Examining Proteasome Assembly with Recombinant Archaeal Proteasomes and Nondenaturing PAGE: The Case for a Combined Approach
Summary
This protocol uses both subunit coexpression and postlysis subunit mixing for a more thorough examination of recombinant proteasome assembly.
Abstract
Proteasomes are found in all domains of life. They provide the major route of intracellular protein degradation in eukaryotes, though their assembly is not completely understood. All proteasomes contain a structurally conserved core particle (CP), or 20S proteasome, containing two heptameric β subunit rings sandwiched between two heptameric α subunit rings. Archaeal 20S proteasomes are compositionally simpler compared to their eukaryotic counterparts, yet they both share a common assembly mechanism. Consequently, archaeal 20S proteasomes continue to be important models for eukaryotic proteasome assembly. Specifically, recombinant expression of archaeal 20S proteasomes coupled with nondenaturing polyacrylamide gel electrophoresis (PAGE) has yielded many important insights into proteasome biogenesis. Here, we discuss a means to improve upon the usual strategy of coexpression of archaeal proteasome α and β subunits prior to nondenaturing PAGE. We demonstrate that although rapid and efficient, a coexpression approach alone can miss key assembly intermediates. In the case of the proteasome, coexpression may not allow detection of the half-proteasome, an intermediate containing one complete α-ring and one complete β-ring. However, this intermediate is readily detected via lysate mixing. We suggest that combining coexpression with lysate mixing yields an approach that is more thorough in analyzing assembly, yet remains labor nonintensive. This approach may be useful for the study of other recombinant multiprotein complexes.
Introduction
Multiprotein complexes carry out numerous critical cellular activities1. For many of these complexes, much more is known about their structure and function than about their assembly2,3. The proteasome is one such complex and is found in all domains of life. In eukaryotes, this molecular machine is at the core of the Ubiquitin/Proteasome System (UPS) and provides the major route of intracellular protein degradation4. The eukaryotic proteasome (referred to as the 26S proteasome) is comprised of two major sub assemblies: a 20S proteasome, or Core Particle (CP)5, that can be capped on one or both ends by a 19S Regulatory Particle (RP)6.
The 20S proteasome is a large compartmentalized protease. Its quaternary structure is absolutely conserved across all domains of life and consists of a stack of four seven-membered rings containing two types of structurally related subunits, α and β5,7,8. In eukaryotes, the two outer rings are each comprised of seven distinct α subunits and the two inner rings are each comprised of seven distinct β subunits; proteolytic activity resides within three of the β subunits. By contrast, the CP rings of archaea and bacteria are usually comprised of only one type of α and one type of β subunit. Archaeal proteasomes have provided an important model system to study proteasome assembly due to both their compositional simplicity and their sharing a common assembly mechanism with their eukaryotic counterparts9-13. In brief, α subunits assemble into α rings first, which serve as a scaffold onto which β subunits assemble. The resulting half-proteasomes (α7β7) dimerize, giving rise to fully assembled CP (α7β7β7α7). During dimerization, the propeptides present on β subunits are autocatalytically removed, exposing the catalytic N-terminal threonines. The use of archaeal proteasomes to model assembly frequently takes advantage of the production of recombinant archaeal proteasome proteins in Escherichia coli. This is a worthwhile approach because it enables the subunits to be produced in various combinations, as both WT and mutant versions, in a host organism that does not produce its own proteasomes.
Monitoring the assembly of multi-protein complexes biochemically requires some kind of fractionation method that separates fully assembled complexes from assembly intermediates and precursors. Due to its superior resolving capacity, nondenaturing Polyacrylamide Gel Electrophoresis (PAGE) has proven to be especially useful in the fractionation of various large multiprotein complexes14-17. The combination of recombinant archaeal proteasome production and nondenaturing PAGE has become a powerful approach in dissecting proteasome assembly9,11,12,18. However, the usual method by which this approach is applied (i.e. via the recombinant coexpression of α and β subunits) has an important drawback. Assembly reactions are cooperative and strongly concentration-dependent3. Given that the protein concentration inside cells is very high19, due to excluded volume effects, assembly reactions proceed rapidly in vivo. Hence it is possible to miss important assembly intermediates when α and β subunits are coexpressed.
Here, we argue for a combined approach in the study of proteasome assembly using recombinant archaeal proteasome subunits. In this approach, both coexpression and lysate mixing methods are employed. The former allows for rapid analysis of assembly because coexpression is less labor intensive. The latter depends on separate expression of α and β subunits, followed by mixing. Though this requires a bit more effort than coexpression, it is more than compensated for by the ability to detect intermediates that are missed during coexpression. Together, these two methods can provide a more complete picture of proteasome assembly.
Protocol
1. Bacterial Expression.
Note: Expression plasmids used in this study are described in Table 1. Solutions, media, and buffers used in this study are described in Table 2. The cloning of archaeal proteasome subunit genes and the generation of expression plasmids are described elsewhere18,20. In brief, plasmids for recombinant coexpression of subunits employ a bicistronic operon strategy which helps in obtaining comparable expression levels of individual subunits18,20. The expression parameters listed below were empirically determined to be optimal for the proteasome subunits in this study. It may be necessary to optimize expression for other proteasome mutants, for proteasomes from other archaeal species, or for other recombinant protein complexes (see Discussion).
- Transform expression plasmid of interest into chemically competent E.coli BL21(DE3) cells.
- Add 1 – 2 μL of plasmid (typically 50 – 100 ng) to an aliquot of DE3 cells that were freshly thawed on ice, and continue to incubate on ice for 20 min.
- Heat shock the cells at 42 °C for 45 s and return tube to ice for additionl 2 min.
- Add 1 mL of LB medium and incubate at 37 °C with shaking (150 rpm) for 1 h.
- Spread 100 – 200 μL of the transformation mixture onto LB-kan plates (plates containing solid LB media, supplemented with kanamycin (all plasmids used in this study encode resistance to this antibiotic)). Incubate plates at 37 ºC O/N.
- Next morning, inoculate a single colony from the LB-kan plate into 3 mL of liquid LB-kan media in a glass culture tube. Incubate at 37 ºC with shaking (150 rpm) for approximately 2.5 – 3 h, or till turbidity is observed.
- Measure the optical density of the culture at 600 nm (OD600) in a spectrophotometer. Dilute the cell culture with an appropriate amount of prewarmed LB-kan media to an OD600 of 0.4 in a final volume of 6 mL. Resume shaking at 37 ºC for 40 min.
- Induce protein expression in the liquid cultures by adding IPTG from a stock solution to a final concentration of 1 mM. Incubate at 37 ºC with shaking (150 rpm) for approximately 6 – 7 h.
- Harvest bacterial cell cultures in their entirety into 1.5 mL microcentrifuge tubes.
- Add 1.5 mL of a culture into microcentrifuge tube. Centrifuge at 10,000 × g for 1 min.
- Discard the supernatant and add another 1.5 mL of the same culture to the pellet. Centrifuge at 10,000 × g for 1 min.
- Repeat step 1.11 until the entire culture is harvested into the microcentrifuge tube.
- Store the induced pellets at -80 °C until lysis.
2. Bacterial Lysis and Lysate Mixing.
Note: For samples studied via coexpression, follow section 2.1 (and its subsections). For samples requiring lysate mixing, follow section 2.2 (and its subsections). The TSP (Total, Soluble, Pellet) analysis that is included in the protocol is useful in optimizing protein expression which can help ensure that approximately equal amounts of α and β subunits are combined during lysate mixing (see Discussion). It also provides an important control if downstream results are not as expected (i.e. it verifies that protein was expressed and soluble). Thus, we highly recommend including TSP analysis initially. Once an optimal protocol has been achieved for a particular subunit combination, TSP analysis is not strictly required which is why it is described as optional below.
- Lysis of bacterial cells and preparation of soluble fractions.
- Thaw the induced cell pellet on ice for 5 min. Resuspend the pellet in 600 µL of lysis buffer.
- Incubate the suspension at 30 ºC with shaking (150 rpm) for 30 min to generate total crude lysate.
Note: The following step and the subsection that follows are optional. - Remove two 25 μL aliquots from the total crude lysate into separate 1.5 mL microcentrifuge tubes and carry out TSP analysis as follows.
- To one of the two 25 μL aliquots, add 5X SDS sample buffer to a final concentration of 1X. Label the tube with "T" for "total crude lysate".
- Incubate the "T" sample at 100 ºC for 5 min to completely denature proteins.
- Meanwhile, take the second 25 μL aliquot and centrifuge it at 10,000 × g for 10 min. Carefully remove the supernatant without disturbing the tiny pellet, and transfer it to a new 1.5 mL microcentrifuge tube. Label this new tube "S" for "soluble fraction".
- To the tiny pellet left behind in the previous step, add 25 μL of lysis buffer and vortex to resuspend. Label this tube "P" for "pellet fraction".
- Add 6 μL of 5X SDS sample buffer to the "S" and "P" tubes and incubate at 100 ºC as described above.
Note: If the TSP samples will not be used that day to analyze expression, they may be frozen at -20 ºC until needed. Once thawed, they must be reincubated at 100 ºC, as described above, immediately prior to loading on a 12% and/or 15% standard SDS-PAGE gel.
- While the TSP analysis is carried out, centrifuge the remaining 550 μL of total crude lysate (or the entire 600 μL of total crude lysate if TSP analysis is not carried out) at 10,000 × g for 10 min. Collect the supernatant in a fresh 1.5 mL microcentrifuge tube. This is the soluble lysate that will be used for purification.
- Lysate mixing.
- Carry out lysis as described in steps 2.1.1 and 2.1.2 above.
- Mix 600 µL of total crude lysate from bacteria expressing the desired α subunit with 600 µL of total crude lysate from bacteria expressing the desired β subunit. Incubate at 37 ºC with slow shaking for 30 min.
Note: It may be necessary to optimize incubation time to achieve maximum assembly during lysate mixing (see Discussion). The time and temperature presented here are optimal for the recombinant proteins in this study and were determined elsewhere18. - Centrifuge the mixed lysate at 10,000 × g for 10 min to separate soluble material from insoluble pellet. Transfer the supernatant to a new 1.5 mL microcentrifuge tube. Use this mixed soluble lysate for protein purification.
3. Protein Purification via Immobilized Cobalt Affinity Resin (ICAR).
- Equilibrate the resin.
- Thoroughly resuspend the resin by inverting the bottle and transfer 50 µL of the slurry to a 1.5 mL microcentrifuge tube. Centrifuge at 700 × g for 2 min to pellet resin. Carefully aspirate supernatant.
Note: We carry out pipette aspiration using a blue 1 mL pipette tip to remove the bulk of the liquid. A white 2 μL tip is used for fine control of removal of the remaining supernatant, leaving a very thin layer of liquid covering the beads. - Add 1 mL of Buffer A. Mix gently to resuspend resin and centrifuge at 700 × g for 2 min to pellet resin. Carefully aspirate the supernatant and repeat this wash step one more time.
- Thoroughly resuspend the resin by inverting the bottle and transfer 50 µL of the slurry to a 1.5 mL microcentrifuge tube. Centrifuge at 700 × g for 2 min to pellet resin. Carefully aspirate supernatant.
- Apply soluble lysate obtained previously in section 2.1 or 2.2 to the equilibrated resin. Incubate the tube with gentle rotation at 4 ºC for 60 min. Centrifuge the tube at 700 × g for 5 min and carefully aspirate the supernatant.
- Wash the resin as follows to remove nonspecifically bound proteins.
- Resuspend resin with 1 mL of Buffer A and incubate with gentle rocking at 4 ºC for 10 min. Centrifuge the tube at 700 × g for 5 min and carefully aspirate the supernatant. Repeat this wash step one more time.
- Resuspend resin with 1 mL of Buffer B (Buffer A with 5 mM Imidazole) and incubate with gentle rocking at 4 ºC for 5 min. Centrifuge the tube at 700 × g for 5 min and carefully aspirate the supernatant. Repeat this wash step one more time.
- Resuspend resin with 1 mL of Buffer C (Buffer A with 10 mM Imidazole) and incubate with gentle rocking at 4 ºC for 5 min. Centrifuge the tube at 700 × g for 5 min and carefully aspirate the supernatant.
- Elute the protein by adding 400 µL of Buffer E (Buffer A with 200 mM Imidazole) to the resin. Incubate with gentle rocking at 4 ºC for 5 min. Centrifuge at 700 × g for 5 min.
- Transfer supernatant containing purified protein to a new 1.5 mL centrifuge tube.
- Desalt the purified protein by serial centrifugation as follows.
- To 400 μL of purified protein, add 100 μL Buffer A (this reduces Imidazole concentration from 200 mM to 160 mM). Apply purified protein to 0.5 mL ultracentrifugal filters with a 10 kDa molecular weight cut-off. Centrifuge at 14,000 × g for 5 min.
- Discard the filtrate and add 400 μL Buffer A to dilute the retentate and centrifuge again. Continue the cycles of centrifugation/dilution until imidazole concentration falls below 4 mM. As an example, if each centrifugation concentrates 500 μl down to ~70 μl (or approximately 7-fold), then two cycles will reduce the starting 160 mM imidazole concentration (7×7 = 49-fold) to approximately 3.3 mM.
- Measure the protein concentration of the desalted sample using the BCA assay21.
Note: Desalting is required to reduce Imidazole levels below the tolerance limit for the BCA assay, as described in the manufacturer's instructions. Our lab prefers the BCA assay because it is sensitive, has a large dynamic range, and exhibits much less protein-to-protein variation. However, other methods to determine protein concentration can be substituted for the BCA assay. The key is to be aware of each method's advantages and limitations.
- Add 5X native sample buffer to a final concentration of 1X and proceed to electrophoresis. Alternatively, store samples at -20 ºC for later analysis.
4. Nondenaturing Polyacrylamide Gel Electrophoresis (PAGE).
Caution: Unpolymerized acrylamide is a neurotoxin. Wear appropriate protective gear.
- Prepare the nondenaturing PAGE gel as follows.
- Prepare the gel cassette for casting using clean glass plates and casting stand. Place gradient maker on top of a small magnetic stirrer and place a tiny stir bar into each chamber.
- Using 40% (w/v) acrylamide and 2% (w/v) bisacrylamide stock solutions, prepare 5% and 10% (w/v) acrylamide gel solutions in a native resolving buffer. Ensure that the ratio of total acrylamide to bisacrylamide is 37.5:1. Chill on ice prior to pouring.
Note: The nondenaturing PAGE system used here is essentially identical to the Laemmli SDS-PAGE system, with SDS omitted22. - Add Ammonium Persulfate (from a 10% (w/v) stock solution prepared in water) to the acrylamide solutions to initiate polymerization. Final concentration of Ammonium Persulfate is 0.1% (w/v).
- Pour the acrylamide solutions into the two chambers of the gradient maker. With the outlet tubing inserted between the plates of the gel cassette, activate the magnetic stirrer and open the gradient maker valves. Pour the 5 – 10% nondenaturing gradient gel.
- Once poured, overlay the gel with a thin layer of Isopropanol and allow the gel to polymerize for 30 min. During this time, prepare fresh 5% acrylamide gel solution in native resolving buffer. After the gel sets, pour off Isopropanol and rinse the top of the gel with deionized water from a squirt bottle.
- To the 5% gel solution prepared above, add Ammonium Persulfate (exactly as described in 4.1.3) and pour on top of the polymerized gel until glass plates are full. Insert gel comb. Allow the overlaid gel to polymerize for an additional 30 min.
- Once gel is polymerized, assemble gel cassette into electrophoresis apparatus. Alternatively, store the gel at 4 ºC, wrapped in moistened paper towels and plastic wrap until ready for use.
- Once nondenaturing PAGE gel is assembled into electrophoresis apparatus, fill the tank with 1X native running buffer prepared fresh from a 10X native running buffer stock.
- Load 10 μg of purified protein, obtained at the end of section 3, into each well using a glass syringe.
- Load 2 μL of high molecular weight native protein standard (diluted in 1X native running buffer) into one of the wells.
- Run gel at 55 V and 4 ºC until the dye front runs off the gel (approximately 4 – 4.5 h).
- In addition to the nondenaturing gel, analyze aliquots of the purified protein by standard SDS-PAGE22.
- Mix aliquots (10 μg) of the purified protein samples, obtained at the end of section 3, with 5X SDS-sample buffer to a final concentration of 1X.
- Incubate at 100 ºC for 5 min and load onto standard 12% and/or 15% SDS-PAGE gels22.
- Run at 80V for 20 min and then at 120 V until the dye front runs off the gel (this is approximately an additional 75 min for a 12% gel, and 120 min for a 15% gel).
5. Visualizing Activity and Protein Staining.
- Following electrophoresis, carefully separate glass plates and transfer the nondenaturing gel to a gel tray containing 50 mL of deionized water. Rinse gel with gentle rocking for 5 min. Discard water and repeat this wash step three times.
- Perform substrate overlay assay as follows.
- Add 1 mL of developing buffer containing the fluorogenic peptide substrate Suc-LLVY-AMC and spread uniformly over the gel. Incubate at 37 ºC for 30 min.
Note: A glass rod or a gel releaser (small wedge of plastic used to separate glass plates) can be used to spread the liquid over the gel. - Carefully transfer the gel onto the UV transilluminator of the gel imaging system and observe fluorescence due to the cleaved peptide substrate. Record image. Carefully transfer the gel back to the gel tray.
- Rinse the gel twice with 50 mL of deionized water for 5 min with gentle rocking.
- Stain the gel with 10 mL of a colloidal coomassie stain reagent for 60 min with gentle rocking. Destain gel with 50 mL water until background becomes clear. This staining step applies to the standard SDS-PAGE gels as well.
- Add 1 mL of developing buffer containing the fluorogenic peptide substrate Suc-LLVY-AMC and spread uniformly over the gel. Incubate at 37 ºC for 30 min.
Representative Results
Proteasome assembly (Figure 1) begins when α subunits combine to form rings9. This can be illustrated when α subunits from the archaeon Methanococcus maripaludis S2 are expressed in E. coli as C-terminally hexahistidine tagged (his-tagged) derivatives (Table 1). When the recombinant α-his protein was purified by ICAR and analyzed by nondenaturing PAGE, two bands were observed (Figure 2A, lane 1). We have previously demonstrated that these correspond to Single α Rings (SR) and Double α-Rings (DR)18. The DR is a dead-end species that is not productive for subsequent assembly9,18.
When α-his subunits were coexpressed with β subunits, representing the usual method by which assembly of recombinant archaeal proteasomes is assayed, a novel species was observed migrating near the 670 kDa size standard (Figure 2A, lane 3). This species was proteolytically active (Figure 2B, lane 3) and contained only completely mature β subunits (mβ) whose propeptides have been removed (Figure 2C, lane 3). This species is the fully mature CP. This sample also contained some SR species, which was expected because SR is a known assembly intermediate, but no DR species. The lack of DR when α-his and β subunits are coexpressed suggests that correct assembly was occurring fast enough such that incorporation of β subunits was able to outcompete the nonproductive formation of DR (for a more detailed analysis see18).
To demonstrate the limitation of the coexpression method, and argue for the utility of a combined approach, α-his and β subunits were expressed separately in E. coli. Following lysis, the soluble fractions were mixed and proteins were purified by ICAR prior to analysis by nondenaturing PAGE. Fully functional proteasomes were also generated via the lysate mixing approach (Figure 2A and 2B, lane 2), and the SR species was also observed as expected. The reappearance of the DR species in the lysate mixing sample indicates that once formed, β subunits are unable to reversibly disassemble it; this underscores the dead-end nature of the DR. Interestingly, a new species also appeared in the lysate mixing sample, migrating just below the CP (termed "half"). Recently, we showed that this species corresponds to the half-proteasome (α7β7)18. To illustrate this here, a β subunit mutant (R166W) was employed. This mutation disrupts β-β ring interaction, leading to impaired half-proteasomes dimerization18. Since half-proteasomes are the immediate precursors to CP, the R166W mutation should lead to both accumulation of half-proteasomes and a decrease in CP formation. When lysate mixing with α-his and β (R166W) subunits was carried out, lower levels of CP and increased levels of the "half" species were observed (Figure 2A, lane 4). This is consistent with a precursor-product relationship for these two bands, and confirms the identity of the "half" species as the half-proteasome. The slightly faster migration of the half-proteasome in the mutant sample (lane 4 versus lane 2) is likely due to the R166W mutation altering the mass-to-charge ratio of the β subunit.
The ability to visualize the half-proteasome during lysate mixing is due mainly to much lower protein concentrations in the lysate as compared to the high protein concentrations inside cells. Lower concentrations result in less efficient (i.e. slower) assembly, which enables intermediates to become more populated and thus detectable. Besides the appearance of the half-proteasome, an additional observation highlights the decreased assembly efficiency during lysate mixing: unprocessed β subunits, which retain their propeptides, become detectable (proβ). Since propeptide processing does not occur until half proteasomes dimerize, the appearance of the immature proβ form correlates with the level of half-proteasome accumulation (compare Figure 2C, lane 3 versus lane 2 versus lane 4). In the case of the R166W mutant, the propeptide processing failure during lysate mixing is absolute even though a small amount of CP does form. This is because propeptide processing not only requires half-proteasome dimerization, but also a properly-formed β-β ring interface23 which the R166W mutation does not afford. A more detailed narrative of coexpression versus lysate mixing, as it pertains to recombinant proteasome assembly, can be found here18.
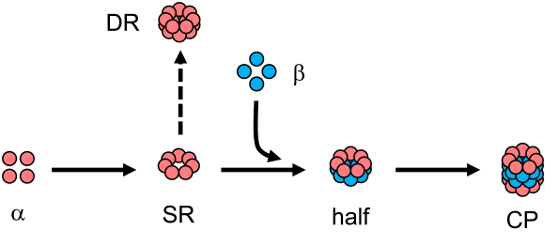
Figure 1: A Simplified Schematic of Core Particle (CP) Assembly.
The α subunits can assemble into a single α-ring first (SR) which serves as a template for the incorporation of β subunits. This leads to the generation of the half-proteasome intermediate (half) which quickly dimerizes. Concurrent with dimerization, the β subunit propeptides (not shown) are autocatalytically removed giving rise to the fully functional core particle (CP). Double α-rings (DR) can arise from SR and are not competent for assembly into CP. Their formation represents a nonproductive assembly pathway (dashed arrow) in contrast to productive assembly events (solid arrows). Please click here to view a larger version of this figure.
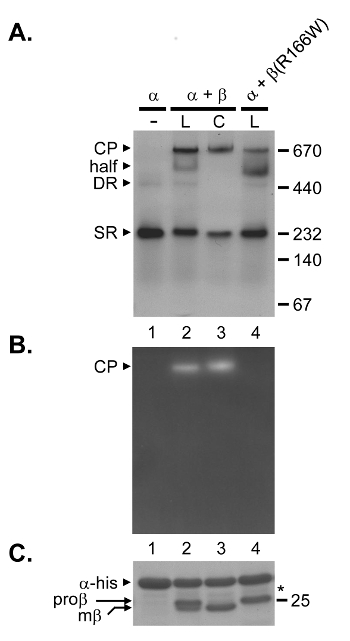
Figure 2: A Combined Approach for Assaying Proteasome Assembly.
Coexpression (C) and lysate mixing (L) were employed to study assembly of recombinant proteasomes from the archaeon M. maripaludis. Proteins were purified by Immobilized Cobalt Affinity Resin (ICAR). (A,B) Purified proteins (10 μg) were loaded onto non-denaturing 5 – 10% gradient gels. Following electrophoresis, peptidase activity was visualized by overlaying the gel with buffer solution containing the fluorogenic peptide substrate Suc-LLVY-AMC (B) prior to staining the gel with colloidal coomassie stain reagent (A). Black arrowheads denote the positions of the assembled 20S core particle (CP), half-proteasome intermediate (half), double α-ring (DR) and single α-ring (SR). The migration of several molecular size standards (in kDa) is indicated at right. (C) Purified proteins (10 μg) from A were also loaded onto 15% SDS-PAGE gels. Following electrophoresis, gels were stained with a colloidal coomassie stain reagent. Migration of α-his subunit and of fully mature (mβ) and immature (proβ) β subunits is indicated. The position of the 25 kDa molecular size standard is shown at right. Asterisk indicates a truncated α-his subunit fragment resulting from nonspecific proteolysis during lysis. Please click here to view a larger version of this figure.
| Plasmid | Genotype | Source |
| AKB191 | pET42 psmA-his | Kusmierczyk et al., (2011) |
| AKB464 | pET42 psmA-his psmB | Kusmierczyk et al., (2011) |
| AKB946 | pET42 psmB | Panfair et al., (2015) |
| AKB952 | pET42 psmB (R166W) | Panfair et al., (2015) |
Table 1: Bacterial Plasmids used in this Study. psmA is the archaeal α subunit gene and psmB is the archaeal β subunit gene.
| Buffer A | 50 mM HEPES-NaOH, pH 7.5, 300 mM NaCl, 5 mM MgCl2. | Buffers B, C, and E, are derivatives of Buffer A containing Imidazole. It is useful to add Imidazole from a 2 M stock prepared in water and stored in the dark. |
| Native resolving buffer | 375 mM Tris-HCl, pH 8.8, 0.1% (v/v) tetramethylethylenediamine (TEMED) and 0.1% (w/v) Ammonium Perulfate (APS). | Prepared fresh no more than an hour before gel is to be polymerized and kept on ice. When preparing acrylamide gel solutions in native resolving buffer, it is useful to add the Tris-HCl from a 4X stock (1.5 M Tris-HCl, pH 8.8). The APS is added immediately prior to polymerization. |
| Native running buffer 10X stock | 250 mM Tris, 1.92 M glycine, do not adjust pH. | |
| 5X native sample buffer | 0.5 M Tris-HCl, pH 8.8, 50% (v/v) glycerol, traces of bromophenol blue. | Traces refers to a very small amount, usually a few grains transferred via spatula. |
| 5X SDS-sample buffer | 0.3 M Tris-HCl, pH 6.8, 600 mM dithiothreitol (DTT), 10% (w/v) SDS, 50% (v/v) glycerol and traces of bromophenol blue. | Traces refers to a very small amount, usually a few grains transferred via spatula. |
| Developing buffer | 50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 1 mM ATP, 50 mM Suc-LLVY-AMC. | Suc-LLVY-AMC is a fluorogenic peptide substrate used to assay proteasome activity. |
Table 2: Solutions, Media, and Buffers used in this study.
Discussion
We demonstrate the benefit of a combined approach to analyzing proteasome assembly by nondenaturing PAGE using recombinant archaeal proteasomes. The usual method9,11 of bacterial coexpression of proteasome subunits allows for rapid analysis but may not reveal key assembly intermediates. We suggest combining coexpression with lysate mixing to develop a broader picture of assembly events.
The advantage of this combined approach is that despite requiring separate expression of the α and β subunits for lysate mixing, it is still relatively labor-friendly. The results can also be semiquantitative if one ensures comparable and consistent expression levels of the individual proteins. To this end, we recommend carrying out the usual optimization of recombinant protein expression to determine conditions that allow comparable levels of expressed proteins prior to lysate mixing. Optimization can include varying induction time, induction temperature, optical density at induction, bacterial expression strain, and so on, until desired levels of soluble protein are achieved. This is especially important when comparing WT and mutant versions of a protein because sometimes the mutant may exhibit comparable expression levels, but decreased solubility. We also highlight the importance of determining protein concentrations of the ICAR-purified samples prior to loading the nondenaturing PAGE. Even with expression optimization in place, and with the best care taken to ensure that parallel samples are processed through the purification steps in the same way, variance can still be inadvertently introduced. A concentration measurement ensures that the same amount of total protein is loaded per sample well. This makes lane-to-lane comparisons of band intensities for a given migrating species more meaningful.
If comparable and consistent levels of protein expression cannot be achieved for lysate mixing, one can always purify all components beforehand and carry out mixing experiments with pure proteins. This has the advantage of allowing more accurate protein determinations and thus makes the approach quantitative. However, the drawback of full purification is that the process becomes considerably more labor intensive, which can preclude rapid analysis of multiple mutants simultaneously18. A final caveat worth mentioning is that sometimes separate expression of α and β subunits may not be possible. This can occur if a mutation causes a subunit to be insoluble in E. coli when expressed on its own but allows solubility to be regained when the mutant is expressed with its binding partner. If this occurs, it will limit analysis to coexpression only. However, this is a caveat that can arise during recombinant protein expression in general, and is not specific to archaeal proteasome subunits24,25.
We chose to generate his-tagged derivatives of our proteasomal proteins due to ease of purification and affordability of the ICAR resin. Other epitope tags are possible, including those for antibody-based purification, and we have successfully expressed and purified Flag-tagged versions of our proteasome subunits (not shown). However, if purification to homogeneity (or increasing the scale of production) is required, the his-tagged versions provide the fastest and most cost-effective means of doing so.
It is a given that results obtained with recombinant proteins, be they archaeal or eukaryotic proteins produced in bacteria, will gain further meaning when followed up with in vivo observations. However, an in vivo approach may not always be immediately accessible experimentally. This is especially true when the subject of study is a large, multisubunit complex such as the proteasome. Recombinant protein approaches provide an important launch point for future experiments. In the case of the proteasome, pairing recombinant archaeal proteasome production with nondenaturing PAGE will continue to be very effective in elucidating key features of proteasome assembly, which are shared between archaeal and eukaryotic species9,11,12,18. Such an approach is likely to be useful for studying the assembly of other large multiprotein complexes as well. Recently, we used this strategy to demonstrate that archaeal proteasomes can assemble via more than one pathway, and that α-rings are not the obligate intermediates during assembly that they were thought to be18. It remains to be determined if the same holds true for eukaryotic proteasomes.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
This work was supported in part by a Research Support Funds Grant (RSFG) from Indiana University-Purdue University, Indianapolis, and in part by an award from the American Heart Association 14GRNT20390154, to A.R.K.
Materials
| Acrylamide (40%) solution | Biorad | 1610104 | Unpolymerized acrylamide is a neurotoxin. Wear proper protective equipment |
| Amicon ultra 0.5ml centrifugal filters | EMDMillipore | UFC501024 | |
| Ammonium persulfate | Sigma | A3678 | |
| ATP | Sigma | A7699 | |
| BCA assay kit | Pierce | 23225 | |
| Bisacrylamide (2%) solution | Biorad | 1610142 | |
| Bromophenol blue | Sigma | B8026 | |
| DNaseI | Sigma | DN25 | |
| Dithiothreitol (DTT) | Thermo Fisher | BP172 | |
| E.coli BL21 competent cells | EMD Millipore | 69450 | |
| GelCode Blue | Thermo Fisher | 24592 | Colloidal coomassie stain reagent for gels |
| Gel doc EZ system | Biorad | 1708270 | Gel documentation system |
| Gel releasers | Biorad | 1653320 | Wedge shaped plastic used to separate gel plates; useful for spreading liquid. |
| Glass rod | Thermo Fisher | 11-380B | |
| Glycerol | Sigma | 49767 | |
| Glycine | Thermo Fisher | BP3865 | |
| Hamilton syringe | Thermo Fisher | 14-813-38 | Glass syringe for loading gels |
| HEPES | US Biologicals | H2010 | |
| HMW Native calibration kit | GE Healthcare | 170445-01 | High molecular weight protein standards |
| Hoefer SG30 | Thermo Fisher | 03-500-277 | Gradient maker |
| Imidazole | US Biologicals | 280671 | |
| IPTG | US Biologicals | I8500 | For induction of protein expression |
| Isopropanol | Thermo Fisher | BP26181 | |
| Kanamycin sulfate | US Biologicals | K0010 | |
| Lysozyme | Sigma | L6876 | |
| MgCl2 | Fluka analytical | 630680 | |
| Mini Protean Tetra Cell | Biorad | 1658002EDU | Gel electrophoresis apparatus |
| NaCl | Thermo Fisher | S640-3 | |
| NaOH | Thermo Fisher | S318-1 | |
| Pefabloc SC | Roche | 11429876001 | Protease inhibitor |
| pET42 | EMD Millipore | 70562 | Expression plasmid |
| Precision plus all blue standard | Biorad | 1610373 | Molecular protein standard for SDS-PAGE |
| Quickchange mutagenesis kit | Agilent technologies | 200521 | |
| Sodium dodecyl sulfate (SDS) | Thermo Fisher | BP166 | |
| Suc-LLVY-AMC | Enzo lifesciences | BML P802-0005 | Fluorogenic substrate |
| Talon Metal Affinity Resin | Clontech | 635502 | Immobilized-cobalt affinity resin |
| TEMED | Sigma | T7024 | |
| Tris | US Biologicals | T8600 | |
| Triton-X100 | Sigma | 93426 | |
| Tryptone | Bacto BD | 211699 | |
| UV sample tray | Biorad | 1708271 | For UV imaging of gels |
| Yeast extract | Bacto BD | 212720 |
Referenzen
- Wan, C., et al. Panorama of ancient metazoan macromolecular complexes. Nature. 525, 339-344 (2015).
- Marsh, J. A., Teichmann, S. A. Structure, dynamics, assembly, and evolution of protein complexes. Annu Rev Biochem. 84, 551-575 (2015).
- Williamson, J. R. Cooperativity in macromolecular assembly. Nat Chem Biol. 4, 458-465 (2008).
- Finley, D., Ulrich, H. D., Sommer, T., Kaiser, P. The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetik. 192, 319-360 (2012).
- Groll, M., et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 386, 463-471 (1997).
- Lander, G. C., et al. Complete subunit architecture of the proteasome regulatory particle. Nature. 482, 186-191 (2012).
- Lowe, J., et al. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 268, 533-539 (1995).
- Hu, G., et al. Structure of the Mycobacterium tuberculosis proteasome and mechanism of inhibition by a peptidyl boronate. Mol Microbiol. 59, 1417-1428 (2006).
- Zwickl, P., Kleinz, J., Baumeister, W. Critical elements in proteasome assembly. Nat Struct Biol. 1, 765-770 (1994).
- Groll, M., Brandstetter, H., Bartunik, H., Bourenkow, G., Huber, R. Investigations on the maturation and regulation of archaebacterial proteasomes. J Mol Biol. 327, 75-83 (2003).
- Frankenberg, R. J., Hsu, T. S., Yakota, H., Kim, R., Clark, D. S. Chemical denaturation and elevated folding temperatures are required for wild-type activity and stability of recombinant Methanococcus jannaschii 20S proteasome. Protein Sci. 10, 1887-1896 (2001).
- Maupin-Furlow, J. A., Aldrich, H. C., Ferry, J. G. Biochemical characterization of the 20S proteasome from the methanoarchaeon Methanosarcina thermophila. J Bacteriol. 180, 1480-1487 (1998).
- Maupin-Furlow, J. A., Ferry, J. G. A proteasome from the methanogenic archaeon Methanosarcina thermophila. J Biol Chem. 270, 28617-28622 (1995).
- Wittig, I., Braun, H. P., Schagger, H. Blue native PAGE. Nat Protoc. 1, 418-428 (2006).
- Langer, T., Pfeifer, G., Martin, J., Baumeister, W., Hartl, F. U. Chaperonin-mediated protein folding: GroES binds to one end of the GroEL cylinder, which accommodates the protein substrate within its central cavity. EMBO J. 11, 4757-4765 (1992).
- McKenzie, M., Lazarou, M., Thorburn, D. R., Ryan, M. T. Analysis of mitochondrial subunit assembly into respiratory chain complexes using Blue Native polyacrylamide gel electrophoresis. Anal Biochem. 364, 128-137 (2007).
- Tomko, R. J., Hochstrasser, M. Incorporation of the Rpn12 subunit couples completion of proteasome regulatory particle lid assembly to lid-base joining. Mol Cell. 44, 907-917 (2011).
- Panfair, D., Ramamurthy, A., Kusmierczyk, A. R. Alpha-ring Independent Assembly of the 20S Proteasome. Sci Rep. 5, 13130 (2015).
- Zimmerman, S. B., Trach, S. O. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J Mol Biol. 222, 599-620 (1991).
- Kusmierczyk, A. R., Kunjappu, M. J., Kim, R. Y., Hochstrasser, M. A conserved 20S proteasome assembly factor requires a C-terminal HbYX motif for proteasomal precursor binding. Nat Struct Mol Biol. 18, 622-629 (2011).
- Smith, P. K., et al. Measurement of protein using bicinchoninic acid. Anal Biochem. 150, 76-85 (1985).
- Laemmli, U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227, 680-685 (1970).
- Chen, P., Hochstrasser, M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell. 86, 961-972 (1996).
- Li, X., Kusmierczyk, A. R., Wong, P., Emili, A., Hochstrasser, M. beta-Subunit appendages promote 20S proteasome assembly by overcoming an Ump1-dependent checkpoint. EMBO J. 26, 2339-2349 (2007).
- Kusmierczyk, A. R., Kunjappu, M. J., Funakoshi, M., Hochstrasser, M. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nat Struct Mol Biol. 15, 237-244 (2008).

