
Positive Control | 1 (GUS only) | 118 | 63% | 25.74 (2.35) | 0.24 (0.07) | 0.89 (0.19) | 15.7 (1.71) | 8.84 (1.4) | 0.07 (0.03) |
| Promoters of Interest | 28 | 1050 | 31% | 14.67 (1.77) | 0.02 (0.01) | 0.05 (0.01) | 13.79 (1.75) | 0.78 (0.21) | 0.03 (0.01) | |
| Populus alba 'pyramidalis' | Positive Control | 1 (GUS only) | 110 | 54% | 12.37 (1.77) | 0.22 (0.07) | 1.85 (0.29) | 5.07 (0.6) | 2.78 (0.75) | 0.29 (0.53) |
| Promoters of Interest | 28 | 990 | 26% | 8.28 (1.18) | 0.16 (0.04) | 0.9 (0.09) | 6.67 (1.16) | 0.27 (0.07) | 0.28 (0.11) |
Table 2: Typical sector ATScm-2 for the positive control and promoters of interest. Figures have been sourced from studies undertaken over two consecutive years in the same eucalypt and poplar clones using a range of promoter sequences and the disc harvesting protocol (step 5.2.2). ATScm-2 values were derived from windows where a sector was identified only. Standard error values in brackets. Please click here to view a larger version of this table.
| Species | Trial | Treatment Description | Number of windows in treatment | Cambial ATScm-2 |
| Eucalyptus globulus | Concentration of A. tumefaciens in Inoculation Media | Resuspended in 25 ml MS | 10 | 1.4 (0.62) |
| Resuspended in 5 ml MS | 10 | 1.6 (0.72) | ||
| Resuspended in 1 ml MS (control) | 10 | 2.4 (1.16) | ||
| Media type in Inoculation Media | LB | 10 | 0.4 (0.22) | |
| MS (control) | 10 | 2.4 (1.16) | ||
| Addition of Acetosyringone to Inoculation Media | With Acetosyringone | 11 | 0.9 (0.37) | |
| Without Acetosyringone (control) | 11 | 0.6 (0.64) | ||
| Age of stem tissue inoculated | 6 months (control) | 11 | 1.1 (0.66) | |
| 18 months | 11 | 1.8 (0.95) | ||
| A. tumefaciens strain | AGL1 (control) | 18 | 0 (0) | |
| C58 | 18 | 0.1 (0.1) | ||
| LBA4404 | 18 | 0.6 (0.4) | ||
| Populus alba 'pyramidalis' | Concentration of A. tumefaciens in media | Resuspended in 25 ml MS | 10 | 2.2 (0.55) |
| Resuspended in 5 ml MS | 10 | 2.7 (0.63) | ||
| Resuspended in 1 ml MS (control) | 10 | 5.1 (1.58) | ||
| Media type used for stem inoculation | LB | 10 | 2.8 (0.71) | |
| MS (control) | 10 | 5.1 (1.58) | ||
| Addition of Acetosyringone to Inoculation Media | With Acetosyringone | 11 | 0.3 (0.14) | |
| Without Acetosyringone (control) | 11 | 0.5 (0.25) | ||
| Age of stem tissue inoculated | 6 months (control) | 11 | 1.5 (0.33) | |
| 18 months | 11 | 1.5 (0.63) | ||
| A. tumefaciens strain | AGL1 (control) | 20 | 8.1 (2) | |
| C58 | 20 | 12.5 (2) | ||
| LBA4404 | 20 | 11.1 (2) |
Table 3: Cambial ATScm-2 values for a range of variables investigated as part of protocol optimization trails. Variables were investigated in a poplar clone and a range of Eucalyptus globulus individuals across a number of years with cambial ATScm-2 data presented. Cambial windows were harvested using the disc harvesting protocol (step 5.2.2). Standard error values in brackets.
| Eucalyptus globulus x camaldulensis clone ID | Number of cambial windows for each month | Cambial ATScm-2 Late Spring (November) Inoculation | Cambial ATScm-2 Early Summer (December) Inoculation | Cambial ATScm-2 Mid Summer (January) Inoculation | Cambial ATScm-2 Late Summer (February) Inoculation | Cambial ATScm-2 Early Autumn (March) Inoculation | Cambial ATScm-2 all months combined |
| SG5 | 10 | 15.7 (7.8) | 50.7 (13.7) | 10.5 (5.1) | 30.7 (4.4) | 16.8 (6.3) | 24.9 (4.3) |
| SG6 | 10 | 6.1 (3.1) | 38.5 (15.1) | 4.5 (1.7) | 14.5 (3.3) | 27.6 (8.4) | 18.3 (4) |
| SG13 | 10 | 28.4 (6.7) | 41.8 (9.1) | 16.1 (7) | 29.3 (5.2) | 19.8 (4.3) | 27.1 (3.1) |
| SG18 | 10 | 6.7 (2.9) | 18.3 (6.7) | 17.4 (3.4) | 19.2 (6.1) | 18.1 (6.3) | 15.5 (2.4) |
| SG21 | 10 | 38.7 (14.3) | 77 (17.5) | 23.9 (5.7) | 27.5 (6.9) | 40.7 (13.5) | 41.6 (6) |
| SG35 | 10 | 23.5 (10.2) | 42.8 (12.2) | 7.6 (2.5) | 17.6 (4.3) | 31.3 (4.6) | 24.6 (3.8) |
| SG37 | 10 | 19.7 (8) | 55.2 (14.5) | 7.4 (1.8) | 35 (10.4) | 15.9 (4.5) | 26.6 (3.8) |
| SG39 | 10 | 12.5 (2.4) | 23.6 (6.3) | 6.9 (3) | 26.3 (8.1) | 7.3 (2.3) | 15.5 (2.6) |
| SG40 | 10 | 24.8 (11) | 24.5 (6.8) | 14 (5.6) | 35.6 (6.1) | 19.9 (4.2) | 23.8 (3.2) |
| SG44 | 10 | 22.3 (3.4) | 63 (29.3) | 9.8 (2.6) | 52.5 (10.3) | 54.3 (15) | 40.4 (7.4) |
| SG46 | 10 | 15.3 (5.2) | 63.9 (20.1) | 23.6 (4) | 22.5 (4.8) | 17.4 (4.9) | 28.5 (5.1) |
| Monthly Cambial ATScm-2 | 19.9 (5) | 45.3 (9.1) | 12.6 (2.8) | 27.8 (4.5) | 24 (5.1) |
Table 4: Influence of genotype and time of inoculation on cambial ATScm-2 in eucalypt clones. Ten windows were introduced each month over the growing season to 10 different Eucalyptus globulus x camaldulensis clones which were all harvested in May. Monthly data is presented for each genotype with an overall average monthly and genotypic ATScm-2 presented also. Cambial windows were harvested using the phloem peel harvesting protocol (step 5.2.1). Standard error values in brackets. Please click here to view a larger version of this table.
| Plant Number | Number of cambial sectors excised | Total mass of all sectors (µg) | Average mass of sectors (µg) |
| 1 | 11 | 750 | 68 |
| 2 | 19 | 1,460 | 77 |
| 3 | 21 | 160 | 8 |
| 4 | 26 | 4,460 | 172 |
| 5 | 15 | 1,320 | 88 |
| 6 | 22 | 2,490 | 113 |
| 7 | 19 | 1,720 | 91 |
| 8 | 30 | 830 | 28 |
| 9 | 25 | 360 | 14 |
| Total | 188 | 13,550 | 72 |
Table 5: Typical mass of a cambial sector. 45 windows across nine plants were investigated in poplar to determine the average mass of a sector and harvested using the disc harvesting protocol (step 5.2.2). Plants were grown for 5 months and the radial growth across a cambial window was between 1-4 mm.
| Morph- ological trait |
GUS only (positive control) | Non-transgenic tissue (GUS only) | P-value | Gene of Interest 1 (FLA1) | Non-transgenic tissue (FLA1) | P-value | Gene of Interest 2 (FLA2) | Non-transgenic tissue (FLA2) | P-value | Gene of Interest 3 (FLA3) | Non-transgenic tissue (FLA3) | P-value |
| Average Cell Wall Thickness (µm)* | 1.81 (0.1) | 1.65 (0.1) | 0.2692 | 1.47 (0.14) | 1.55 (0.12) | 0.6403 | 1.65 (0.13) | 1.78 (0.13) | 0.4839 | 1.59 (0.12) | 1.63 (0.11) | 0.8254 |
| Average Cell Wall Area (µm2)* | 69.1 (4.74) | 60.7 (2.03) | 0.1229 | 64.1 (15.68) | 59.9 (9.79) | 0.5004 | 61.6 (3.4) | 65 (3.96) | 0.5182 | 61.9 (3.56) | 61.5 (3.16) | 0.9438 |
| Average Cell Area (µm2)* | 115.9 (11.01) | 99.9 (5.1) | 0.2041 | 124.4 (6.1) | 108 (4.36) | 0.0445 | 110.8 (8.45) | 111.3 (9.06) | 0.9671 | 108.5 (4.43) | 103 (3.07) | 0.3204 |
| Average Lumen Area (µm2)* | 46.9 (7.55) | 39.2 (4.89) | 0.407 | 60.2 (5.28) | 48.1 (4.46) | 0.0991 | 49.2 (7.56) | 46.3 (7.5) | 0.7882 | 46.6 (3.31) | 41.5 (3.9) | 0.3264 |
| Average Microfibril Angle (O)# | 24.3 (0.75) | 24.2 (0.45) | 0.8648 | 22.3 (0.82) | 22.7 (0.7) | 0.5664 | 23.7 (0.55) | 26.6 (0.5) | 0.0001 | 26 (0.88) | 27.2 (0.72) | 0.0903 |
Table 6: ISSA Fiber morphology measurements derived from gene of interest study. Comparison of fiber cell wall, cell size and MFA measurements taken from transgenic fibers sourced from Eucalyptus globulus x camaldulensis clones stems transformed with Eucalyptus grandis FLAs (EgrFLA1, 2, 3) and positive control (GUS only) with the adjacent non transgenic control fibers. Data sourced from MacMillan et al.9. * indicates trial involved the measurement of 10 fibers in 10 sector (total of 100 fibers). # indicates trial involved the measurement of 5 fibers in 20 sectors (total of 100 fibers). α <0.05 shown in bold. Please click here to view a larger version of this table.
| EgCESA1 | EgCESA2 | EgCESA3 | EgCESA4 | EgCESA5 | EgCESA7 | GUS only (positive control) | |
| EgrCESA1 | 0.708 | 2.839 | 8.856 | 9.976 | 9.18 | 29.165 | |
| EgrCESA2 | 1.11 | 11.05 | 12.008 | 11.363 | 30.234 | ||
| EgrCESA3 | 15.151 | 16.123 | 15.488 | 37.791 | |||
| EgrCESA4 | 4.852 | 0.108 | 46.858 | ||||
| EgrCESA5 | 3.275 | 11.278 | |||||
| EgrCESA7 | 36.082 | ||||||
| GUS only (positive control) |
Table 7: Chi2 values derived from comparison of promoter expression patterns in cambial derivatives. Chi2 analysis comparing the presence/absence of GUS staining in P, X1 and X2 region of cambial derivatives between six Eucalyptus grandis CESA gene promoters sequences (EgrCESA 1, 2, 3, 4, 5, 7) and positive control (GUS only). Data sourced from Creux et al.10. α <0.05 shown in underline.
Discussion
The ISSA protocol is a relatively simple and efficient method for the creation of transgenic stem tissues in tree species in the space of a few months for analysis of genes and promoters of interest involved in wood and stem formation. Little effort, beyond keeping plants alive, is required to grow transgenic stem tissue following inoculation which stands in contrast to in vitro methods where extensive culturing is required to maintain tissue or plants, where wood production can take up to years to commence or where a true secondary stem is not created1. ISSA has also been shown to be applicable to a broad range of tree species, including species in the genus of Populus, Eucalyptus and Pinus6 as well as Acacia and Corymbia (data not shown) where very little, if any, optimization is required to derive transgenic tissue. Using this method, it is therefore possible to investigate a number of genes or promoters of interest in most species of interest. Further, the protocol can be used on its own or in conjunction with more widely used in vitro techniques where multiple genes can be investigated using ISSA in the first instance to identify potential candidates to take forward to more involved in vitro methods.
The described examples show that sufficient sectors can be created to undertake downstream analysis from only a small number of transgenic cambial windows. With ATScm-2 values of 46.1 in eucalypt and 14.6 in poplar only one or two cambial windows would be sufficient to investigate a single trait as it is possible to add numerous windows to a single plant stem or replicate on numerous plants. In practice, however, not all cambial windows lead to the creation of a cambial sector that can be utilized (see Table 1, Table 2) as the size of sectors can vary, they are generally small and in cases can be located in close proximity to each other limiting their use for downstream analysis. In addition, the gene of interest may impact on cell viability12 or other aspects of cell development potentially resulting in the complete absence of transgenic sectors for a specific gene of interest. It is therefore advisable to always aim for a greater number of windows than might be needed for analysis. Similar caution should be applied when using ISSA for promoter studies as significant variability can be expected with little/weak or no expression observed in some cases (Table 2). However, a smaller number of sectors, as little as 6, have been shown to be successfully used for analysis10. These results show that quite modest experiments involving only a small number of windows can lead to significant insights into gene function and promoter activity.
From an analysis perspective, the close proximity of transgenic and non-transgenic tissue provides a powerful tool to identify small but meaningful differences in traits between genes of interest. Cells and tissues sampled directly adjacent to each other are of the same genotypic background and have been influenced by the same environmental conditions reducing the number of independent variables and simplifying the statistical analysis required. From a sampling perspective, power analysis on ISSA derived data from multiple experiments (e.g., Table 6), has shown that a smaller number of measurements per sector, e.g., cell wall thickness of 3-5 cells per sector, and measuring a larger number of sectors, e.g., 20-25 sectors for a gene of interest, provides a more efficient and statistically robust sampling strategy. Each sector represents an independent transformation event or 'line' of cells with the statistical analysis undertaken determining the average effect of multiple transformation events involving the gene of interest. Equally, the ability to transform all stem tissues provides an opportunity to study the expression of promoters of interests across the whole stem and more specifically during cambium differentiation. ISSA protocols described here provide a reliable and validated pipeline from the creation of transgenic tissue through to the harvesting of samples, collection and analysis of data for tissue and cell morphology and for promoter expression patterns.
In most instances, sectors are the result of the transformation of a single cell, which through cell division may have given rise to a number of derivative cells and tissues. For example, a cambial sector arises from transformation of a single cell that post wound recovery becomes a cambial initial which in turn divides periclinally and anticlinally to create a sector extending all the way from the original wound into the phloem. In these cases, the transformed initial was maintained within the cambium until the time of harvest and a 'full' variant is observed, however, where this initial is lost from the cambial layer during radial growth and replaced by a non-transformed neighboring cell through 'cell-invasion', then a 'lost initial' variant will be the result. In order to ensure correct classification of sector types and to assist with the interpretation of results it is strongly recommended that familiarization with anatomical features of secondary stem development and wound responses be prerequisites for the analysis of ISSA results. Also, regular testing of the pH of the GUS reagent is advised (see step 5.6). A low pH (e.g., below 6) can lead to activation of endogenous β-glucuronidases (GUS) in the plant stem which can mask the true extent of a sector or lead to the identification of false positives. Where this is suspected, it is recommended that these samples be excluded from further analysis. The GUS reagent protocol reliably applied in the studies outlined here in order to alleviate potential problems with endogenous GUS activity was adapted from Hawkins et al. (2002)16 and utilizes additional key steps including two rinses with phosphate buffer to equilibrate samples to pH7, and heat treatment at 55 oC for 10 minutes to destabilize endogenous GUS activity17,18. GUS reagent permeability is also limited and is responsible for fewer sectors being identified using the transverse disc protocol compared to the phloem peel protocol.
All species trialed have shown to be susceptible to transformation of cambial tissue. However, significant differences in the ATScm-2 both between and within genus and species have been observed. Similarly the strain of A. tumefaciens and time/season of inoculation can influence ATScm-2. It is suggested that undertaking some preliminary testing of these variables in the species and genotypes under investigation prior to larger scale adoption of the protocol. Theoretically there is no limit to the age or size of stems to which ISSA could be applied as long as there is an actively growing cambium. However, for ease of downstream processing of transgenic tissue sectors it is recommended inoculating stems of about 1 cm diameter.
ISSA is not without its limitations. The relatively small size of transgenic tissue sectors currently restricts downstream phenotyping methods and consequently only a limited number of such methods have been routinely used to date. These methods are mainly focused on morphological characterization as outlined above as well as semi quantitative estimate of monosaccharides composition in the secondary cell wall as noted in introduction 9,11. With the advent and refinement of new technologies for fine scale analysis of biological materials there is further potential to develop additional phenotyping pipelines for ISSA sectors, for instance in cell wall compositional analysis. It remains difficult, however, to perform quantitative, sector by sector, Ribonucleic Acid (RNA) based gene expression and protein studies and/or identify and propagate ISSA derived transgenic cells through in vitro culturing for additional analysis due the destructive nature of the GUS assay. Some fluorescent reporter genes and tissue printing have been trialed to quantify RNA from individual sectors but to date these approaches have not been successfully used for medium to high throughput screening in routine operation. Further, transgenic techniques not trialed to date such as RNAi could complicate analysis where GUS expression and down-regulation of target gene may not co-localize.
While there are some current limitations, incorporation of new and emerging technologies and analytical methods are likely to overcome these limitations, further enhancing the role of ISSA as a medium to high throughput protocol for dissecting the complex molecular nature of cambial differentiation, wood development and stem formation.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
The authors would like to acknowledge funding support for aspect of the work through Linkage Grants LP0776563 (GB, AS) and LP0211919 (GB) and industry partners Sappi and Mondi as well as Australian Postgraduate Award (EM) from the Australian Research Council and the Young Innovators and Scientist Award through the Australian Department of Agriculture (LT). We also like to thank the Zander Myburg, Qing Wang, Colleen MacMillan and Simon Southerton for the many discussions and ideas they put forward during the development of this protocol and to Martin Ranik, Minique De Castro, Julio Najera, Valerie Frassiant, Angelique Manuel and Noemie Defaix for assistance in laboratory related work.
Materials
| Plants | NA | NA | Please consult local nursery suppliers for plants as needed |
| Agrobacterium strain | NA | NA | There are many possible avenues to obtain Agrobactrium strains. We suggest you follow up within your local research community as there may be restrictions in obtaining the bacteria in your country and region. |
| Binary vector (gene and promoter) | NA | NA | We have developed a range of vectors to suite the ISSA protocol using a the Gateway Recombinase system. This include overexpression, RNAi knockouts and promoter fusion vectors based on modified pCAMBIA vectors and happy to provide as needed. In addition, there are many vectors avialable to the research community. |
| LB media | Sigma | L3022 | The same product could be sourced from another company |
| LB media with agar | Sigma | L2897 | A like product could be sourced from another company |
| Antibiotics | Sigma | NA | The catalog number will be dependent on the antibiotic you require as a range of antibiotic are used for bacterial selection in binary vectors. This product could be sourced from a range of companies |
| 50 ml Screw top tubes | Fisher Scientific | 14-432-22 | The same product could be sourced from another company |
| 2 ml Microtube | Watson Bio Lab | 132-620C | The same product could be sourced from another company |
| MS Media | Sigma | M9274 | The same product could be sourced from another company |
| Scalpel blade no 11 | Sigma | S2771 | The same product could be sourced from another company |
| Parafilm "M" | Bemis | PM996 | This is the best product to use to bind the cambial window post creation |
| 14 ml round bottom tubes | Thermo Scientific | 150268 | The same product could be sourced from another company |
| EDTA | Sigma | E6758 | The same product could be sourced from another company |
| Triton | Sigma | X100 | The same product could be sourced from another company |
| X-Gluc | X-GLUC direct | You will need to go to the website to order – http://www.x-gluc.com/index.html | |
| Potassium Ferricyanide (III) | Sigma | 244023 | The same product could be sourced from another company |
| Potassium Ferrocyanide (II) | Sigma | P9387 | The same product could be sourced from another company |
| Litmus paper | Sigma | WHA10360300 | The same product could be sourced from another company |
| Single edge razor blade | ProSciTech | L055 | The same product could be sourced from another company |
| Double edge razor blade | ProSciTech | L056 | The same product could be sourced from another company |
| SEM Pin Stub | ProSciTech | GTP16111 | The same product could be sourced from another company |
| Sample vial with screw cap | ProSciTech | L6204 | The same product could be sourced from another company |
| Ethanol | sigma | E7023 | The same product could be sourced from another company |
| LR white | ProSciTech | C025 | The same product could be sourced from another company |
| Embedding Mould | ProSciTech | RL090 | We recommend this variety, however there are plenty of options available |
| Water Soulable mounting media | ProSciTech | IA019 | One example of a mounting media that could be used however other options do exist and could be explored. |
| Hydrogen peroxide | Sigma | 216763 | A like product could be sourced from another company |
| Glacial acetic acid | Sigma | A9967 | A like product could be sourced from another company |
| Safranin O | ProSciTech | C138 | A like product could be sourced from another company |
| Quanta Environmental Scanning Electron Microscope | FEI | This is the instrument used at part of this study but any other SEM that has a low vacuum mode could be utilised | |
| Image J imaging software | can be sourced from the following URL http://rsbweb.nih.gov/ij/ |
Referenzen
- Spokevicius, A. V., Tibbits, J. F. G., Bossinger, G. Whole plant and plant part transgenic approaches in the study of wood formation – benefits and limitations. TPJ. 1 (1), 49-59 (2007).
- Chaffey, N. Wood formation in forest trees: from Arabidopsis to Zinnia. Trends Plant Sci. 4 (6), 203-204 (1999).
- Moller, R., McDonald, A. G., Walter, C., Harris, P. J. Cell differentiation, secondary cell-wall formation and transformation of callus tissue of Pinus radiata D. Don. Planta. 217 (5), 736-747 (2003).
- Spokevicius, A. V., Van Beveren, K., Leitch, M. M., Bossinger, G. Agrobacterium-mediated in vitro transformation of wood-producing stem segments in eucalypts. Plant Cell Rep. 23 (9), 617-624 (2005).
- Plasencia, A., et al. Eucalyptus hairy roots, a fast, efficient and versatile tool to explore function and expression of genes involved in wood formation. Plant Biotech J. , (2015).
- Van Beveren, K. S., Spokevicius, A. V., Tibbits, J., Wang, Q., Bossinger, G. Transformation of cambial tissue in vivo provides efficient means for Induced Somatic Sector Analysis (ISSA) and gene testing in stems of woody plants species. Funct Plant Biol. 33 (7), 629-638 (2006).
- Spokevicius, A. V., Van Beveren, K., Bossinger, G. Agrobacterium-mediated transformation of dormant lateral buds in poplar trees reveals developmental patterns in secondary stem tissues. Funct Plant Biol. 33 (2), 133-139 (2006).
- Spokevicius, A. V., et al. beta-tubulin affects cellulose microfibril orientation in plant secondary fiber cell walls. Plant J. 51 (4), 717-726 (2007).
- MacMillan, C. P., et al. The fasciclin-like arabinogalactan protein family of Eucalyptus grandis contains members that impact wood biology and biomechanics. New Phytol. 206 (4), 1314-1327 (2015).
- Creux, N. M., Bossinger, G., Myburg, A. A., Spokevicius, A. V. Induced somatic sector analysis of cellulose synthase (CesA) promoter regions in woody stem tissues. Planta. 237 (3), 799-812 (2013).
- Hussey, S. G., et al. SND2, a NAC transcription factor gene, regulates genes involved in secondary cell wall development in Arabidopsis fibers and increases fiber cell area in Eucalyptus. BMC Plant Biology. 11, (2011).
- Melder, E., Bossinger, G., Spokevicius, A. V. Overexpression of ARBORKNOX1 delays the differentiation of induced somatic sector analysis (ISSA) derived xylem fiber cells in poplar stems. Tree Genet. Genomes. 11 (5), (2015).
- Baldacci-Cresp, F., et al. PtaRHE1, a Populus tremula x Populus alba RING-H2 protein of the ATL family, has a regulatory role in secondary phloem fiber development. Plant J. 82 (6), 978-990 (2015).
- Sambrook, J., Russell, D. W. . Molecular cloning: A laboratory manual. , (2001).
- Murashige, T., Skoog, F. A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiol. Plant. 15 (3), 473-497 (1962).
- Hawkins, S., Pilate, G., Duverger, E., Boudet, A., Grima-Pettenati, J., Chaffey, N. The use of GUS histochemistry to visualise lignification gene expression in situ during wood formation. Wood formation in trees: Cell and Molecular Biology Techniques. , 271-295 (2002).
- Hodal, L., Bochardt, A., Nielsen, J. E., Mattsson, O., Okkels, F. T. Detection, expression and specific elimination of endogenous beta-glucuronidase activity in transgenic and nontransgenic plants. Plant Sci. 87 (1), 115-122 (1992).
- Hansch, R., Koprek, T., Mendel, R. R., Schulze, J. An improved protocol for eliminating endogenous beta-glucuronidase background in barley. Plant Sci. 105 (1), 63-69 (1995).

