Multimer-PAGE: A Method for Capturing and Resolving Protein Complexes in Biological Samples
Summary
A method for stabilizing and separating native protein complexes from unmodified tissue lysate using an amine-reactive protein cross-linker coupled to a novel two-dimensional polyacrylamide gel electrophoresis (PAGE) system is presented.
Abstract
There are many well-developed methods for purifying and studying single proteins and peptides. However, most cellular functions are carried out by networks of interacting protein complexes, which are often difficult to investigate because their binding is non-covalent and easily perturbed by purification techniques. This work describes a method of stabilizing and separating native protein complexes from unmodified tissue using two-dimensional polyacrylamide gel electrophoresis. Tissue lysate is loaded onto a non-denaturing blue-native polyacrylamide gel, then an electric current is applied until the protein migrates a short distance into the gel. The gel strip containing the migrated protein is then excised and incubated with the amine-reactive cross-linking reagent dithiobis(succinimidyl propionate), which covalently stabilizes protein complexes. The gel strip containing cross-linked complexes is then cast into a sodium dodecyl sulfate polyacrylamide gel, and the complexes are separated completely. The method relies on techniques and materials familiar to most molecular biologists, meaning it is inexpensive and easy to learn. While it is limited in its ability to adequately separate extremely large complexes, and has not been universally successful, the method was able to capture a wide variety of well-studied complexes, and is likely applicable to many systems of interest.
Introduction
Normal cellular function is dependent on protein-protein interactions1,2. As a result, human diseases are often marked by perturbations in the assembly and behavior of various protein complexes3. The ability to characterize such interactions is therefore critical. Current means of detecting these interactions require purification of target proteins, often followed by pull-down of their interacting partners. Conventional purification is accomplished by differential centrifugation, precipitation, and/or chromatography4. These methods are time-consuming, must be altered for each target protein, and often result in low yields. Modern purification methods involve the fusion of peptide tags to target proteins, followed by immunoprecipitation or extraction on columns loaded with beads bound to a capture molecule5,6. While this is extensible to many proteins, it requires sequence modification of the target, potentially resulting in constructs that differ in affinity for their usual binding partners. The delicate nature of some protein-protein interactions means this method may not be applicable to many scenarios. Additionally, the pull-down assays used to map protein-protein interactions may not capture an accurate cellular picture, due to restricted degrees of freedom and non-native levels of the bait protein.
Ideally, protein complexes could be detected in their native states, without the need for purification or pull-down. Blue native polyacrylamide gel electrophoresis (BN-PAGE) was developed as a less-denaturing alternative to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and allows for the separation of some proteins and complexes from biological samples7. However, proteins in BN-PAGE separate based on a large number of variables, including size, charge, three-dimensional structure, and association with other molecules. The interactions of these factors often result in co-separation of proteins, aggregate formation, and poor protein band resolution. Two-dimensional native-polyacrylamide gel electrophoresis resolves some, but not all, of these problems8.
To circumvent the complications associated with native separation, some authors use amine-reactive cross-linking reagents, such as dithiobis(succinimidyl propionate) (DSP), to capture protein complexes in tissue lysates4. These treated lysates can then be denatured and separated by SDS-PAGE, while preserving the native size and makeup of protein complexes. However, since cross-linking reagents react based on proximity of one molecule to another, and proteins in tissue lysates have many degrees of freedom and can stochastically interact, nonspecific background cross-linking can be high, especially in concentrated samples. This can lead to difficult-to-interpret results.
Here, we demonstrate the use of a hybrid BN-PAGE/SDS-PAGE method, termed multimer-polyacrylamide gel electrophoresis (multimer-PAGE), to separate and detect protein assemblies in complex mixtures. Initially, cell lysate is suspended in polyacrylamide gel via BN-PAGE. The lysate-containing gel is then reacted with the cross-linking reagent DSP. Pseudo-immobilized and slightly separated on the gel, proteins are much less likely to react nonspecifically, meaning background cross-linker reactivity is reduced. After cross-linking, the gel bands are excised and separated via SDS-PAGE. The resultant gel can then be analyzed by any means typically associated with polyacrylamide gel electrophoresis. This method allows for the separation and detection of native protein complexes in unmodified tissue lysate, without the need for additional purification or pull-down.
Protocol
1. Tissue Preparation
- Prepare 10 mL of 4x BN-PAGE sample buffer (200 mM Bis(2-hydroxyethyl)amino-tris(hydroxymethyl)methane (Bis-Tris), 200 mM NaCl, 40% w/v glycerol, 0.004% Ponceau S, pH 7.2).
NOTE: This stock solution may be made in advance and stored at 4 °C. - Dilute 250 µL of 4x sample buffer in 750 µL dH2O containing 1x commercial protease inhibitor cocktail. Vortex and chill on ice.
- Homogenize 20 mg of target tissue in the 1 mL 1x ice-cold BN-PAGE sample buffer with 30 strokes of a clean dounce homogenizer.
NOTE: For this demonstration experiment, the target tissue is whole rat brain tissue. After homogenization, samples may be treated with mild detergent such as 2% digitonin to solubilize and permit electrophoresis of membrane proteins. - Transfer the homogenate to a 1.5 mL microcentrifuge tube, and centrifuge at 14,000 x g for 30 min to pellet insoluble cellular contents. After centrifugation, decant the supernatant into a clean tube on ice.
- Follow manufacturer instructions to measure supernatant protein concentration using bicinchoninic acid (BCA) or similar protein quantitation assay.
- If the homogenate sample(s) contain detergent, add a quantity of 5% Coomassie Blue G-250 in aqueous solution sufficient to bring the homogenate solution to 0.25% Coomassie.
2. BN-PAGE
- Dilute 25 mL 20x blue native (BN) electrophoresis buffer stock (1.0 M Bis-Tris, 1.0 M tricine, pH 6.8) into 475 mL dH2O containing 0.002% Coomassie Blue to make 500 mL of 1x running buffer.
- If samples contain detergent, instead make 250 mL of 1x running buffer containing 0.02% Coomassie and another 250 mL containing none.
- Chill buffer(s) to 4 °C.
- Clean and assemble a polyacrylamide gel pouring cassette according to manufacturer instructions.
- Pour BN polyacrylamide gel (3% T stacking layer, 6% T resolving layer) according to a standard recipe7, and place the well comb.
- After the gels have polymerized, rinse the cassette with dH2O and assemble the electrophoresis apparatus according to manufacturer instructions.
- Remove the well comb and fill the wells with 1x BN-PAGE running buffer. Avoid filling the entire inner chamber with buffer until samples are loaded.
- If samples contain detergent, fill the wells with the buffer containing 0.02% Coomassie Blue.
NOTE: Perform steps 2.7-2.9 at 4 °C.
- If samples contain detergent, fill the wells with the buffer containing 0.02% Coomassie Blue.
- Using gel-loading tips, pipette a volume of homogenate containing 20 µg of protein into the desired wells. Pipette an equivalent volume of 1x BN-PAGE sample buffer into any unused wells.
NOTE: Use the protein concentration determined from the BCA assay to calculate the appropriate volume of homogenate to add to the wells. - After samples are loaded, fill the inner chamber with 1x BN-PAGE running buffer. Be sure the gel is entirely submerged in buffer. Next, fill the outer chamber with 1x running buffer to the level indicated by the manufacturer.
- If samples contain detergent, fill the inner chamber with the 1x buffer containing 0.02% Coomassie Blue, and the outer chamber with dye-free 1x running buffer.
- Connect the electrodes to the power supply, and electrophorese the proteins in the gel at 150 V until the dye band progresses ~2 cm into resolving layer. Stop and disconnect the power supply.
3. Cross-linking
NOTE: Perform Steps 3.1-3.6 at 4 °C.
- Disassemble the electrophoresis apparatus, and separate the glass panes of the gel cassette. Remove and discard the stacking layer.
- Carefully cut the gel just below the bottom edge of the dye front. Take care to make this cut as smooth and straight as possible, and then discard the unused piece of gel. This will leave a ~2 cm wide, horizontal strip of polyacrylamide gel, which will contain all the protein in the homogenate sample.
- Trim away any unused portions along the edges of the gel strip.
- Carefully place the strip into 10 mL phosphate buffered saline (PBS) in a small container, and gently mix by nutation for 30 min to equilibrate.
- After equilibration, discard and replace the PBS with another 10 mL. Pipette 500 µL of 25 mM DSP dissolved in dimethyl sulfoxide into the PBS, and continue mixing as above (step 3.4) for 30 min.
- Pour off the DSP solution. Add 10 mL of 0.375 M tris(hydroxymethyl)aminomethane hydrochloride (Tris-HCl), pH 8.8, containing 2% sodium dodecyl sulfate (SDS) to quench the unreacted DSP. Continue nutation for 15 min.
4. SDS-PAGE
- While gel strip is quenching, prepare SDS-PAGE gel solutions according to standard methods9. Do not add polymerization reagents.
- After quenching, return the ΒΝ gel strip to room temperature, and cast the strip into a new gel cassette.
- To do this, carefully pick up the gel and place it onto a clean gel cassette spacer plate.
- Orient the strip so the bottom of the dye front is nearest to the top of new cassette (i.e., the side that travelled furthest during BN-PAGE should be closest to the top of the new cassette; the gel strip should be flipped from its prior orientation). See Figure 3.
- Place the strip such that its top edge lies even with where the top edge of the cover plate will be (i.e., it will be at the top of the new gel). Make sure the dye front is parallel to the horizontal edges of the glass plate.
- Push one side of the excised strip against one of the spacer walls, leaving room on the other side for gel to be poured and a protein standard or ladder to be loaded.
- If the bottom edge of the gel strip contains any jagged or uneven areas, carefully cut them away. If present, they will trap bubbles during gel pouring.
- Once the gel strip is positioned correctly, lay the cover plate over the spacer plate. Apply gentle pressure to push out any trapped air bubbles.
- Continue to assemble the gel-pouring apparatus according to manufacturer instructions.
- Add polymerization reagents to the resolving gel buffer, and pour it into the prepared gel cassette, using a serological pipette. Fill the gel cassette to ~2 cm below the excised BN-PAGE gel strip, to leave room for the stacking layer.
- Add 100 µL butanol over the top of the poured gel, and allow 30 min for polymerization of the resolving layer. Pour off the butanol.
- Add polymerization reagents to the stacking gel solution. Using a serological pipette, pour the stacking layer to fill all remaining empty space in the gel cassette.
- Tilt the gel cassette as the stacking layer is poured so it fills the space below the gel strip, and air bubbles are not trapped.
- As the stacking gel buffer fills the empty space below the gel strip, gradually return the gel cassette to level footing.
- Continue to fill the empty space next to the excised gel strip with the stacking gel buffer, until it nearly overflows.
- Allow the stacking layer to polymerize for 30 min.
NOTE: Make 10x SDS-PAGE running buffer (250 mM tris(hydroxymethyl)aminomethane (tris), 1.9 M glycine, 1% SDS) while the gel is polymerizing. This can also be made beforehand and stored at room temperature. - Dilute 50 mL 10X SDS-PAGE running buffer stock into 450 mL dH2O to make 1x working buffer.
- After the stacking layer has polymerized, remove the gel cassette from the pouring apparatus, rinse with dH2O, and assemble the electrophoresis apparatus according to manufacturer instructions.
- Fill the inner chamber completely with 1x SDS-PAGE running buffer, then fill the outer chamber to the level indicated by the manufacturer.
- Load the space next to the excised gel strip with a molecular weight ladder or appropriate protein standard.
- Attach the electrodes to the power supply, and electrophorese the samples at 120 V. When the Coomassie dye runs off the gel, stop and disconnect the power supply.
- Analyze the gel using standard methods of electroblotting and protein detection by antibody binding10.
Representative Results
In this demonstration experiment, multimer-PAGE was performed on whole rat brain lysate. The resultant separated proteins were blotted onto polyvinylidene diflouride (PVDF) membranes, and then probed with antibodies against proteins that are known to form complexes. Figure 1 shows a validation of the protocol by two means. First, we demonstrate that the cross-linked proteins are cleavable by addition of a reducing agent, meaning the observed higher molecular weight species are formed by the cross-linking reagent, and are not due to any other component of the protocol (Figure 1A). Second, we demonstrate that cross-linking is sensitive to addition of detergents (Figure 1B), meaning the cross-linking is specific to complexed proteins and that random reactivity due to close proximity on the gel is minimized. Figure 2 demonstrates that the method fails to capture respiratory complex II, but otherwise has wide applicability for capturing both soluble and membrane-bound complexes.
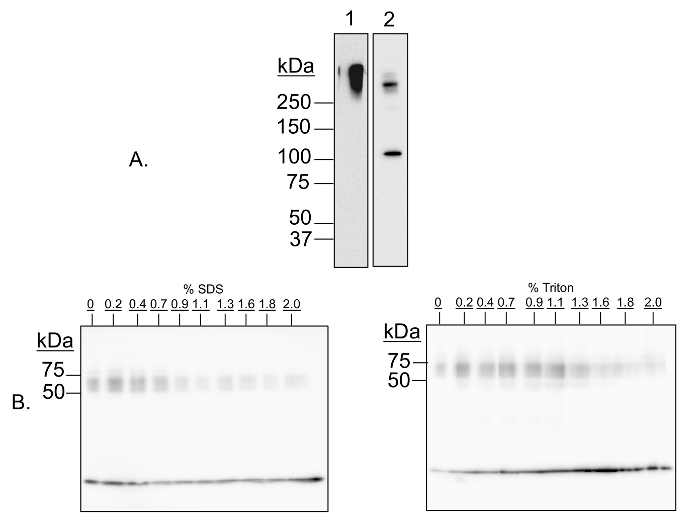
Figure 1: Validation of multimer-PAGE method. (A) Capture of protein complexes by in-gel cross-linking with DSP is sensitive to cleavage by dithiothretol (DTT). Multimer-PAGE was performed on whole rat brain lysate without (A1) or with (A2) 5 mM dithiothretol included in the Tris/SDS quenching solution. The gel was electroblotted onto PVDF membranes, and then probed for kinesin heavy chain. A high molecular weight band is observed on both membranes, likely corresponding to all or part of the kinesin motor protein complex. There is an additional band occurring at 126 kDa on the DTT-treated blot, the molecular mass of the kinesin heavy chain peptide. Dithiothreitol is a reducing agent, and is able to reverse cross-linking by cleaving the disulfide bond present in DSP. The kinesin aggregate is therefore sensitive to cleavage by DTT. (B) Protein complex capture is sensitive to addition of denaturing detergents. Multimer-PAGE was performed on whole rat brain lysate as described in the text, except increasing amounts (0 to 2%) of SDS (left) or Triton X-100 (right) were added to the lysate samples, and then incubated on ice for 30 min before loading the first gel. The final gels were blotted onto PVDF membranes, and probed for α-synuclein. At least three bands of increasing molecular weight are observed, corresponding to α-synuclein monomer, tetramer, and octamer, respectively. The signal intensities of the oligomeric bands decrease with increasing detergent concentration, indicating that cross-linking efficacy is dependent of native protein conformation. Please click here to view a larger version of this figure.
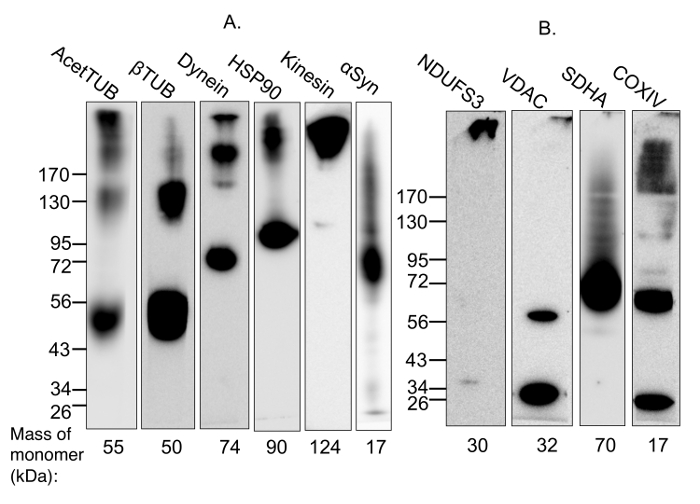
Figure 2: Demonstration of the capture of protein complexes using multimer-PAGE. (A) Soluble and membrane-bound protein complexes were captured from (A) whole rat brain lysate or (B) lysate incubated with 2% digitonin for 1 h at 4 °C, by performing multimer-PAGE as described in the text. The gels were then blotted and the PVDF membranes probed for components of various complexes. High molecular weight species are observed on the membrane probed for acetylated tubulin, which is known to stabilize microtubules. Dynein and kinesin are multi-subunit motor protein complexes with molecular weights of 1.5 MDa and 380 kDa, respectively. The membranes blotted for components of these complexes show high-molecular weight species. In addition, multimer-PAGE successfully captured the HSP90 dimer. α-Synuclein forms tetramers and octamers, as well as neurotoxic high-molecular weight oligomers. The octamer and a streak of higher-weight species are seen on the blotted membrane. VDAC dimerization is also observed. High molecular weight species are detected on the membranes blotted for components of mitochondrial complexes I and IV. However, the membrane blotted for SDHA, a member of complex II, does not demonstrate any appropriate high-molecular weight band. AcetTUB: acetylated α-tubulin; βTUB: β-tubulin; Dynein: dynein heavy chain; HSP90: heat shock protein 90; Kinesin: kinesin heavy chain; NDUFS3: iron-sulfur center 3 of mitochondrial complex I; VDAC: porin; SDHA: succinate dehydrogenase of mitochondrial complex II; COXIV: cytochrome C oxidase of mitochondrial complex IV. Please click here to view a larger version of this figure.
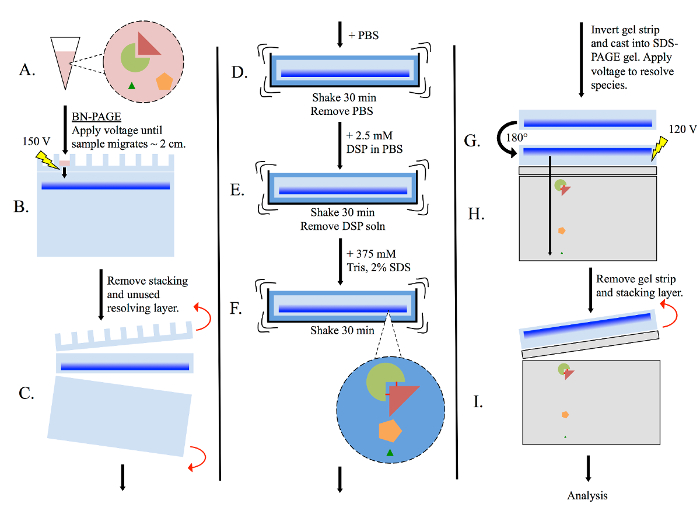
Figure 3: General multimer-PAGE flowchart. (A) Prepare tissue lysate using non-denaturing methods, such as homogenizing in BN-PAGE sample buffer. (B) Pipette lysate into BN-PAGE gel, then perform electrophoresis (150 volts) in BN-PAGE running buffer. Allow sample to migrate until the dye front has migrated ~2 cm into the resolving gel. (C) Remove the stacking layer and unused portion of the resolving layer. (D) Submerge the gel strip in PBS, and equilibrate with shaking for 30 min. (E) Remove the PBS, and re-submerge the gel strip in PBS containing 2.5 mM DSP. Shake for 30 min. (F) Remove DSP solution, then re-submerge the gel strip in 375 mM Tris containing 2% SDS, and shake for 30 min. Complexes present in the gel strip are now stabilized by cross-linking. (G) Rotate the gel strip 180° and cast into a new SDS-PAGE gel. Include both stacking and resolving layers. (H) Resolve sample by electrophoresis (120 volts). (I) Remove gel strip and stacking layer, then analyze resolving layer as necessary. Please click here to view a larger version of this figure.
Discussion
Protein-protein interactions are important for every task living things carry out. Because of this, they are the subject of intense scrutiny and research. Multimer-PAGE is a novel method for capturing, separating, and analyzing a wide range of protein complexes. We have previously demonstrated its applicability to studying oligimerization of the disease-associated protein α-synuclein11. However, it is extensible to many protein complexes, as demonstrated in Figure 2. When compared to other methods of studying protein complexes, multimer-PAGE is advantageous because of its simplicity and brevity. The time from tissue sample to fully separated SDS gel is approximately 8 h. Since detection can be done by typical western blot, the protocol can be performed on unmodified tissue lysate, with detection limits much lower than those associated with pull-down experiments. Additionally, most well-equipped molecular biology laboratories will be able to perform the technique with little upfront investment. As is discussed later, most of the challenge associated with multimer-PAGE comes with troubleshooting and optimizing the method for each protein complex studied.
While multimer-PAGE should be accessible to most molecular biologists, its success is dependent on the researcher's skill and experience. Critically, the cutting and re-casting of the BN gel strip into the new SDS gel must be done with care. It is important to orient the gel strip so that electrophoresis causes proteins to migrate into the SDS gel in parallel bands. If the gel strip is cast crooked, the protein bands will migrate into one another and data will be lost. Especially for large complexes, rotating the gel strip before casting it into the SDS-PAGE gel is similarly important. This gives larger complexes, which may not have migrated very far into the BN-PAGE gel, more time to be pulled into the resolving layer of the SDS-PAGE gel. Importantly, the changes to protein size and physical characteristics caused by cross-linking should also be considered during analysis. For example, some low molecular weight proteins, such as α-synuclein, are not well retained on PVDF membranes, but their cross-linked oligomers are12. This can confound comparative analysis, and must be taken into account.
When separating a complex by multimer-PAGE for the first time, it is important to validate the method. We suggest three validation/troubleshooting tests. First, ensure that the peptide of interest migrates into the BN gel by following the multimer-PAGE protocol until excision of the gel strip, then stop, blot the strip onto PVDF membrane, and probe for the subunit of interest. If there is no signal, the complex likely did not migrate into the BN gel, meaning it should be made more porous, or the sample should be allowed to migrate further into the resolving layer. Presence of a signal confirms that at least unbound subunit migrated into the gel. At this stage, it will be difficult to confirm the presence of the full complex. If evidence of the subunit is detected in the BN gel strip, move on to the second test. Perform multimer-PAGE without a cross-linking step, then blot onto PVDF membrane and probe for the subunit. The subunit should denature in the presence of SDS and migrate normally into the SDS gel. If this does not occur, there is likely some problem with the researcher's technique. The BN gel strip may have been poorly cut, leaving some protein behind, or the SDS gel may have been cast incorrectly. Finally, perform multimer-PAGE with an included cross-linker cleavage step, then blot and probe, as discussed in Figure 1. This confirms aggregation of the complex is due to addition of DSP, and not an unexpected consequence of some other part of the multimer-PAGE protocol. The cleavage should result in a reduced-intensity aggregate band and an increased-intensity subunit monomer band upon imaging. If no aggregate band is seen from normal multimer-PAGE, but the monomer band increases in intensity when DSP is cleaved, the complex has likely made it to the SDS gel, but failed to migrate into the resolving layer.
In its present state, multimer-PAGE is applicable to many protein complexes, but is not a one-size-fits-all protocol. It likely needs to be modified for each complex of interest. One common problem is demonstrated in Figure 2: captured complexes are often so large that they barely migrate on SDS-PAGE. This may be managed by changing the porosity of the SDS gel or performing electrophoresis for longer periods of time. An additional complication is the occasional presence of more than two bands on the gel. Incomplete reaction with the cross-linker can result in a distribution of intermediate molecular weight bands13. This can make it difficult to determine whether multiple bands are representative of physiologically important oligomeric intermediates, or simply unintended consequences of partial cross-linking. For membrane proteins that must be solubilized before analysis, it is also important to consider the impact that the choice of detergent has on the behaviors of protein complexes. The detergent used to solubilize membrane proteins affects their conformations, associations with binding partners, and functions14,15. Detergent choice also impacts the efficacy of cross-linking16. Thus, it is likely that the ideal solubilizing conditions will vary with the complex being studied.
Multimer-PAGE occasionally fails to capture a complex, as is shown on the SDHA blot in Figure 2. Succinate dehydrogenase is part of the 124 kDa mitochondrial Complex II, which is small enough that it should move easily into the gel. That it was not detected indicates that multimer-PAGE was unable to capture it. Cases such as these have many possible explanations. Most cross-linking reagents contain two reactive ends, which form covalent linkages with amino acid side chains. The efficiency with which cross-linking reagents capture protein complexes is dependent on their accessibility to reactive residues. In the case of DSP, cross-linking is accomplished by acylation of solvent-exposed primary and secondary aliphatic amino groups, usually the ε-amino groups of lysine17. Lysine residues are normally found on the protein surface, but their occasional sequestration in hydrophobic centers decreases the efficiency of cross-linking13. The subunits of complex II are buried in the mitochondrial membrane, and may remain inaccessible to DSP, even after solubilization with digitonin18. Very large and densely-packing complexes, such as amyloid oligomers, may also limit the availability of surface lysine residues to cross-linking reagents. As a result, these types of protein assemblies may not be compatible with multimer-PAGE. It is also possible that Coomassie Blue, which binds to hydrophobic regions and cationic residues, lingers after equilibration in PBS and blocks cross-linking in some cases19. This might decrease or eliminate the detection of affected oligomers by multimer-PAGE. Thus, the method is not a universal assay for characterizing all protein associations, and should not be considered where quantitation in desired. However, multimer-PAGE is an inexpensive, relatively swift tool for analyzing protein complexes, and may be considered alongside other established means.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
Supported by the NIH/NIDA DA034783. We thank Bryan A. Killinger for technical assistance with the multimer-PAGE.
Materials
| Chemicals | |||
| ε-Aminocaproic acid | Sigma | A2504 | |
| Acrylamide | Acros Organics | 164855000 | Toxic. |
| Acrylamide/bisacrilamide 37.5:1 (40%T stock solution) | BioRad | 161-0148 | Toxic. |
| Ammonium persulfate | Sigma | A3678 | |
| Anti rabbit IgG-HRP from goat | Santa Cruz Biotechnology | sc-2004 | |
| Bicinchoninic acid assay kit | Thermofisher | 23225 | |
| Bis-tris | Sigma | B9754 | |
| Bovine serum albumin | Sigma | A9647 | |
| Butanol | Fisher Scientific | A399-1 | |
| Chemiluminescence substrate kit | ThermoFisher | 24078 | |
| Coomassie blue G-250 | Sigma-Aldrich | B0770 | |
| Digitonin | Sigma | D141 | Toxic. |
| Dimethyl sulfoxide | Fisher Scientific | D128-1 | |
| Dithiobis(succinimidylpropionate) | Thermo Scientific | 22585 | |
| Dry nonfat milk | LabScientific | M0841 | |
| Glycerol | Sigma | G9012 | |
| Glycine | Fisher Scientific | BP381-5 | |
| Halt Protease Inhibitor Cocktail | Thermofisher | 78430 | |
| Hydrochloric acid | Fisher Scientific | A144SI-212 | For titration. Caustic. |
| Methanol | Fisher Scientific | A412-4 | For PVDF membrane activation. Toxic. |
| Monoclonal anti α-synuclein IgG from rabbit | Santa Cruz Biotechnology | sc-7011-R | |
| N,N,N',N'-tetramethylethylenediamine | Sigma | T9281 | |
| N,N'-methylenebisacrylamide | Acros Organics | 16479 | Toxic. |
| NP40 | Boston Bioproducts | P-872 | |
| Polysorbate 20 (tween-20) | Fisher Scientific | BP337-500 | |
| Polyvinylidene fluoride transfer membranes | Thermo Scientific | 88518 | |
| Ponceau S | Sigma | P3504 | |
| Potassium chloride | Fisher Scientific | P217-3 | |
| Potassium phosphate monobasic | Sigma | P9791 | |
| Protease inhibitor cocktail | Thermo Scientific | 88265 | |
| SDS solution (10% w/v) | BioRad | 161-0416 | |
| Sodium chloride | Fisher Scientific | BP358-212 | |
| Sodium dodecyl sulfate | Sigma | L37771 | |
| Sodium phosphate monobasic | Fisher Scientific | BP329-1 | |
| Tricine | Sigma | T0377 | |
| Tris base | Fisher Scientific | BP152-500 | |
| Tris-HCl (0.5 M buffer, pH 6.8) | BioRad | 161-0799 | |
| Tris-HCl (1.5 M buffer, pH 8.8) | BioRad | 161-0798 | |
| Name | Company | Catalog Number | Comments |
| Instruments | |||
| GE Imagequant LAS 4000 | GE Healthcare | 28-9558-10 | |
| ImageJ software | NIH | ||
| Synergy H1 microplate reader | BioTek | ||
| Gel Former + Stand | Biorad | ||
| Microfuge 22R centrifuge | Beckman Coulter | ||
| 2 mL dounce homogenizer | |||
| Vortex mixer | Fisher Scientific | ||
| Ultrasonic tissue homogenizer | Fisher Scientific | FB120220 |
Referenzen
- Alberts, B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 92 (3), 291-294 (1998).
- Pawson, T., Nash, P. Protein-protein interactions define specificity in signal transduction. Genes Dev. 14 (9), 1027-1047 (2000).
- Rual, J. F., et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 437 (7062), 1173-1178 (2005).
- Phizicky, E. M., Fields, S. Protein-protein interactions: methods for detection and analysis. Microbiol Rev. 59 (1), 94-123 (1995).
- Ho, Y., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 415 (6868), 180-183 (2002).
- Krogan, N. J., et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 440 (7084), 637-643 (2006).
- Wittig, I., Braun, H. P., Schagger, H. Blue native PAGE. Nat Protoc. 1 (1), 418-428 (2006).
- Schagger, H., Cramer, W. A., von Jagow, G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem. 217 (2), 220-230 (1994).
- Laemmli, U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227 (5259), 680-685 (1970).
- Beisiegel, U. Protein Blotting. Electrophoresis. 7 (1), 1-18 (1986).
- Killinger, B. A., Moszczynska, A. Characterization of alpha-Synuclein Multimer Stoichiometry in Complex Biological Samples by Electrophoresis. Anal Chem. 88 (7), 4071-4084 (2016).
- Newman, A. J., Selkoe, D., Dettmer, U. A new method for quantitative immunoblotting of endogenous alpha-synuclein. PLoS One. 8 (11), e81314 (2013).
- Wong, S. S. . Chemistry of protein conjugation and cross-linking. , (1991).
- Seddon, A. M., Curnow, P., Booth, P. J. Membrane proteins, lipids and detergents: not just a soap opera. Biochim Biophys Acta. 1666 (1-2), 105-117 (2004).
- Tanford, C., Reynolds, J. A. Characterization of membrane proteins in detergent solutions. Biochim Biophys Acta. 457 (2), 133-170 (1976).
- Peters, K., Richards, F. M. Chemical cross-linking: reagents and problems in studies of membrane structure. Annu Rev Biochem. 46, 523-551 (1977).
- Lomant, A. J., Fairbanks, G. Chemical probes of extended biological structures: synthesis and properties of the cleavable protein cross-linking reagent [35S]dithiobis(succinimidyl propionate). J Mol Biol. 104 (1), 243-261 (1976).
- Sun, F., et al. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell. 121 (7), 1043-1057 (2005).
- Wittig, I., Schagger, H. Native electrophoretic techniques to identify protein-protein interactions. Proteomics. 9 (23), 5214-5223 (2009).

