Maturation of Human Stem Cell-derived Cardiomyocytes in Biowires Using Electrical Stimulation
Summary
The cardiac biowire platform is an in vitro method used to mature human embryonic and induced pluripotent stem cell-derived cardiomyocytes (hPSC-CM) by combining three-dimensional cell cultivation with electrical stimulation. This manuscript presents the detailed setup of the cardiac biowire platform.
Abstract
Human pluripotent stem cell-derived cardiomyocytes (hPSC-CMs) have been a promising cell source and have thus encouraged the investigation of their potential applications in cardiac research, including drug discovery, disease modeling, tissue engineering, and regenerative medicine. However, cells produced by existing protocols display a range of immaturity compared with native adult ventricular cardiomyocytes. Many efforts have been made to mature hPSC-CMs, with only moderate maturation attained thus far. Therefore, an engineered system, called biowire, has been devised by providing both physical and electrical cues to lead hPSC-CMs to a more mature state in vitro. The system uses a microfabricated platform to seed hPSC-CMs in collagen type I gel along a rigid template suture to assemble into aligned cardiac tissue (biowire), which is subjected to electrical field stimulation with a progressively increasing frequency. Compared to nonstimulated controls, stimulated biowired cardiomyocytes exhibit an enhanced degree of structural and electrophysiological maturation. Such changes are dependent upon the stimulation rate. This manuscript describes in detail the design and creation of biowires.
Introduction
Cell-based therapy is one of the most promising and investigated strategies to achieve cardiac repair/regeneration. It has been aided by cardiac tissue engineering and the co-delivery of biomaterials1,2. Most available cell sources have been studied in animal models for their potentially beneficial effects on damaged, diseased, or aged hearts3. In particular, significant efforts have been made to use human pluripotent stem cell (hPSC)-derived cardiomyocytes (hPSC-CM), a potentially unlimited autologous cell source for cardiac tissue engineering. hPSC-CMs can be produced using several established protocols4,5,6. However, the obtained cells display fetal-like phenotypes, with a range of immature characteristics compared to adult ventricular cardiomyocytes7,8. This can be an obstacle to the application of hPSC-CMs as models of adult heart tissue in drug discovery research and in the development of adult cardiac disease models9.
In order to overcome this limitation of phenotypic immaturity, new approaches have been actively investigated to promote cardiomyocyte maturation. Early studies revealed effective pro-maturation properties in neonatal rat cardiomyocytes via cyclic mechanical10 or electrical stimulation11. Gel compaction and cyclic mechanical stimulation were also shown to improve some aspects of hPSC-CM maturation12,13, with minimal enhancement of the electrophysiological and calcium handling properties. Therefore, a platform system called "biological wire" (biowire) was devised by providing both structural cues and electrical field stimulation to enhance the maturation of hPSC-CMs14. This system uses a microfabricated platform to create aligned cardiac tissue that is amenable to electrical field stimulation. This can be used to improve the structural and electrophysiological maturity of hPSC-CMs. Here, we describe the details of making such biowires.
Protocol
1. Master Design and Fabrication
NOTE: Use soft lithography for device fabrication. Make a two-layer SU-8 master for polydimethylsiloxane (PDMS) molding.
- Design the device using a design and drafting software (Figure 1A, left). Draw each layer of the master separately. Print the device design on two photomasks at 20,000 dpi, corresponding to the two layers of the master15. Set the device pattern as transparent and the surrounding area as dark; the masks will serve as the template to optically transfer the device pattern onto SU-8 by lithography (as described by others15).
- Dispense about 4 mL of SU-8 50 onto the center of a 4 inch silicon wafer. Coat the SU-8 50 evenly on the wafer using a spin coater by spinning at 2,000 revolutions per minute (RPM) for 30 s. Bake the wafer on a 95 °C hotplate for 10 min. Expose the wafer to ultraviolet (UV) light at 200 mJ/cm2.
- Pour about 4 mL of SU-8 2050 onto the center of the wafer and spin at 2,500 RPM for 30 s. Bake on the 95 °C hotplate for 15 min. Cool to room temperature (RT).
- Repeat step 1.3 two more times.
- Cover the coated wafer with the first-layer transparency photomask. Expose the wafer to UV light at 270 mJ/cm2 to make the first layer. Bake on the 95 °C hotplate for 15 min.
- Make the second layer of SU-8 2050 by repeating step 1.3 twice.
- Align the second-layer mask to the features on the first layer using a mask aligner. Then, expose to UV light at 240 mJ/cm2. Bake at 95 °C for 15 min.
NOTE: The exposure time is determined by the required total UV dose and the UV light output intensity measured on the day prior to use. - Develop the wafer by submerging it in propylene glycol monomethyl ether acetate (PGMEA) and shaking at 200 RPM for 30 min on an orbital shaker.
- Place the master into a Petri dish and carefully pour the PDMS mixture (base to crosslinker at a ratio of 10:1 by weight) on the center of the wafer. Cover all the features and avoid air bubbles.
- Heat in an oven at 70 °C for 2 h. Use a sharp blade to cut around the edge of the PDMS biowire template (Figure 1A, right). Peel the PDMS from the master. Trim the device with the blade. Put the PDMS biowire template in a sterilization pouch and steam autoclave at 121 °C for 20 min.
- In a biosafety cabinet (BSC), place the PDMS biowire template into a Petri dish. Use tweezers to hold and mount a piece of sterile surgical silk suture (6-0) in the center of the channel of the PDMS microwell by seating it in the grooves at both ends of the channel (Figure 1B-I).
2. Creation of the Electrical Stimulation Chamber
- Make a pair of rectangular polycarbonate pieces (length x width x thickness: 2 cm x 0.85 cm x 0.35 cm) and use them as the frame of the electrical stimulation chamber (Figure 2).
- Drill two 3 mm wide holes 2 cm apart along the center line of each polycarbonate frame piece.
- Cut two pieces of 1.5 cm long carbon rods. Drill a 1 mm-wide hole through the carbon rod about 3 mm from the end. Thread a platinum wire through the carbon rod hole and tighten the wire by wrapping it around the carbon rod. Attach a clip to the other end of the platinum wire for later connection with the electric stimulator.
- Insert the carbon rods into the polycarbonate frame pieces. Place the assembled frame with the carbon rods into a well of a six-well plate.
- Pour 2 mL of PDMS into the well. Submerge the bottom of the polycarbonate frames in PDMS while keeping the PDMS surface level close to but not reaching the bottom of the carbon rods. Heat in an oven at 70 °C for 2 h.
- Take the electrical stimulation chamber out of the six-well plate and put it in a sterilization pouch for steam sterilization at 121 °C for 20 min.
3. Enzymatic Dissociation of hPSC-CMs
- Induce hPSC to differentiate into cardiomyocytes using an established protocol4.
NOTE: Briefly, generate cardiomyocytes via established embryoid body (EB) protocols in stem pro medium through the sequential supplementation with bone morphogenetic protein 4, basic fibroblast growth factor, activin A, vascular endothelial growth factor, and Dickkopf homolog 14. Use EBs from day 20-34 of differentiation to make biowires. - Collect the EBs from the low attachment plate with a P1,000 pipette tip and transfer to a conical tube. Pellet by centrifugation for 5 min at 125 x g and RT. Discard the supernatant and resuspend the pellet with pre-warmed, 37 °C collagenase type I containing 1% deoxyribonuclease I (DNAse I). Incubate at 37 °C for 2 h for digestion.
- Add 5 mL of pre-warmed, 37 °C wash medium and centrifuge for 5 min at 125 x g and RT. Discard the supernatant. Resuspend the pellet with 2 mL of trypsin/ethylenediaminetetraacetic acid (EDTA) solution and incubate at 37 °C for 5 min.
- Add 1 mL of stop medium containing 3% (v/v) DNase I.
- Attach a 20 G (0.9 mm) needle to a 5 mL syringe and pass the EB suspension through it 3-5 times.
- Add 7 mL of wash medium and centrifuge at 280 x g for 5 min. Discard the supernatant. Resuspend the pellet in Iscove's modified Dulbecco's medium (IMDM) and store on ice. Determine the cell number with a hemocytometer. Take 0.5 x 106 cells and pellet by centrifugation at 280 x g for 5 min. Remove the supernatant.
- Pre-cool all the gel components on ice before mixing them in a sterile tube. Mix the following collagen gel components (final concentration): 2.1 mg/mL rat tail collagen type I, 24.9 mM glucose, 23.8 mM NaHCO3, 14.3 mM NaOH, 10 mM HEPES, and 10% extracellular matrix in 1x M199 medium. Add the collagen and extracellular matrix last. Mix by pipetting up and down.
- Resuspend 0.5 x 106 cells with 3.5 µL of collagen gel and mix well.
- Use a P10 pipette tip to add 4 µL of the cell suspension into the 0.5 cm-long channel (Figure 1B-II). Adjust the position of the suture with sterile forceps if necessary.
- Cover the bottom of the Petri dish with sterile phosphate-buffered saline (PBS), making sure not to touch the gel. Incubate at 37 °C for 30 min.
- Replace the PBS with 12 mL of culture medium (e.g., stem pro or medium recommended by the cell manufacturer), enough to cover the cells (Figure 1B-III). Culture in an incubator at 37 °C for 1 week. Change the medium every other day.
4. Electrical Stimulation for Biowire Cultivation
- In the BSC, add 2 mL of PBS to each well of a six-well plate that will house an electrical stimulator chamber. Use sterile tweezers to gently place the autoclaved electrical stimulation device into the well. Discard the PBS from the well.
- Cover the carbon rods with 5 mL of pre-warmed culture medium. Use sterile tweezers to mount the biowires in the electrical stimulation chamber and orient them perpendicular to the carbon rods (Figure 2).
- Place the lid on top of the six-well plate, but leave sufficient platinum wires reaching outside the plate to connect with the electrical stimulator. Place the six-well plate in a cell culture incubator at 37 °C and connect the platinum wire to the electrical stimulator.
- Stimulate the biowires using an electrical stimulator with the following setting: biphasic repeating pulse, 1-ms pulse duration, 3 V/cm, and 1 pulse per second (PPS). Raise the PPS every 24 h to the following values: 1.83, 2.66, 3.49, 4.82, 5.15, and 6. Change the medium every other day.
- Observe the cardiomyocyte contractility in response to electrical stimulation under a microscope at 10X magnification.
NOTE: After one week of stimulation, the biowires becomes mature (see the results section).
Representative Results
The rational for the use of a suture in the biowires is to serve as a template for the formation of 3D constructs that align in one axis and mimic the shape of cardiac fibers. We show that after seven days of culture in the biowire, cells remodeled the gel around the suture (Figure 3A). The cells assembled along the axis of the suture to form aligned cardiac tissue (Figure 3). After 7 days of preculture, the biowires were subjected to 7 days of electrical field stimulation and further exhibited properties compatible with structural and functional maturation of cardiomyocytes.
Human PSC-CMs in biowires exhibited strong expression of cardiac contractile proteins, sarcomeric α-actinin, and actin (Figure 4A). We observed sarcomeric banding of the hPSC-CM contractile apparatus and alignment of the myofibrilar strictures along the axis of the suture. This is qualitatively similar to the structure in the adult heart (Figure 4A). Second, compared to those of EBs, these biowired hESC-CMs displayed lower proliferation rates (Figure 4B). Third, the biowired hESC-CMs exhibited improved calcium handling properties (Figure 5) and cardiomyocyte electrophysiology maturation (Figure 6). In mature cardiomyocytes, treatment with caffeine induces an abrupt release of Ca2+ from the sarcoplasmic reticulum8. Such Ca2+ transients were found in electrically stimulated hESC-CMs, but not in cells from non-stimulated controls (Figure 5A-C). The response to caffeine in the electrically stimulated cells, compared to non-stimulated controls, revealed a significantly higher amplitude of intensity in a stimulation frequency-dependent manner (Figure 5D and E). Meanwhile, hPSC-CMs in biowires showed improved hERG current and inward rectifier current (Ik1) densities, which was further enhanced by electrical stimulation (Figure 6A and B). Such improvements in Ik1 current are in accordance with an induction in potassium inwardly rectifying channel gene (KCNJ2) protein expression in the biowires (Figure 6C).
Another important parameter regarding maturation is the ability of the cardiomyocytes to beat spontaneously, also known as automaticity, which is a sign of immaturity in working cardiomyocytes (atrial and ventricular)16. Automaticity was significantly higher in EB-derived cardiomyocytes compared to control biowires (Figure 6D), which was comparable to that in biowires subjected to the 6-Hz regimen.
Taken together, these results indicate that culture in biowire with electrical stimulation promoted the structural and electrophysiological maturation of hPSC-CMs14.
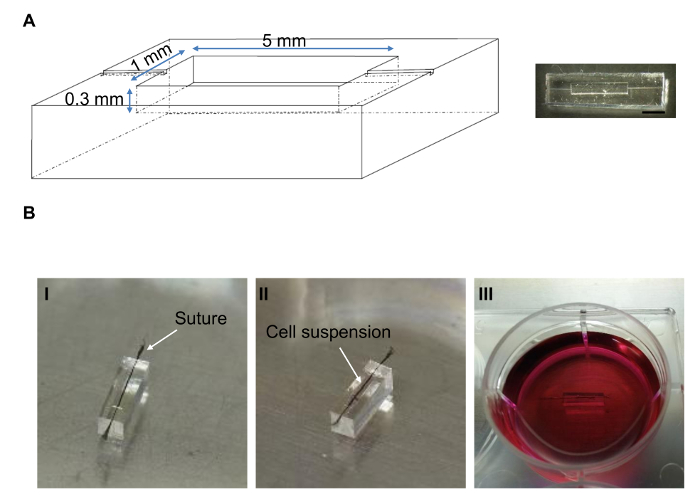
Figure 1: Biowire Culture Platform. (A) Schematic (left) and actual (right) PDMS mold of biowire. Scale bar = 2 mm. (B) To set up the biowire, a suture was mounted centrally in the channel (I). A hESC-CM suspension in collagen type I gel was seeded around the suture in the channel (II) and incubated in medium (III). Please click here to view a larger version of this figure.
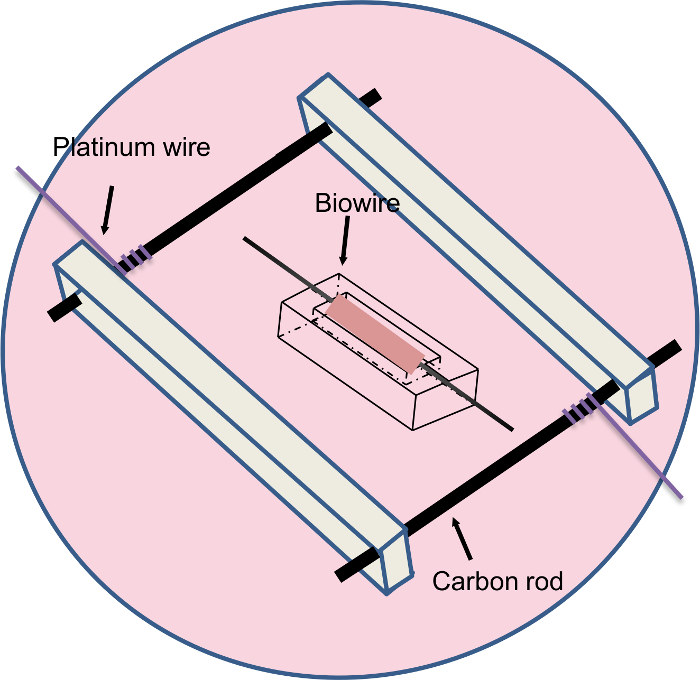
Figure 2: The Biowire was Cultured in the Electrical Stimulation Chamber during the Second Week of Cultivation. The biowire was placed perpendicular to the carbon electrodes. Please click here to view a larger version of this figure.
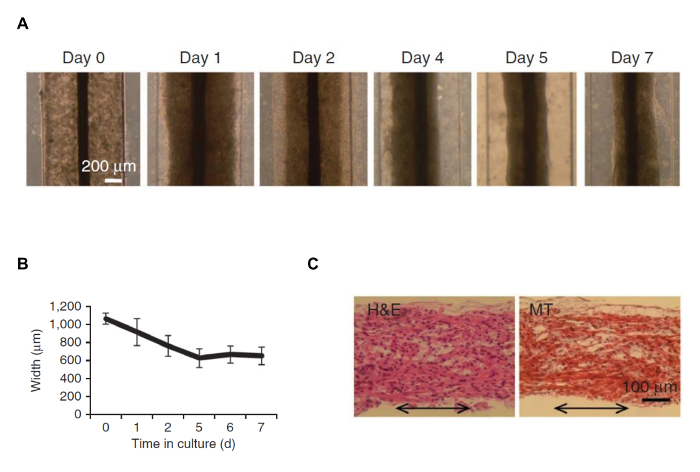
Figure 3: Cell Remodeling and Gel Contracting around the Suture after 7 day of Preculture in Biowire. (A) Brightfield images of hESC-CMs on indicated days of preculture in the biowire template. Scale bar = 200 µm. (B) Quantification of gel compaction on the indicated days of culture (average ± s.d.). (C) Hematoxylin and eosin (H&E) and Masson's trichrome (MT) staining of biowire sections (the double-headed arrows represent the suture axis). Scale bar = 100 µm. This figure has been modified from reference14. Please click here to view a larger version of this figure.
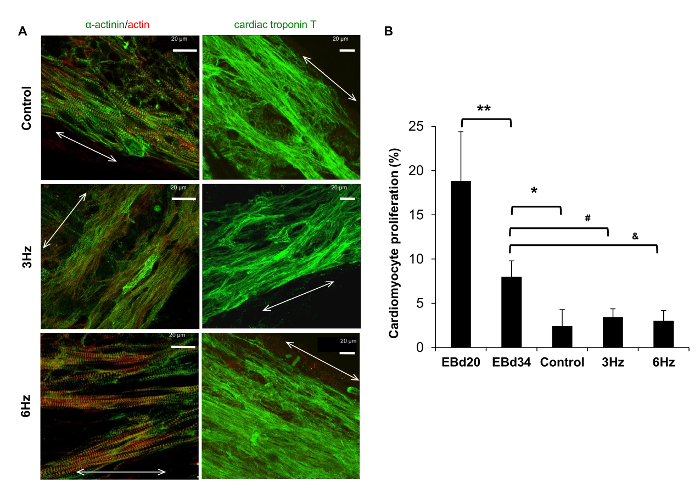
Figure 4: Physiology of Cardiomyocytes Cultured in the Biowire Platform. (A) Representative confocal images of non-stimulated (control) and electrically stimulated biowires (3 Hz and 6 Hz regimens) showing cardiomyocyte alignment and frequent Z-disks (the double-headed arrows represent the suture axis). Green, α-actinin; red, actin. Scale bar = 20 µm. (B) Cardiomyocyte proliferation in the biowires was lower than in the EBs. Proliferation was assessed by double staining for sarcomeric α-actinin and Ki67 (n = 3-4 per condition, average ± s.d.). *, **, #, and & represent statistically significant differences compared to EBd34 (Student's t-test). This figure has been modified from reference14. Please click here to view a larger version of this figure.
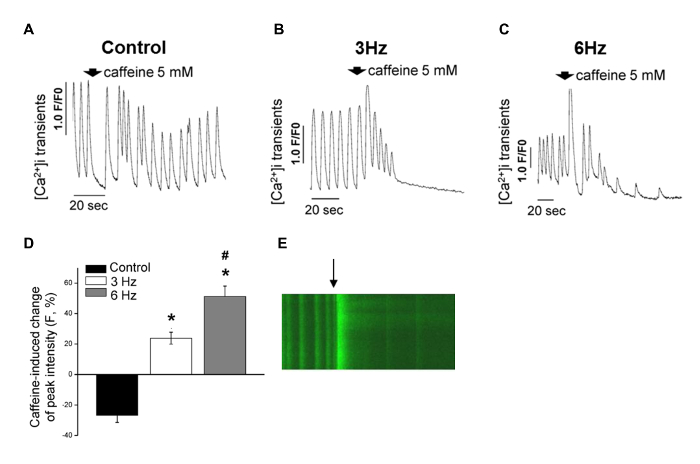
Figure 5: Electrical Stimulation Promoted Improvements in Ca2+ Handling Properties. (A-C) Representative traces of Ca2+ release in response to caffeine in nonstimulated control cells (A) and cells subjected to the 3-Hz regimen (B) and the 6 Hz regimen (C). (D) Caffeine-induced change of peak fluorescence intensity among experimental groups (mean ± s.e.m. after normalizing the peak fluorescence intensity before the administration of caffeine; control versus 3 Hz, P = 1.1 x 10−6; control versus 6 Hz, P = 2.1 x 10−7; 3 Hz versus 6 Hz, P = 0.003; n = 10 (control), n = 6 (3 Hz), and n = 9 (6 Hz) (Student's t-test). (E) Representative fluorescent recording of Ca2+ transients before and after the administration of caffeine at 5 mM (arrow) in cells subjected to the 6 Hz regimen. This figure has been modified from reference14. Please click here to view a larger version of this figure.
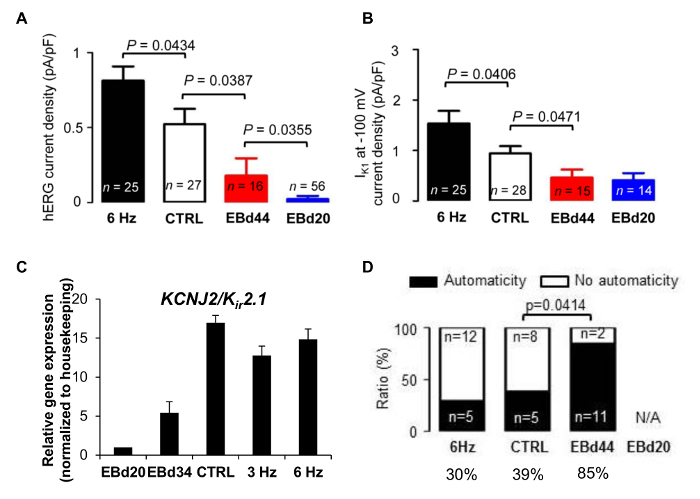
Figure 6: Electrical Pacing Improves Electrophysiological Properties and Automaticity. Electrophysiological properties in single cardiomyocytes isolated from biowires or embryoid bodies (EBs) were recorded with a patch clamp. (A) hERG tail current density. (B) IK1 current density measured at -100 mV. (C) Upregulation of potassium inwardly rectifying channel gene (KCNJ2). (D) Ratio of cells displaying spontaneous beating (automaticity) or no spontaneous beating (no automaticity). The data in A-D represent the average ± S.E.M. The differences between the experimental groups were analyzed by Student's t-test, chi-squared test, and ANOVA (pairwise multiple comparisons, Holm-Sidak method). This figure has been modified from reference14. Please click here to view a larger version of this figure.
| Protocol step # | Critical Steps | ||||
| 2.4 | Place the carbon rods 1 – 2 cm apart or at the smallest possible distance to avoid high-energy stimulation and cell death. | ||||
| 2.6 | Test the connections between the platinum wire and the carbon once the stimulation chamber is made for the first time. Link the stimulation chamber to the electrical stimulator and then set a desired voltage. Place a volt lead onto each of the two carbon rods. The output voltage from the stimulation chamber should be the same voltage read on the voltmeter. Any incorrect readings indicate the possible coating of the platinum wire with PDMS or another defective connection in the system. | ||||
| 3.1 | Single cells were used to seed the biowire. For dissociation, the incubation time with collagenase is critical and should be determined by the age of the cells and the differentiation protocol (EBs versus monolayers). In this protocol, an incubation time of 2 h is recommended for day 21 EBs and 30 min for monolayers. For cells of other ages, the collagenase digestion time might need optimization by monitoring cell viability and contractile function post-digestion. | ||||
| 3.3 | Trypsin can damage the cells; thus, limit the incubation duration to no longer than 5 min. | ||||
| 3.5 | Avoid introducing contaminants by holding only the end of the syringe to attach the syringe to the needle tip. The number of plunges is determined by the absence of large clumps of cells. Minimize needle disruption to avoid cell death. Avoid bubble formation. In case of visible clumps after needle disruption, prolong the incubation time in collagenase in subsequent isolations instead of increasing the incubation time in trypsin or the number of plunges. | ||||
| 3.6 | Remove the supernatant completely prior to the addition of gel. Residual medium may dilute the ratio of cell:gel and affect gel polymerization. | ||||
| 3.7 | Maintain the concentrations of the collagen gel components when scaling up or down. Keep in mind that stock concentrations may vary according to the batch or preparation of the reagents. | ||||
| 3.11 | To ensure that PDMS microwells stay at the bottom of the Petri dish, make sure that the Petri dish is dry and use a pair of sterile forceps to push the PDMS microwells down. | ||||
| 4.2 | Ensure that all biowires are in place. Cover the biowires completely with medium in the electrical stimulation chamber. Arrange all biowires perpendicular to the carbon rods. | ||||
| 4.3 | Avoid any damage to the platinum wire by covering the lid gently. Use tape to secure the lid properly on the plate in case the lid does not close fully. When closing the incubator doors, place the thinner portion of the wires from the electrical stimulator at the door closing points to ensure proper sealing of the incubator. | ||||
| 4.4 | Disconnect the electrical stimulator for the medium changes. Use a P1000 to remove only the top portion of the old medium and then add fresh medium. Ensure that the biowires are always submerged under medium. | ||||
Table 1: Critical Steps in the Protocol.
Discussion
This manuscript describes the setup and implementation of the engineered platform, biowire, to improve the maturation of hPSC-CMs. The device can be made in standard microfabrication facilities, and biowires can be produced with common cell culture techniques and an electrical stimulator.
To our knowledge, there is no reported method similar to biowires. This strategy reveals that improved maturation properties were dependent on the electrical stimulation regimen, as evidenced by the greater maturation level obtained in the biowires subjected to the higher stimulation rate ramp-up regimen (6 Hz versus 3 Hz). Cardiomyocytes in biowires demonstrated less automaticity and higher hERG currents and IK1 compared to EBs cultivated for 20 days (EBd20) or for 40-44 days (EBd44). EBd20 cardiomyocytes represented the cell properties before their incorporation into biowires, whereas EBd44 cardiomyocytes were cultured 10 days longer than the biowire culture time and shows that even with longer time in culture in EB format, cardiomyocytes could not reach equivalent maturation properties as those induced by electrical stimulation in biowires. However, the cardiomyocytes obtained via biowires are still not as mature as adult cardiomyocytes.
Mechanical stimulation has been suggested to be effective in the induction of structural proteins of hPSC-CMs. However, mechanical stimulation did not lead to electrophysiological maturation12. This might suggest the need to combine electrical stimulation and mechanic stress, sequentially or concurrently, to induce terminal differentiation in hPSC-derived cardiomyocytes. The combination of electrical stimulation with other methods, including the delivery of biochemical factors17 and cell alignment in 3D tissues18, might be able to achieve a more effective pro-maturation strategy for future in vivo studies.
The dimensions of the biowire device used here are 5 mm x 1 mm x 0.3 mm (length x width x height), but it is possible to modify the device to make longer biowires. However, the height of the biowires is limited to 0.3 mm (with a radius of ~300 µm upon gel compaction), given the limitations in oxygen delivery in larger tissues. To overcome this limitation of generating thicker tissues, a perfusion method would be required19. Nevertheless, it is recommended to maintain the ratio of 0.5 x 106 cells/0.5 cm of wire in length. For longer biowires, collagen gel polymerization may take longer, and more frequent medium changes may be needed due to the increased number of cells. Moreover, the current setup of the biowire using silk suture does not allow for the measurement of the force of contraction generated by the seeded cardiomyocytes. However, the use of a biodegradable suture may make this possible in the future.
The cell type used to make the biowire described here is hESC-CM using the Hes2 cell line20. The protocol could also use hiPSC cell lines (e.g., CDI-MRB and HR-I-2Cr-2R)14. The cardiomyogenic ability of different human stem cell lines might be different, and suitability for this platform should be experimentally determined and the conditions optimized as needed. There are several critical steps within the protocol (see Table 1).
Additionally, the biowires were designed to fit in a 6-well plate, and the example here showed the making of one biowire. However, it is possible to culture and electrically stimulate multiple biowires simultaneously in one well.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
This work was supported by a grant-in-aid from the Heart and Stroke Foundation of Canada (G-14-0006265), operating grants from the Canadian Institutes of Health Research (137352 and 143066), and a J.P. Bickell foundation grant (1013821) to SSN.
Materials
| L-Ascorbic acid | Sigma | A-4544 | hPSC-CM culture media componet |
| AutoCAD | Autodesk, Inc | Software to design device | |
| Carbon rods, Ø 3 mm | Electrical stimulator chamber component | ||
| Collagen, type 1, rat tail | BD Biosciences | 354249 | Collagen gel: 2.1 mg/ml of rat tail collagen type I in 24.9 mM glucose, 23.8 mM NaHCO3, 14.3 mM NaOH, 10 mM HEPES, in 1xM199 media with 10 % of growth factor-reduced Matrigel. |
| Collagenase type I | Sigma | C0130 | 0.2% collagenase type I (w/v) and 20% FBS (v/v) in PBS with Ca2+ and Mg2+. Sterilize with 0.22 μm filter and make 12 ml aliquots. Store at -20 °C. |
| Deoxyribonuclease I (DNase I) | EMD Millipore | 260913-25MU | Make 1 mg/ml DNase I stock solution in water. Filter sterile and store 0.5 ml aliquots at −20 °C |
| Drill & drill bits (Ø 1mm and 2 mm) | Dremel | Drill holes in polycarbonate frames | |
| Electrical stimulator | Grass | s88x | |
| Fetal bovine serum (FBS) | WISENT Inc. | 080-450 | |
| D-(+)-Glucose | Sigma | G5767 | Collagen gel component |
| L-Glutamine | Thermo Fisher Scientific | 25030081 | |
| H2O | MilliQ | 18.2 MΩ·cm at 25 °C, ultrapure, to make all solutions | |
| HEPES | Sigma | H4034 | Collagen gel component |
| Hot plate | Torrey Pines | HS40 | |
| Iscove's Modified Dulbecco's Medium(IMDM) | Thermo Fisher Scientific | 12440053 | |
| Mask aligner | EVG | EVG 620 | |
| Matrigel, growth factor reduced | Corning | 354230 | Collagen gel component |
| Medium 199 (M199) | Thermo Fisher Scientific | 11150059 | Collagen gel component |
| Monothioglycerol (MTG) | Sigma | M-6145 | hPSC-CM culture media componet |
| Orbital shaker | VWR | 89032-088 | |
| Penicillin/Streptomycin (P/S) | Thermo Fisher Scientific | 15070063 | |
| Phosphate-buffered saline (PBS) with Ca2+ and Mg2+ | Thermo Fisher Scientific | 14040133 | |
| Plate (6-well) | Corning | 353046 | |
| Plate (6-well), low attachment | Corning | 3471 | |
| Platinum wires, 0.2 mm | Electrical stimulator chamber component | ||
| Polydimethylsiloxane (PDMS) | Dow Corning | Sylgard 184 | |
| Propylene glycol monomethyl ether acetate (PGMEA) | Doe & Ingalls Inc. | To develop the wafer | |
| Pouch, peel-open | Convertors | 92308 | For steam sterilization |
| Silicon wafer, 4-inch | UniversityWafer Inc. | ||
| Sodium bicarbonate (NaHCO3) | Sigma | S5761 | Collagen gel component |
| Sodium hydroxide | Sigma | S8045 | Collagen gel component |
| Sprin coater | Specialty Coating Systems | G3P-8 | |
| StemPro-34 culture medium | Thermo Fisher Scientific | 10639011 | hPSC-CM culture medium. To make 50 ml, add 1.3 ml supplement, 500 μl of 100× L-Glutamine, 250 μl of 30 mg/ml transferrin, 500 μl of 5 mg/ml ascorbic acid, 150 μl of 26 μl /2 ml monothioglycerol (MTG), and 500 μl (1 %) penicillin/streptomycin. |
| Stop media | Wash medium:FBS (1:1) | ||
| SU-8 50 | MicroChem Corp. | photoresist, master component | |
| SU-8 2050 | MicroChem Corp. | photoresist, master component | |
| Transferrin | Roche | 10-652-202 | hPSC-CM culture media componet |
| Trypsin/EDTA, 0.25% | Thermo Fisher Scientific | 25200056 | hPSC-CM culture media componet |
| Wash medium | IMDM containing 1% Penicillin/Streptomycin |
Referenzen
- Sun, X., Nunes, S. S. Overview of hydrogel-based strategies for application in cardiac tissue regeneration. Biomed Mater. 10 (3), 034005 (2015).
- Sun, X., Altalhi, W., Nunes, S. S. Vascularization strategies of engineered tissues and their application in cardiac regeneration. Adv Drug Deliv Rev. 96, 183-194 (2016).
- Hastings, C. L., et al. Drug and cell delivery for cardiac regeneration. Advanced Drug Delivery Reviews. 84, 85-106 (2015).
- Yang, L., et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 453 (7194), 524-528 (2008).
- Zhang, J., et al. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method. Circ Res. 111 (9), 1125-1136 (2012).
- Lian, X., et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A. 109 (27), E1848-E1857 (2012).
- Snir, M., et al. Assessment of the ultrastructural and proliferative properties of human embryonic stem cell-derived cardiomyocytes. Am J Physiol Heart Circ Physiol. 285 (6), H2355-H2363 (2003).
- Dolnikov, K., et al. Functional properties of human embryonic stem cell-derived cardiomyocytes: intracellular Ca2+ handling and the role of sarcoplasmic reticulum in the contraction. Stem Cells. 24 (2), 236-245 (2006).
- Yang, X., Pabon, L., Murry, C. E. Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ Res. 114 (3), 511-523 (2014).
- Zimmermann, W. H., et al. Tissue engineering of a differentiated cardiac muscle construct. Circ Res. 90 (2), 223-230 (2002).
- Radisic, M., et al. Functional assembly of engineered myocardium by electrical stimulation of cardiac myocytes cultured on scaffolds. Proc Natl Acad Sci U S A. 101 (52), 18129-18134 (2004).
- Schaaf, S., et al. Human engineered heart tissue as a versatile tool in basic research and preclinical toxicology. PLoS One. 6 (10), e26397 (2011).
- Tulloch, N. L., et al. Growth of engineered human myocardium with mechanical loading and vascular coculture. Circ Res. 109 (1), 47-59 (2011).
- Nunes, S. S., et al. Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat Methods. 10 (8), 781-787 (2013).
- Lake, M., et al. Microfluidic device design, fabrication, and testing protocols. Protocol Exchange. , (2015).
- Shiba, Y., Hauch, K. D., Laflamme, M. A. Cardiac applications for human pluripotent stem cells. Curr Pharm Des. 15 (24), 2791-2806 (2009).
- Yang, X., et al. Tri-iodo-l-thyronine promotes the maturation of human cardiomyocytes-derived from induced pluripotent stem cells. J Mol Cell Cardiol. 72, 296-304 (2014).
- Zhang, D., et al. Tissue-engineered cardiac patch for advanced functional maturation of human ESC-derived cardiomyocytes. Biomaterials. 34 (23), 5813-5820 (2013).
- Radisic, M., et al. Oxygen gradients correlate with cell density and cell viability in engineered cardiac tissue. Biotechnol Bioeng. 93 (2), 332-343 (2006).
- Reubinoff, B. E., Pera, M. F., Fong, C. Y., Trounson, A., Bongso, A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 18 (4), 399-404 (2000).

