Improved Protocol for Chromatin Immunoprecipitation from Mouse Skeletal Muscle
Summary
A novel protocol for the preparation of chromatin from adult mouse skeletal muscle adapted to the study of gene regulation in muscle fibers by chromatin immunoprecipitation is presented.
Abstract
We describe an efficient and reproducible protocol for the preparation of chromatin from adult mouse skeletal muscle, a physically resistant tissue with a high content of structural proteins. Dissected limb muscles from adult mice are physically disrupted by mechanical homogenisation, or a combination of mincing and douncing, in a hypotonic buffer before formaldehyde fixation of the cell lysate. The fixed nuclei are purified by further cycles of mechanical homogenisation or douncing and sequential filtrations to remove cell debris. The purified nuclei can be sonicated immediately or at a later stage after freezing. The chromatin can be efficiently sonicated and is suitable for chromatin immunoprecipitation experiments, as illustrated by the profiles obtained for transcription factors, RNA polymerase II, and covalent histone modifications. The binding events detected using chromatin prepared by this protocol are predominantly those taking place in the muscle fiber nuclei despite the presence of chromatin from other fiber-associated satellite and endothelial cells. This protocol is therefore adapted to study gene regulation in the adult mouse skeletal muscle.
Introduction
Chromatin immunoprecipitation (ChIP) coupled to quantitative polymerase chain reaction (qPCR) and high throughput sequencing (ChIP-seq) have become the methods of choice to study transcription and epigenetic regulation of gene expression in various tissues and cell-types1. This technique allows genome-wide profiling of covalent chromatin modifications, histone variant occupancy, and transcription factor binding2,3.
While performing ChIP from cultured cells is well established, ChIP from mammalian tissues remains more challenging. Preparing chromatin for ChIP involves several critical steps that need to be optimized for every tissue and cell type. Chromatin can be prepared from the cellular lysates of cultured cells or following purification of their nuclei. In the case of mammalian tissues, efficient lysis, formaldehyde fixation, and purification of nuclei are critical in order to ensure optimal recovery of the chromatin. Moreover, the choice of whether to fix the nuclei before or after their purification has to be experimentally determined. Despite these hurdles, ChIP has been successfully performed from tissues such as the liver, testis, or brain4,5,6. In the case of skeletal muscle, disruption of such a physically resistant tissue, which exhibits a high content of structural proteins, and isolation of its nuclei is particularly challenging. Given this specificity, protocols optimized for other tissues do not give satisfactory results for skeletal muscles.
Here we describe a protocol to isolate ChIP-grade chromatin from mouse skeletal muscle tissue that involves physically disrupting the tissue, formaldehyde fixation, and then the isolation of nuclei and sonication. The efficiency of this method to prepare chromatin suitable for ChIP from this tissue was demonstrated by performing ChIP-qPCR and ChIP-seq for various transcription factors, RNA polymerase II, and covalent histone modifications7.
This new technique is much faster than a previously reported protocol8 comprising long collagenase digestion steps during which time, changes in genomic localisation of transcription factors and alterations in gene expression may take place. The rapidity with which the nuclei are isolated and fixed makes the method described here particularly attractive to prepare chromatin that more faithfully captures the native genomic occupancy state. Moreover, although other methods for chromatin isolation or protein extraction from muscle tissue have have been described8,9,10 and used for ChIP-qPCR experiments on selected genes, no ChIP-seq data has been reported using them. The method reported here is suitable for transcription factor and histone modification ChIP-seq, and hence should be also suitable for 3/4C or HiC chromatin conformation capture applications.
Protocol
Mice were kept in accordance with the institutional guidelines regarding the care and use of laboratory animals and in accordance with National Animal Care Guidelines (European Commission directive 86/609/CEE; French decree no.87-848). All procedures were approved by the French national ethics committee.
1. Isolation of Muscle Tissue
- Sacrifice one adult 6 to 8-week-old mouse by cervical dislocation. Sterlise the limb by rinsing it with 70% ethanol and dissect the hind limb muscles (Gastrocnemius, Tibialis Anterior, Quadriceps) using fine point scissors and forceps.
NOTE: One mouse should yield around 500 mg of tissue. - Mince the muscles in a 2 mL test tube containing 1 mL of ice-cold hypotonic buffer to a homogenous preparation of small (<2 – 3 mm3) pieces using fine scissors and leave the tube shaking on a bench top agitator at 4 °C for 5 – 10 min.
2. Tissue Lysis
NOTE: Please refer to Table 1 for all buffer compositions.
- Transfer the homogenate to a 14 mL round-bottom tube and resuspend in 5 mL cold hypotonic buffer (EDTA-free protease inhibitor cocktail and PMSF).
- Homogenise the minced muscle tissue using a loose dounce (20 – 30 strokes within 3 min) or a mechanical tissue homogenizer for 15 – 30 s in round-bottom 14 mL tubes.
NOTE: The efficiency of lysis can be assessed at this stage by light microscopy, and if required, additional disruption can be performed.
- Homogenise the minced muscle tissue using a loose dounce (20 – 30 strokes within 3 min) or a mechanical tissue homogenizer for 15 – 30 s in round-bottom 14 mL tubes.
- Transfer the homogenate to 15 mL tubes and fill volume to 10 mL using cold hypotonic buffer. Fix the homogenate as described below before proceeding with the next mouse.
3. Fixation
- Add formaldehyde to 1% final concentration and shake for 10 min at room temperature.
- Add glycine to a final concentration of 0.125 M in each tube to stop fixation and shake for 5 – 10 min at room temperature.
4. Nuclei Preparation
NOTE: Please refer to Table 1 for all buffer compositions.
- Homogenise the fixed lysate using a loose dounce (5 – 10 strokes).
- Transfer to a 15 mL tube and centrifuge at 1,000 x g for 5 min at 4 °C to obtain nuclei and cellular debris.
- Remove the supernatant and resuspend the pellet in fresh 5 mL hypotonic buffer. Filter the lysate through a 70 µm cell strainer into a 50 mL tube. Re-filter the filtrate through a 40 µm cell strainer.
- Transfer the filtrate to a 15 mL tube and centrifuge at 1,000 x g for 5 minutes at 4 °C to obtain the nuclear pellet.
NOTE: At this stage, the nuclei can be immediately sonicated, or snap frozen as a dry pellet in liquid nitrogen and stored at -80 °C.
5. Sonication
- Assess the volume of the nuclear pellet (around 50 µL for both mechanical and dounce homogenisation) and resuspend it in sonication buffer up to 3 to 4 times of packed nuclear volume and sonicate for 10 – 15 min at 4 °C using a sonicator.
NOTE: The chromatin can be analysed immediately as described below (6) or frozen and stored at -80 °C.
6. Assessing the Chromatin Fragment Size following Sonication
NOTE: Please refer to Table 1 for all buffer compositions.
- To de-crosslink, take 30 µL of chromatin in a 1.5 mL test tube and add 20 µL 5 M NaCl. Complete the volume to 500 µL and incubate at 65 °C overnight.
- The next day, perform a treatment with 1 µL proteinase K (20 mg/mL stock), 10 µL 2 M Tris pH 6.8, and 10 µL of 0.5 M EDTA for 1 h at 42 °C. Perform a phenol-chloroform/chloroform extraction and precipitate the DNA with 1 volume of sodium acetate (3 M) and 2 volumes of 100% ethanol for one h at -80 °C or overnight at -20 °C.
- Pellet the DNA by centrifugation at 13,500 x g for 15 min. Decant the supernatant and wash the pellet thoroughly with cold 70% ethanol and centrifuge again for 5 min. Decant the supernatant and air-dry the pellet.
- Resuspend the pellet in the original volume (30 µL) of TE buffer. Measure the DNA concentration in a spectrophotometer by absorbance at 260 nm/280 nm. Pipette 500 – 1,000 ng of DNA together with a DNA size ladder on separate lanes of a 1.5% agarose gel and perform electrophoresis to assess the fragment size of chromatin.
NOTE: Fragment size should be between 200 – 500 base pairs, and there should be no detectable high molecular weight fragments corresponding to non- or poorly-fragmented DNA. If required, the chromatin solution (step 5) can be re-sonicated for a longer time and then re-analysed as described above until the optimum size is obtained.
7. Chromatin Immunoprecipitation (ChIP)
NOTE: Please refer to Table 1 for all buffer compositions.
- Block the Protein G sepharose beads by aliquoting 1 mL of the 50% slurry in ethanol. Pellet the beads by centrifugation at 400 x g for 1 min in a benchtop centrifuge and wash with 1 mL of TE buffer centrifuge at 400 x g and repeat the wash.
- After the second centrifugation, re-suspend in 1 mL of ChIP dilution buffer with 25 µL of BSA (stock 20 mg/mL) and 20 µL yeast tRNA (stock 10 mg/mL). Block the beads for at least 2 h prior to use by rotation (40 – 60 rpm) at 4 °C.
- Dilute 50 µg of chromatin (step 5) 8 to 10 times using ChIP dilution buffer. Add 50 µL of blocked bead slurry and incubate for 2 h rotating (40-60 rpm) at 4°C. Centrifuge at 400 x g at 4 °C to obtain pre-cleared chromatin and transfer it to a fresh tube.
- For immunoprecipitation, add the appropriate amount of primary antibody (e.g., 1-2 µg per 10 µg of chromatin) and incubate overnight with rotation at 4 °C.
NOTE: The optimal amount of antibody may be recommended by the supplier or can be determined empirically by performing ChIP-qPCR with different quantities of antibody until optimum enrichment is observed. Also Protein G sepharose may be replaced by Protein A sepharose depending on the subtype of antibody used. - Add the blocked beads the next day and incubate for 1 h with rotation (40 – 60 rpm) at 4 °C. Centrifuge at 400 x g for 20 – 30 s to pellet the beads, remove the supernatant, and start the ChIP washes.
- ChIP washes: Perform the following washes with the buffers: once with Low Salt Buffer, and then twice with High Salt Buffer, twice with LiCl Buffer, and finally twice with TE Buffer.
- Perform each wash for 10 min with rotation (40 – 60 rpm) at 4 °C and pellet the beads between each wash by centrifugation at 400 x g for 20 – 30 s in a bench-top centrifuge.
- For elution, remove the last wash and resuspend the beads in 250 µL elution buffer (freshly prepared) for 15 min at room temperature while shaking.
- Centrifuge at 400 x g and collect the eluate in a fresh tube. Perform this step twice and then de-crosslink the eluate overnight with 20 µL NaCl 5 M and 1 µL RNase A (10 mg/mL), treat with proteinase K, and phenol-chloroform/chloroform extract as in Step 6.
- Resuspend in 50 µL of TE buffer and use aliquots for ChIP-qPCR as per standard protocols15 to check the quality of the ChIP.
Representative Results
To isolate nuclei, we performed mechanical homogenization of the dissected and minced muscle tissue for either 15 or 45 s, at 18,000 and 22,000 rpm (see Table of Materials). In all conditions, nuclei could be separated from the tissue debris, but the yield was optimal using the lower speed (Figure 1A-B). Nuclei could also be prepared by douncing for 3 – 5 min (Figure 1A, and data not shown), but mechanical homogenization is the method of choice, as optimal yields of nuclei can be achieved in as little as 15 s, allowing an important gain of time especially if multiple samples have to be processed (Figure 1B). Using this protocol, up to 2.7 x 107 nuclei from 500 mg of tissue (equivalent to 1 mouse's tissue) can be isolated to generate 100 µg of chromatin.
To optimize chromatin fragmentation from fixed nuclei, we sonicated for 5 to 20 min. Under the settings (see Table of Materials), a 10 min sonication was optimal to generate chromatin with an average fragment size of 250 bp (Figure 2). This step should be optimized experimentally taking into account the type of sonicator used.
To assess the quality of the muscle chromatin prepared by the protocol described above, ChIP-qPCR and ChIP-seq experiments were performed against transcription factors, a histone modification or RNA polymerase II (Pol II). ChIP-qPCR against the glucocorticoid receptor (GR) in the presence or absence of dexamethasone (Dex) treatment revealed strong Dex-dependent GR enrichment at regulatory elements of the Redd1 and Murf1 genes that are known to be Dex and GR regulated in muscle fibers11, while no signal was seen at the testis specific Prm1 promoter used as a negative control (Figure 3A).
ChIP-qPCR against acetylated lysine 27 of histone H3 (H3K27ac), a covalent modification found at active enhancers and promoters, showed strong enrichment at the Desmin (Des) promoter that is active in muscle fibers, compared to a negative intergenic control region (Figure 3B). H3K27ac ChIP-seq revealed a high level of this mark throughout the Des locus, typical of a 'super-enhancer' regulated tissue identity gene12 (Figure 4A). Similarly, Pol II ChIP-seq showed high levels of transcribing Pol II at this locus.
ChIP-qPCR for transcription factor Tead4, which plays an important role in myogenic differentiation13,7, revealed its enrichment on previously described binding sites at the Ccnd1 and Ifrd1 genes (Figure 3C). Importantly, enrichment was reduced using chromatin prepared from mice where Tead4 was selectively inactivated in muscle fibers (Tead4skm-/-)7 (Figure 3C). Analyses of ChIP-seq data for both Tead1 and Tead4 showed their binding to a site in the promoter of the Kdm5a gene using chromatin from wild-type muscle, whereas the Tead4 signal, but not that of Tead1, H3K27ac, or Pol II, was lost using the chromatin from muscles of the Tead4skm-/- mice (Figure 4B). Similarly, Tead1 and Tead4 binding to a regulatory element downstream of the Adssl1 gene is seen with chromatin from wild-type muscle, whereas Tead4 binding was selectively lost using chromatin from the Tead4skm-/- muscle (Figure 4C). These observations demonstrate that the Tead4 signal seen in the ChIP-seq data came from binding in the muscle fiber.
To determine the contribution of other cell types associated with the muscle fiber to the ChIP-seq results, we assessed the H3K27ac and Pol II signals at the Vcam1 gene that is active in muscle satellite cells and Pecam1, a marker of endothelial cells of blood vessel that irrigate the muscles. Compared to the high levels seen at the Des or Adssl1 genes, the levels of H3K27ac and Pol II seen at both the Vcam1 and Pecam1 genes were much lower (Figure 4A and Figure 4C). The large differences in signal for muscle fiber expressed genes compared to those expressed in associated tissues indicated that the signal seen in ChIP from the chromatin prepared using the protocol described above comes predominantly from binding events in the muscle fiber.
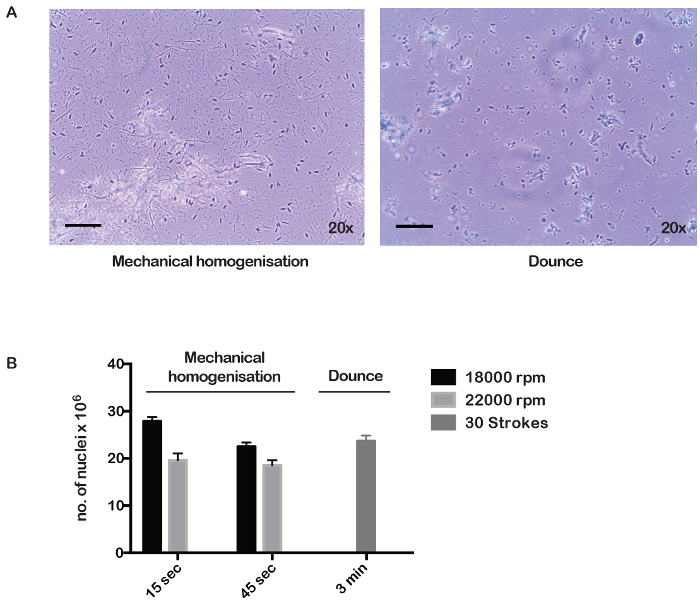
Figure 1: Purification of nuclei from adult mouse muscle. (A) Tissue lysates obtained by mechanical homogenisation (18,000 rpm, 45 s) and dounce homogenization (30 strokes/3 min) were stained with trypan blue and observed under a digital microscope and images were captured at 20X magnification. Scale bar = 100 µm. (B) Number of nuclei obtained from 500 mg of tissue (equivalent of 1 mouse) after preparation under the indicated conditions, 18,000 and 22,000 rpm for 15 and 45 s or 3 min of douncing. Error bars represent mean ± S.E.M. for n = 3 mice for mechanical homogenization, and n = 3 mice for douncing. Please click here to view a larger version of this figure.
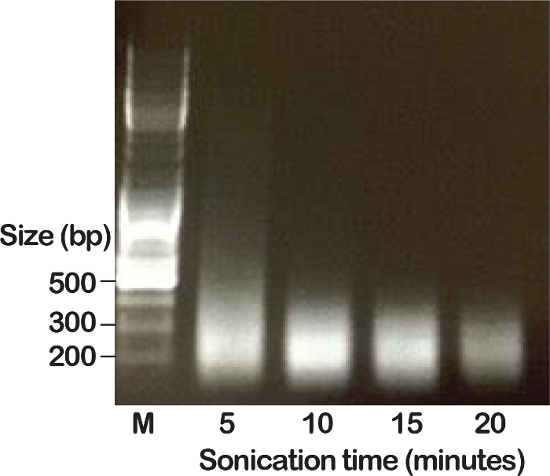
Figure 2: Verification of chromatin fragment size. Isolated nuclei were sonicated for 5, 10, 15 or 20 min as indicated. After de-crosslinking and purification, the DNA fragment size was assessed by agarose gel electrophoresis in the presence of ethidium bromide. M is DNA size marker. Please click here to view a larger version of this figure.
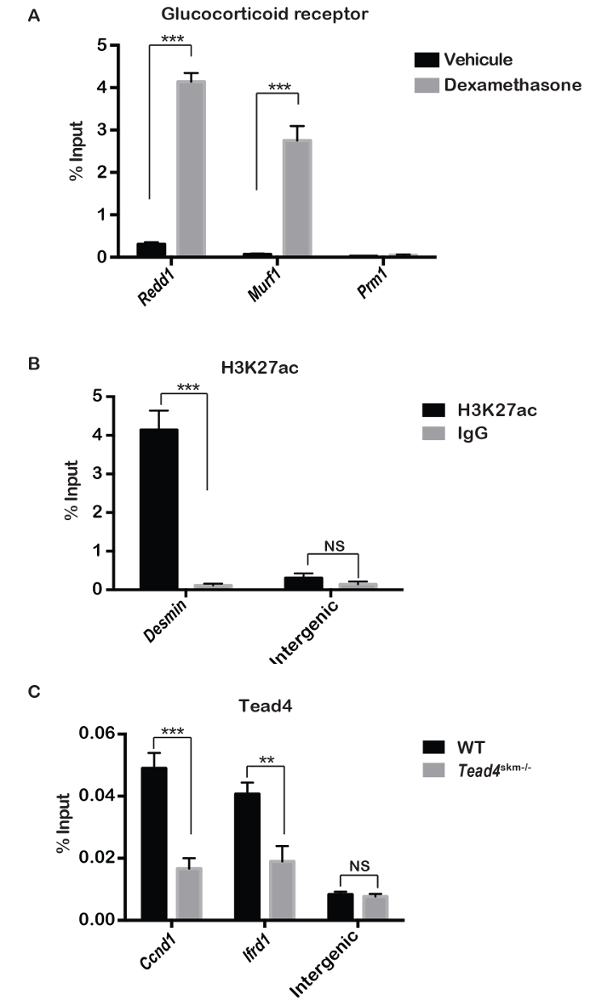
Figure 3: Use of chromatin for ChIP-qPCR. (A) Mice were subjected to intra-peritoneal injections of vehicle or dexamethasone (10 mg/kg) and after 1.5 h, chromatin was prepared from hind limb muscles. The enrichment in ChIP at GR target genes Redd1 and Murf1, compared to the negative control protamine promoter (Prm1), is indicated as a % of input precipitated. (B) ChIP was performed for H3K27ac and IgG control antibodies and analysed at the Desmin promoter and compared to an intergenic negative control region. (C) Tead4 ChIP was performed on muscle chromatin obtained from wild type and muscle-specific Tead4 knockout mice (Tead4skm-/-) and analysed at Tead4 binding sites in the Ccnd1 and Ifrd1 promoters, compared to the control intergenic region. In all experiments, chromatin from 3 mice was pooled and error bars represent mean ± S.E.M in a Student's T-test for three technical replicates. P value <0.001**, <0.0001 ***; NS, non-significant. Please click here to view a larger version of this figure.
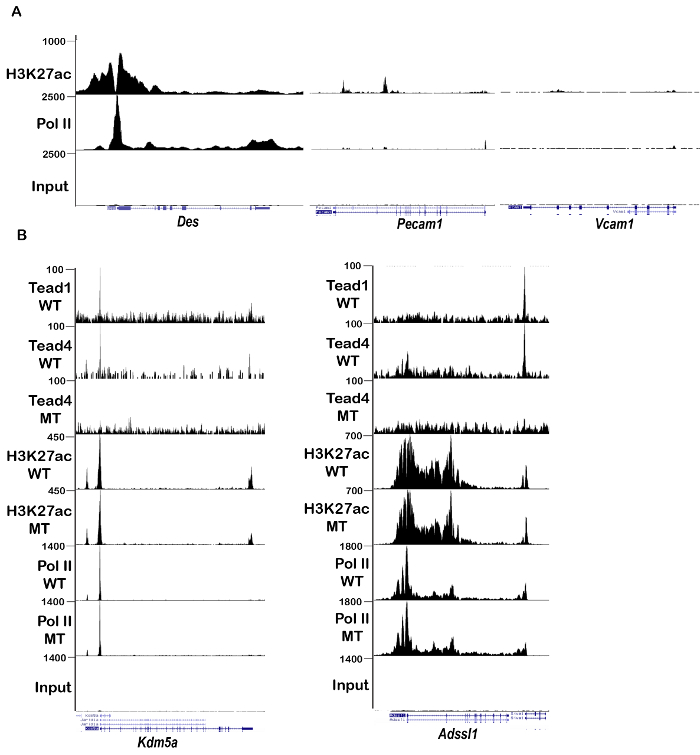
Figure 4: Genomic profiles of muscle fiber chromatin. (A) Screenshots from the University of California at Santa Cruz genome browser of the indicated loci illustrating the much higher H3K27ac and Pol II levels at the Desmin gene, highly expressed in muscle fiber, compared to the Vcam1 and Pecam1 genes expressed in satellite cells and endothelial cells, respectively. (B) UCSC screenshots of the indicated loci illustrating binding of Tead1 and Tead4, as well as H3K27ac and Pol II, in chromatin from wild-type (WT) muscle compared to that from Tead4skm-/- (MT) muscle, where selective loss of Tead4 binding is observed. The ChIP-seq data illustrated is available as GSE82193 in the GEO database. Please click here to view a larger version of this figure.
| Buffer Name | Composition | ||
| Hypotonic buffer | 10 mM HEPES-KOH (pH 7.3) | ||
| 10 mM KCl | |||
| 5 mM MgCl2 | |||
| 0.1% NP-40 | |||
| 0.1 mM PMSF (freshly added) | |||
| Protease Inhibitor cocktail (PIC) 1x (freshly added) | |||
| Sonication buffer | 1% SDS | ||
| 10 mM EDTA | |||
| 20 mM Tris-HCl pH 8 | |||
| 150 mM NaCl | |||
| 0.1% Sodium Deoxycholate | |||
| 1% Triton X-100 | |||
| Freshly added 0.1 mM PMSF | |||
| PIC 1x | |||
| ChIP Dilution buffer | 0.01% SDS | ||
| 1.1% Triton X-100 | |||
| 1.2 mM EDTA | |||
| 16.7 mM Tris HCl pH8 | |||
| 167 mM NaCl | |||
| Freshly added 0.1 mM PMSF | |||
| PIC 1x | |||
| Low Salt buffer | 0.1% SDS | ||
| 1% Triton X-100 | |||
| 500 mM EDTA | |||
| 20 mM Tris HCl pH8 | |||
| 150 mM NaCl | |||
| High Salt buffer | 0.1% SDS | ||
| 1% Triton X-100 | |||
| 500 mM EDTA | |||
| 20 mM Tris HCl pH8 | |||
| 500 mM NaCl | |||
| LiCl Buffer | 250 mM LiCl | ||
| 1% NP-40 | |||
| 1 mM EDTA | |||
| 10 mM Tris HCl pH 8 | |||
| Elution buffer | 1% SDS | ||
| 100 mM NaHCO3 | |||
| Tris-EDTA (TE) buffer | 50 mM Tris HCl pH 8 | ||
| 1 mM EDTA | |||
Table 1: Buffer compositions.
Discussion
Here we describe a novel protocol for preparing chromatin from adult mouse skeletal muscles and show that this chromatin is suitable for ChIP experiments that detect transcription factor binding and covalent histone modifications in the muscle fiber nuclei. This protocol involves several critical steps. The first is tissue disruption that can be performed either by dounce or by mechanical shearing. Mechanical shearing is faster and more reproducible, and is therefore the method of choice. Nevertheless, if no suitable apparatus is available, disruption can also be performed by douncing, where assessment of the efficiency of lysis under the microscope is recommended before proceeding to the fixation step. Fixation of whole-tissue lysates, rather than fixation of isolated nuclei, allowed more reproducible sonication in smaller volumes and for short durations. We also chose to fix the lysates after disruption rather than fix the tissue before disruption, as described by Thomas et al.10 Pre-fixing the tissue resulted in reduced efficiency of homogenisation. This protocol based on mechanical shearing is also faster than other proposed solutions, such as collagenase digestion, which involve prolonged incubation of the dissected tissue at 37 °C8. During this period, changes in gene expression and transcription factor binding can occur. Fixing the nuclei as fast as possible is therefore essential to more faithfully capture the normal physiological state.
A second critical step is the purification of nuclei from the fixed lysate. For this, sequential filtering of the fixed lysate solution, first through a 70 µm cell strainer to eliminate the bigger debris and then through a 40 µm cell strainer to remove the fine debris, avoids clogging the strainers and maximizes the yields of nuclei obtained. Once purified by filtration, nuclei are sonicated and the DNA fragment size determined after de-crosslinking. A typical preparation from the two hind limbs of one mouse yields 100 µg of chromatin. Pooling the lysates from up to 3 mice may improve the yield by reducing the losses during preparation. This procedure is efficient as it allows the recovery of up to 75% of the genomic DNA as chromatin (data not shown).
ChIP experiments designed to assess transcription factor binding and epigenetic modifications demonstrated the suitability of this protocol for studying gene regulation in the muscle fiber nuclei. Strong and robust ChIP-seq signals for Pol II and H3K27ac were generated from the chromatin, allowing identification of muscle fiber identity genes by their distinctive H3K27ac and Pol II profiles7. The much higher levels of H3K27ac and Pol II on genes strongly expressed in fiber nuclei, compared to genes expressed in satellite or endothelial cells, showed that the signal came predominantly from the fiber nuclei. This was also confirmed by the loss of Tead4 ChIP signal using chromatin from mice where it was specifically inactivated in muscle fibers. Nevertheless, weaker, but clearly above background signals, for satellite cell markers such as Vcam1, Pax7 (data not shown) can be detected showing that chromatin from these cells is also present. We anticipate that our protocol should be suitable for other techniques that use formaldehyde-fixed chromatin such as 3C/4C and HiC chromatin conformation capture techniques. Use of our protocol for combining ChIP-seq for transcription factors and chromatin modifications with chromatin conformation capture should in the future allow a detailed understanding of gene regulatory mechanisms in muscle fiber and how these may be regulated or upset by physiological stimuli, such as exercise, fasting, a high fat diet, denervation, or in disease, by using chromatin prepared from genetically modified mice reproducing diseases like X-linked centronuclear myopathy14.
In summary, this low-cost and time-efficient protocol allows the isolation of muscle fiber nuclei that can be sonicated immediately or frozen at -80 °C for ulterior use. Use of chromatin prepared by this protocol has provided the first genome wide ChIP-seq data from muscle fiber7 and will facilitate future studies on gene regulation in this tissue.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
We thank all the staff of the IGBMC high throughput sequencing facility, a member of "France Génomique" consortium (ANR10-INBS-09-08) and all IGBMC general services in particular the staff of the IGBMC animal facility. This work was supported by grants from the CNRS, the INSERM, the AFM, the Ligue Nationale contre le Cancer, the French state fund through the ANR under the programme Investissements d'Avenir labelled ANR-10-IDEX-0002-02, the Labex INRT ANR-10-IDEX-0001-02 and the ANR-AR2GR-16-CE11-009-01. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. S.J was supported by the Ligue Nationale contre le Cancer and the ANR-AR2GR-16-CE11-009-01, and V.U by the Ministère de l'Enseignement et de la Recherche. ID is an 'équipe labellisée' of the Ligue Nationale contre le Cancer.
Materials
| T 25 digital ULTRA-TURRAX | T 18 ULTRA-TURRAX | 0009022800 | |
| PMSF | SIGMA-ALDRICH CHIMIE SARL | P7626-25G | |
| Protease Inhibitor Cocktail-EDTA Free | Roche Diagnostics | 11873580001 | |
| Protein G Sepharose beads | SIGMA-ALDRICH CHIMIE | P-3296 | |
| Proteinase K | SIGMA-ALDRICH CHIMIE | P-2308 | |
| Cell Strainers 70um | Corning BV | 431751 | |
| Cell Strainers 40um | Corning BV | 352340 | |
| Formaldehyde EM grade | Euromedex | 15710-S | |
| Rnase A | Fischer Scientific | 12091039 | |
| Phenol:Chloroform:Isoamyl Alcohol 25:24:1 | SIGMA-ALDRICH CHIMIE | P3803 | |
| BSA | SIGMA-ALDRICH CHIMIE | B4287-25G | |
| yeast tRNA | SIGMA-ALDRICH CHIMIE | R5636 | |
| Glycogen Blue | AMIBION | AM9516 | |
| Pol II ChIP Antibody | Santa Cruz | SC-9001 | |
| H3K27ac ChIP Antibody | Active Motif | 39133 | |
| Tead4 ChIP Antibody | Aviva Systems Biology | (ARP38276_P050) | |
| IGEPAL | SIGMA-ALDRICH CHIMIE | I-3021 | |
| E220 Focused Ultrasonicator | Covaris E220 | ||
| Name | Company | Catalog Number | Comments |
| Additional items | |||
| Scissors | |||
| Loose Dounce | |||
| 15 ml and 50 ml falcon tubes | |||
| 14 ml round bottom tubes | |||
| 1.5 ml and 2 ml Eppendorf tubes | |||
| 70 μm and 40 μm cell strainers (Corning) |
Referenzen
- Johnson, D. S., Mortazavi, A., Myers, R. M., Wold, B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 316 (5830), 1497-1502 (2007).
- Furey, T. S. ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat Rev Genet. 13 (12), 840-852 (2012).
- Wade, J. T. Mapping Transcription Regulatory Networks with ChIP-seq and RNA-seq. Adv Exp Med Biol. 883, 119-134 (2015).
- Alpern, D., et al. TAF4, a subunit of transcription factor II D, directs promoter occupancy of nuclear receptor HNF4A during post-natal hepatocyte differentiation. Elife. 3, e03613 (2014).
- Martianov, I., et al. Cell-specific occupancy of an extended repertoire of CREM and CREB binding loci in male germ cells. BMC Genomics. 11, 530 (2010).
- Achour, M., et al. Neuronal identity genes regulated by super-enhancers are preferentially down-regulated in the striatum of Huntington’s disease mice. Hum Mol Genet. 24 (12), 3481-3496 (2015).
- Joshi, S., et al. TEAD transcription factors are required for normal primary myoblast differentiation in vitro and muscle regeneration in vivo. PLoS Genet. 13 (2), e1006600 (2017).
- Ohkawa, Y., Mallappa, C., Dacwag-Vallaster, C. S., Imbalzano, A. N., DiMario, J. . Myogenesis: Methods and protocols. 798, 517-531 (2012).
- Dimauro, I., Pearson, T., Caporossi, D., Jackson, M. J. A simple protocol for the subcellular fractionation of skeletal muscle cells and tissue. BMC Res Notes. 5, 513 (2012).
- Thomas, J. L., et al. PAK1 and CtBP1 Regulate the Coupling of Neuronal Activity to Muscle Chromatin and Gene Expression. Mol Cell Biol. 35 (24), 4110-4120 (2015).
- Kuo, T., et al. Genome-wide analysis of glucocorticoid receptor-binding sites in myotubes identifies gene networks modulating insulin signaling. Proc Natl Acad Sci U S A. 109 (28), 11160-11165 (2012).
- Whyte, W. A., et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 153 (2), 307-319 (2013).
- Benhaddou, A., Keime, C., Ye, T., Morlon, A., Michel, I., Jost, B., Mengus, G., Davidson, I. Transcription factor TEAD4 regulates expression of myogenin and the unfolded protein response genes during C2C12 cell differentiation. Cell Death and Differentiation. 19 (2), 220-231 (2011).
- Cowling, B. S., et al. Reducing dynamin 2 expression rescues X-linked centronuclear myopathy. J Clin Invest. 124 (3), 1350-1363 (2014).
- . ChIP Analysis Available from: https://www.thermofisher.com/fr/fr/home/life-science/epigenetics-noncoding-rna-research/chromatin-remodeling/chromatin-immunoprecipitation-chip/chip-analysis.html (2017)

