Zygotic Fluorescence Recovery After Photo-bleaching Analysis for Chromatin Looseness That Allows Full-term Development
Summary
Chromatin looseness appears to be involved in the developmental potential of blastomeres. However, it is not known whether chromatin looseness can be used as a reliable index for the developmental potential for embryos. Here, an experimental system in which chromatin looseness-evaluated zygotes can develop to full term has been described.
Abstract
Live imaging is a powerful tool that allows for the analysis of molecular events during ontogenesis. Recently, chromatin looseness or openness has been shown to be involved in the cellular differentiation potential of pluripotent embryonic stem cells. It was previously reported that compared with embryonic stem cells, zygotes harbor an extremely loosened chromatin structure, suggesting its association with their totipotency. However, until now, it has not been addressed whether this extremely loosened/open chromatin structure is important for embryonic developmental potential. In the present study, to examine this hypothesis, an experimental system in which zygotes that were analyzed by fluorescence recovery after photo-bleaching can develop to term without any significant damage was developed. Importantly, this experimental system needs only a thermos-plate heater in addition to a confocal laser scanning microscope. The findings of this study suggest that fluorescence recovery after photo-bleaching analysis (FRAP) analysis can be used to investigate whether the molecular events in zygotic chromatin are important for full-term development.
Introduction
After fertilization, the chromatin structure is altered dynamically, and zygotic chromatin structure is then eventually established1,2. During this period, in paternal pronuclei, the dominant chromatin protein is changed from protamine into histone. The resulting chromatin is extremely different from that of sperms and female oocytes in several points (e.g., histone variant composition, histone modification). Thus, formed zygotic chromatin is thought to be important for subsequent embryonic development. However, despite efforts to reveal the details of zygotic chromatin structure over long periods, methods to evaluate the quality of zygotes or to predict their full-term development at the one-cell stage by analyzing their chromatin structure have never been established.
In the previous study, it is discovered that zygotes have an extremely loosened chromatin structure3. Currently, chromatin looseness or openness is believed to be an important factor for cellular differentiation potential in embryonic stem (ES) cells4. ES cells do not exhibit homogeneity in nature, but are rather heterogeneous; in ES cell colonies, some transiently acquire a higher differentiation potential comparable to blastomeres of two-cell stage embryos. During this transition into the two-cell like state, chromatin looseness in ES cells changes into what is comparable with two-cell stage embryos5. Thus, chromatin looseness seems to be important for cellular differentiation potential and it is possible that extensively open chromatin in zygotes is useful for the evaluation of zygotic developmental potential.
Live imaging is a powerful tool that allows for the analysis of molecular events during ontogenesis since this method allows for subsequent development and even full-term development6. As one of the live imaging methods, FRAP analysis has been used to examine chromatin looseness in preimplantation embryos and ES cells3,4,5. If zygotic chromatin looseness can be analyzed without a detrimental effect on full-term development by FRAP analysis, it may be a valuable tool for the evaluation of the quality of embryos at the one-cell stage. However, the effects on full-term development by this experimental method have not been examined. Recently, an experimental system using FRAP to evaluate zygotic chromatin looseness was developed. Because this was a new observation system for zygotes, it was termed as zygotic FRAP (zFRAP). zFRAP did not critically affect full-term development and has been reported elsewhere7. In this report, the protocol of this experimental method is described.
Protocol
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the University of Yamanashi. The protocol was approved by the Committee on the Ethics of Animal Experiments of the University of Yamanashi (Permit Number: A24-50). All surgeries were performed under tribromoethanol anesthesia, and all efforts were made to minimize suffering. An illustrated overview of the procedures and time-table are shown in Figure 1 and Table 1, respectively.
1. Preparation of Messenger RNA (In Vitro Transcription of eGFP-H2B mRNA)
- Linearize 5 µg of template DNA pTOPO-eGFP-H2B3,7 with 30 U of Not I in 50 µL at 37 °C overnight.
- Collect 1.5 µL of digested DNA for confirmation whether completely digested by electrophoresis.
- Apply 1.5 µL and 3 – 5 µL of undigested DNA with loading dye to the well of 2% agarose gel, respectively, and subject to electrophoresis.
NOTE: If completely digested, a single band (approx. 5 kb) is observed. Undigested DNA is used as a control, which appears at a different position.
- Apply 1.5 µL and 3 – 5 µL of undigested DNA with loading dye to the well of 2% agarose gel, respectively, and subject to electrophoresis.
- Add 151.5 µL of ddH2O and 200 µL of phenol/chloroform/isoamyl alcohol (25:24:1) to the remaining digested DNA.
- Mix vigorously by vortex.
- Centrifuge at the maximum speed for 15 min.
- Recover the supernatant and then add 200 µL of chloroform.
- Mix vigorously by vortex.
- Centrifuge at maximum speed for 15 min.
- Recover the supernatant and then add 20 µL of 3M sodium acetate and 1.5 µL of ethachinmate.
- Mix vigorously by vortex.
- Add 550 µL of 100% ethanol and then mix vigorously by vortex.
- Centrifuge at maximum speed for 15 min.
- Discard the supernatant and then wash the pellet with 70% ethanol.
- Dry the pellet by evaporation for 5 – 10 min.
- Dissolve precipitated plasmid DNA in 8 µL of nuclease-free water.
- Assess plasmid concentration (expected concentration is about 300 – 500 ng/µL) with nanodrop by following manufacturer's instruction.
- Perform in vitro transcription for mRNA encoding eGFP-H2B with 1 µg of purified DNA as a template in 20 µL of total reaction volume by following the manufacturer's instructions.
- After in vitro transcription, add 36 μL of water then recover 1.5 µL from 56 µL of synthesized mRNA for analysis by electrophoresis (as without poly A).
- Perform poly A tailing for synthesized mRNA in a 100 µL reaction volume by following the manufacturer's instructions.
- Add 60 µL of lithium chloride precipitation solution to the poly A tailed mRNA of eGFP-H2B and then mix vigorously.
- Cool at -30 °C for 30 min.
- Centrifuge at maximum speed for 15 min.
- Discard the supernatant and then wash the pellet with 70% ethanol.
- Dry the pellet by evaporation for 5 – 10 min.
- Dissolve the pellet with 20 µL of nuclease-free water by heating at 55 °C for 30 min.
- Assess the concentration of the precipitated mRNA with nanodrop. Dilute to 500 ng/µL with nuclease-free water and store at -80 °C until use.
- Confirm preferred mRNA production; subject 1 µL of mRNA with/without (obtained in 1.8.1 and 1.12 steps, respectively) poly A tail mixed with 1.5 µL of 0.1 mg/mL ethidium bromide and 3 µL of sample loading dye to the electrophoresis analysis. Applied volume was 5.5 µL.
NOTE: If Poly A tailing was successful, the shifted band compared to the mRNA without poly A tailing should be observed (Figure 2A).
2. Preparation of Vasectomized Male Mice
- Weigh the ICR male mice at 8 – 12 months using a weight scale.
- Intraperitoneally inject male mice with 0.7 – 0.9 mL 1.2% tribromoethanol anesthesia.
NOTE: The amount of anesthesia depends on the size of the mouse. For example, a mouse weighing 40 g is injected with 0.8 mL of tribromoethanol anesthesia. - Wet the berry with 70% ethanol.
- Make a small cut (3 – 5 mm) on the berry and then make an incision on the muscle wall with the help of scissors.
NOTE: Scissors and forceps should be cleaned with 70% ethanol before starting the surgery. - Grip a fat pad with forceps and gently pull it out. Then, the testis and vas deferens appear.
NOTE: Vas deferens is a U-shaped tube and with a red blood vessel. - Tie off the vas deferens at two points with surgical sutures and then cut out the middle part between the tied points.
- Gently push back the fat pad and testis into the berry.
- Repeat the surgery with the remaining testis.
- Sew the muscle wall with two stitches with surgical suture.
3. IVF
- Inject 8 – 10-week old female B6D2F1 (BDF1) mice intraperitoneally with 7.5 IU equine chorionic gonadotropin (eCG) and human chorionic gonadotropin (hCG) at 46 – 50 h interval.
- Prepare drops of 300 µL human tubal fluid (HTF)8 media for capacitation and insemination 1 day before IVF in a 30-mm dish, cover these drops with the mineral oil on the laboratory bench and then preincubate in an incubator overnight.
- Sacrifice matured male ICR mice by cervical dislocation. Dissect cauda epididymis with a 27-G needle and collect spermatozoon. Culture them for capacitation in 300 µL of HTF media for 1 – 2 h at 38 °C.
- 16 h after hCG injection, sacrifice super ovulated female mice by cervical dislocation. Transfer the oviduct ampulla into mineral oil next to HTF media and then draw out cumulus cells and oocytes complex from this oviduct ampulla by a 30-G needle into the preincubated insemination-HTF medium.
NOTE: Some female mice do not often show ovulation despite super ovulation treatment. Enlarged oviduct ampulla is a sign of ovulation. - After capacitation, transfer 2 – 3 µL of the capacitated spermatozoon into insemination-HTF media in which the ovulated oocytes were collected.
- 1 h after insemination, collect the fertilized zygotes and treat them with hyaluronidase at 100 µg/mL for 10 min in CZB9 media.
NOTE: When insemination is performed properly, 1 h after insemination, the fertilized zygotes with the 2nd polar body are observed. If less or low-quality sperm is used, only little zygotes with the 2nd polar body are observed. Also, if many sperms are used for insemination, polyspermy occurs. Proper insemination leads to zygotes with male and female pronuclei. By using these criteria, it is possible to find whether insemination is done well or not.- After washing in CZB media, subject the zygotes with a second polar body to microinjection with eGFP-H2B mRNA.
4. Preparation of Chamber and Microinjection Pipettes
- Dilute RNA encoding eGFP-H2B using nuclease-free water to obtain a concentration of 250 ng/µL.
- Prepare 2 – 3 drops of PVP-HTF and eGFP-H2B drops mRNA, and 5 – 6 drops of Hepes buffered CZB (H-CZB) drops on the 60-mm dish. Cover these drops with mineral oil.
NOTE: These preparations can be performed on the laboratory bench. There is no requirement for using laminar airflow cabinet. - Pull borosilicate glass with a micropipette puller (e.g., P = 500, heat = 825, pull = 30, vel = 120, and time = 200)
- After breaking the tip of the pipette, bend the pulled glass microinjection pipette close to the tip (−300 µm back) at around 30° using a microforge.
5. Microinjection
- Turn off the plate heater on the stage of micromanipulator to prevent the killing of the zygotes with a piezo during mRNA injection.
- Wash and coat the inside of injection needles in HEPES-buffered-HTF containing 10% PVP (PVP-HTF).
- Collect the fertilized zygotes with a 2nd polar body (Figure 2B) and transfer 20 – 25 zygotes onto the chamber of the microscope stage attached with a micromanipulator.
- Fill the diluted eGFP-H2B mRNA into borosilicate glass capillaries from the tips.
- Hold the zygote with a holding pipette and then break the zona pellucida and the cytosolic membrane with a piezo drive.
- Inject about 10 pL of mRNA into the cytoplasm of the zygotes. Finish mRNA-injection within 10 min to prevent damage caused by longer culturing the zygotes outside the incubator.
NOTE: The injection volume should be as large as the 2nd polar body. If the amount of mRNA injected is too large, it is possible to cause the free eGFP-H2B fraction, which may affect the mobile fraction. - 10 min after microinjection, wash in at least 3 drops and then culture the injected zygotes in CZB at 38 °C until zFRAP analysis.
6. zFRAP Analysis
- 1 h prior to zFRAP analysis, turn on the laser and hot plate attached to the stage of the confocal microscope. Set the temperature at 38 °C.
- Put 6 – 10 µL of HEPES-buffered CZB media covered with mineral oil on the 60-mm glass bottom dish and warm on the heater until zygote collection.
- 8 – 12 h after insemination, collect 5-10 eGFP-H2B-expressing zygotes and transfer into 6 – 10 µL of pre-warmed HEPES-buffered CZB medium drop.
NOTE: To avoid longer culturing outside incubator, 5 – 10 eGFP-H2B-expressing zygotes were used at each analysis. 5 to 10 zygotes can be analyzed within 1 h. - Set the bleaching and imaging conditions according to the manufacturer's instructions. The case a confocal laser scanning microscope (Table of Materials) is shown below:
- Use 60X oil lens (60X 60X/1.35 oil).
NOTE: Lenses with higher magnification (higher numerical aperture) show higher sensitivity. - Set scanning speed as 4.0 µs/Pixel and size as "512" on "Acquisition Setting" (Supplemental Figure 1).
- Increase Digital zoom to "5" (Supplemental Figure 1).
NOTE: A higher zoom is required to recognize whether focus drift occurs or not. - Open dye list (Supplemental Figure 1) and then chose "EGFP". Set observation time as 1.6 s (interval is set as "free run" setting) (Supplemental Figure 1)
NOTE: More observation points may cause more detailed data, but it may also cause phototoxicity. In the zFRAP analysis, a 1.6 s observation time seems to be enough to detect the parental asymmetry of chromatin looseness. - Set the number of pictures as 12 in total (Supplemental Figure 1).
- Start "Stimulus setting" panel and then click "Main" on UseScanner. Set scanning speed as 10.0 µs/Pixel. Click "Activation in Series" and then set PreActivation as "3 frames" and Activation Time as "5 sec" (Supplemental Figure 1).
- Click "Focus x 2" and set the focus and then "Stop".
- Set bleaching and imaging laser power (477 nm) at 110 and 15 µW, respectively, by adjusting "laser gage power" (Supplemental Figure 1).
NOTE: Very strong bleaching may cause a larger bleached area, which causes difficulties for the quantitative analysis. For measurement of the laser power, use a power meter. It is important to confirm the laser power to avoid the effect of the time-related deterioration of the laser power. - Click "Clip Rect" (Supplemental Figure 1) in Stimulus Setting panel. Set the region of interest (ROI; red; ROI #1B) as 40 x 40 pixels (7.6 µm2). Prepare a reference region (REF; green; ROI #2), and a background (BG; blue; ROI #3) using "copy ROI" and "paste ROI" (right-click button on the mouse).
NOTE: Pixel size can be set through "ROI manager" which can be called by clicking the right button of the mouse when the cursor is on the ROI. Larger ROIs are thought to be better since they can standardize the dispersion of the zFRAP score. However, too large of an ROI cannot be used for this analysis because it makes it difficult to avoid the nucleolus precursor body (NPB). eGFP-H2B in this compartment was thought to be free from chromatin and showed remarkable higher mobility3. ROI and REF areas should be separated as much as possible. If these two regions are close, bleaching may also affect the region of REF. - Click "Live" on the tool bar and then start "Live Plot" (Supplemental Figure 1).
NOTE: Live plot indicates fluorescence signal intensity within each ROI and is used for adjusting laser power using HV. Note that if ROI is not selected, no intensity would be detected on Live plot panel. - Determining the ROI position
NOTE: ROI can be moved by dragging into the desired position in pronuclei. (1) Be sure to avoid the nucleolus, and (2) fit the entire ROI into the nucleoplasm. Do not contain the region outside the area of the nuclear membrane (cytoplasm) in ROI. The localization of eGFP-H2B is highly restricted into the nucleoplasm so that it is easy to find whether ROI contains cytoplasm or not by the fluorescence of eGFP-H2B. Male pronuclei tend to be larger than those of females, which are often localized near the 2nd polar body. While using these criteria, judge the male or female pronuclei. - Click "Focus x 2" (Supplemental Figure 1) and then adjust the HV power gage of the channel for EGFP into fluorescence intensity to around 2000 (fluorescence intensity can be seen in the "live plot" window).
- Use 60X oil lens (60X 60X/1.35 oil).
- Click "Time" and then "XY" (Supplemental Figure 1) to start the zFRAP analysis.
NOTE: If "Time" button is suitably selected, "t" appears on the "XY" button.- Click "Series Done" (Supplemental Figure 1), which appears after FRAP imaging near the "XY button" and then obtain FRAP-analyzed data.
- Save the file "2D View-Image XXXXX" as a preferred named file (oib. file format) by "save as" (Ctrl + Shift + S can be also used).
- Select ROI, REF, and BG and then click (Ctrl+A can be also used) "Series Analysis" in "2D View Image" window.
- Obtain row FRAP data and then click "save" button in the live plot window.
NOTE: Row FRAP quantitative data is saved as a csv file.
- For no zFRAP control, put some of the zygotes injected with mRNA on the drop, which is near to the rim of the glass bottom dishes to avoid the effect from fluorescence observation.
7. Data Calculation
- While using the obtained quantitative data, make the graph of the "recovery curve" and "mobile fraction" by Excel as shown in the references3,7 with some modifications (see below and supplemental excel file).
- Calculate the relative intensity by subtracting the value of BG from those of ROI and REF in 12 pictures for each time point.
- Divide the value of ROI by that of the REF. As a result, 12 standardized relative intensity scores at each time point can be obtained.
- Calculate the average of the relative score for ROI in 3 images taken before photo bleaching.
- Calculate the recovery rate. Divide the 12 obtained relative scores for ROI in the images taken at each time point (obtained at 7.1.2.) by the average of the relative score before photo bleaching (obtained at 7.1.3.). Draw recovery curve with using these scores (Figure 2E).
- Calculate the bleaching rate.
NOTE: Bleaching rate (%) = 100 – recovery rate of 4th image. - Calculate the mobile fraction (MF) by using the formula as follows10,11,12
MF (%) = 12th recovery rate (at the end point) – 4th recovery rate/ bleaching rate × 100.
8. Embryo Transfer
- Mate matured ICR females at 8 – 12 months with vasectomized male ICR mice the night before the embryo transfer by letting them be in the same cage overnight.
- Collect the embryos that were zFRAP-analyzed and determined to develop to the two-cell stage.
- Intraperitoneally inject female mice with 0.7 – 0.9 mL 1.2% tribromoethanol anesthesia or with 0.35 – 0.45 mL of 0.75/4/5mg/kg medetomidine/midazolam/butorphanol mixed agents prior to embryo transfer.
NOTE: The volume of anesthesia is determined by the body weight of mice. Tribromoethanol: 0.2 mL/10 g of body weight; medetomidine/midazolam/butorphanol mixed agents: 0.1 mL/ 10 g of body weight.- Transfer these two-cell stage embryos into the oviducts of pseudopregnant female ICR mice with a vaginal plug at 0.5 days post coitum (dpc).
- After the embryo transfer, put the female mice on the heater set at 37 °C until they wake up.
- At 18.5 dpc, sacrifice the surrogate mother by cervical dislocation and recover the pups derived from zFRAP-analyzed zygotes by cesarean section. Some of the recovered pups were delivered to the foster mother and their body weights were assessed each week.
Representative Results
zFRAP analysis with eGFP-H2B
Properly produced mRNA encoding eGFP-H2B, which is seen as a shifted band caused by poly A tailing (Figure 2A), was injected into the cytoplasm of zygotes with a 2nd polar body at 1 – 3 h after insemination (Figure 2B). Eight to 12 h after insemination, zygotes with 2 pronuclei showing the expression of eGFP-H2B were collected and subjected to zFRAP analysis (Figure 2C). The FRAP analysis consists of bleaching and recovery steps. At the pre-bleaching phase, the signal of eGFP-H2B could be observed (Figure 2D-1), but once bleached, the signal declined to a negligible level (Figure 2D-2). If bleaching was successful, a dark hole as large as the ROI appeared soon after bleaching. After bleaching, the intensity of the eGFP-H2B fluorescence signal recovered slightly; the fluorescent signal gradually increased because of the inflow of the unbleached fraction of eGFP-H2B into the ROI from the unbleached area in the nucleoplasm (Figure 2D-3). For confirmation of the success of zFRAP analysis of chromatin looseness, it is recommended to use parental asymmetry of chromatin looseness; male pronuclei showed a faster recovery curve and more mobile fractions than those of females (Figure 2E and F). After zFRAP analysis, washed and cultured zygotes in CZB media, could develop to the blastocyst stage without significant damage (Figure 2G and H). Even if strongly bleached for a longer time, the zygotes could still develop into the blastocyst stage as well7.
Analysis of the full-term development of zFRAP-analyzed zygotes
To analyze the full-term development of embryos derived from zFRAP-analyzed zygotes, the embryos at the two-cell stage were transferred into the oviducts of pseudopregnant mothers. As controls, intact embryos, and one injected with mRNA but not zFRAP-analyzed were prepared. The birth rate of zFRAP-analyzed embryos (41.0%) was slightly but significantly lower than that of intact embryos (62.2%), but was the same as that of the no zFRAP control (52.6%) (Table 2). Thus, zFRAP-analysis seems to be slightly detrimental to full-term embryonic development. However, importantly, the pups derived from zFRAP-analyzed zygotes seemed healthy and fully developed (Figure 3).
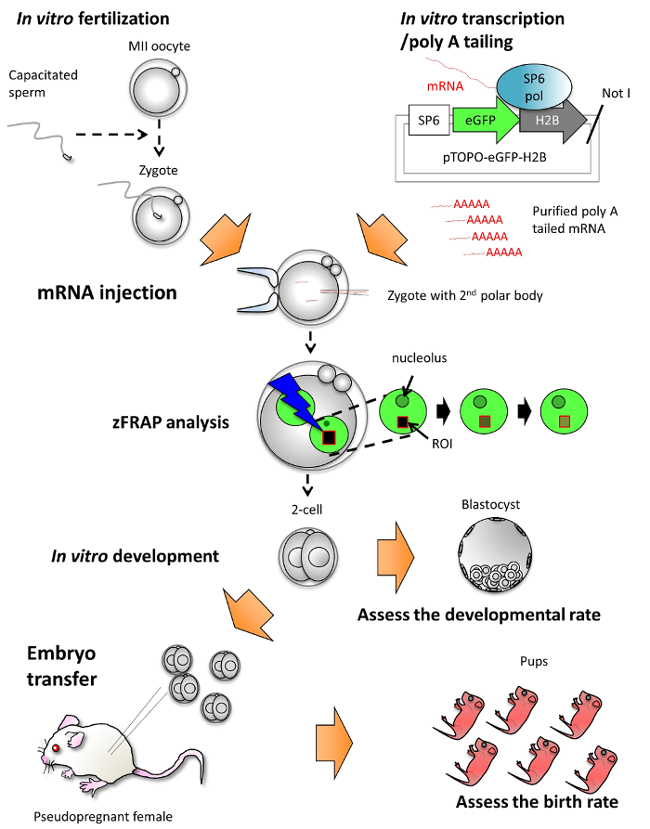
Figure 1: A schematic illustration of the flow of procedures for the experiments. In vitro fertilization: Freshly collected metaphase II (MII) oocytes were inseminated with capacitated sperm. In vitro transcription: Messenger RNA (mRNA) encoding eGFP-fused histone H2B (eGFP-H2B) were transcribed from the SP6 promoter of the linearized pTOPO-eGFP-H2B3 with Not I. The eGFP-H2B mRNA subjected poly A tailing and then purification. The purified eGFP-H2B mRNA is used in mRNA injection. mRNA injection: The prepared eGFP-H2B mRNA is injected into the cytoplasm of the zygotes with 2nd polar body. zFRAP analysis: The eGFP-H2B-expressing zygotes were subjected to zFRAP analysis. The region of interests (ROI; red rectangle) was bleached with a strong laser and then the eGFP-H2B signal declined to a negligible level. After bleaching, the intensity of the eGFP-H2B signal gradually increased. In vitro development: After zFRAP analysis, the zygotes could develop into the blastocyst stage. Embryo transfer: At two-cell stage, the zFRAP-analyzed embryos were transferred into the oviducts of pseudopregnant female mice. 18 days later, healthy live pups could be obtained from zFRAP-analyzed embryos. Please click here to view a larger version of this figure.
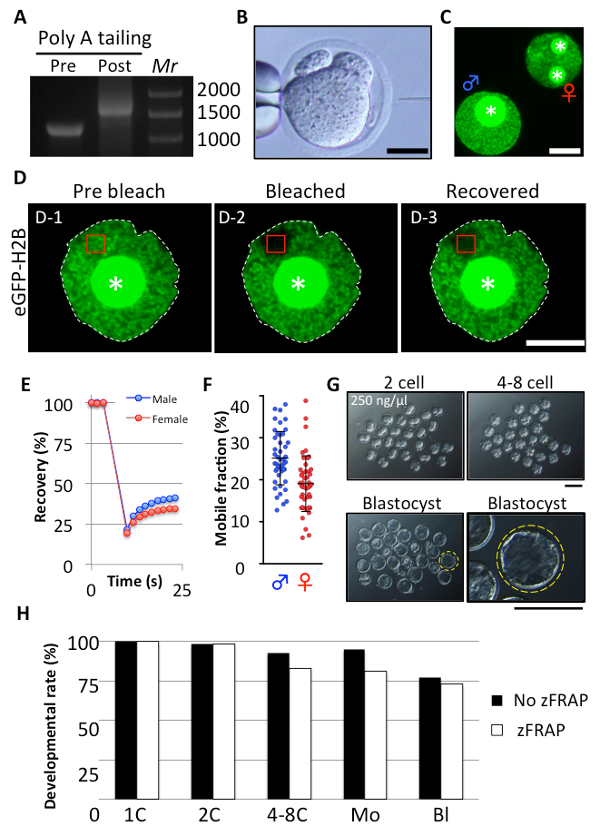
Figure 2: zFRAP analysis of zygotes with eGFP-H2B. (A) Representative image of properly prepared mRNA by in vitro transcription and poly A tailing. Lane 1: pre-poly A tailing; lane 2: post poly A tailing. (B) mRNA encoding eGFP-H2B was injected into the cytoplasm of zygotes with a 2nd polar body 1 – 3 h post insemination (hpi). Scale bar = 25 µm (C) eGFP-H2B-expressing zygotes were collected at 8 – 12 hpi. White asterisks show nucleolus precursor bodies (NPB). Scale bar = 10 µm (D) eGFP-H2B expression was detected in the entire nucleoplasm even in NPB at the pre-bleaching phase (D-1), soon after bleaching (D-2), and after recovery (D-3). The nuclear membranes (white dotted lines) and NPB (asterisks) are indicated. Scale bar = 10 µm. (E, F) Recovery curve and mobile fractions were obtained from 45 zygotes in 5 independent experiments. Blue and red indicate male and female, respectively. In recovery curves, single circles indicate the measuring point. In scatter plots, single dots indicate the score of mobile fractions obtained from pronuclei. (G) Representative images of the preimplantation development of zFRAP-analyzed zygotes are shown. Two-cell, 4-8 cell, and blastocyst stage embryos at 24, 48, and 96 hpi, respectively. The yellow circle indicates the well-developed blastocyst. This blastocyst is enlarged and shown on the right panel. Scale bar = 100 µm. (H) Bar graph of developmental rates of zFRAP-analyzed embryos. Bars indicate zFRAP-analyzed (zFRAP) embryos (white) and control embryos (black) injected with mRNA but no zFRAP analysis (No zFRAP). Data shown are from three independent experiments, examining at least 57 embryos in total. Two-cell, 4-8 cell, morula (Mo), and blastocyst (Bl) stages were observed at 24, 48, 72, and 96 hpi, respectively. This figure has been modified from [Ooga et al 2017]7. Please click here to view a larger version of this figure.
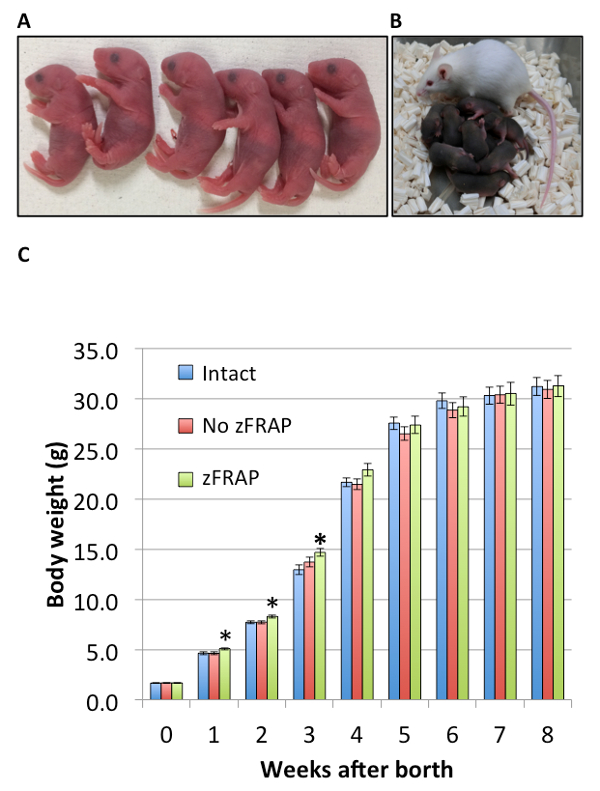
Figure 3: The healthy growth of pups derived from the FRAP-analyzed zygotes. (A) The pictures of pups derived from zFRAP-analyzed zygotes are shown. (B) Normal growth was observed during the nursing of the zFRAP-analyzed pups. (C) The graph indicates the weight of pups derived from intact embryos (blue), unbleached control embryos (red), and zFRAP-analyzed embryos (green). Body weights of 29 pups derived from intact embryos, 24 from no-zFRAP embryos, and 15 from zFRAP-analyzed embryos over an 8-week period. Values indicated by asterisks are significantly different from the intact control. This figure has been modified from [Ooga et al 2017]7. Please click here to view a larger version of this figure.
Supplemental Figure 1: Graphical user interface for FRAP analysis. (A) An overview of the graphical user interface was shown. (B) Enlarged images for each window were shown. (1) The Acquisition Setting window is used for setting the image acquisition condition. (2) The Stimulus Setting window is used for setting the bleaching condition. This window can be opened by clicking the button on the Image acquisition window. (3) The Image Acquisition Control window is used for the FRAP analysis. (4) The Live View shows the present image (the present image appears after clicking Focus x2 or XY button). The red, green, and light blue rectangles show the ROI (region of interest), REF (reference), and BG (background), respectively. (5) The 2D View shows the FRAP-analyzed image and appears after clicking the "Series Done" button. In this case, a FRAP-analyzed male pronucleus is shown as an example. Three rectangles with 8 dots indicate that these regions are selected. (6) The Live Plot shows the present fluorescence intensity at each region. The X and Y axes indicate time (ms) and fluorescence intensity, respectively. (7) The Series Analysis shows the tracks of fluorescence intensity at each region and appears after clicking Series Analysis button on the 2D View window. Please click here to download this file.
Supplemental Excel File 1: Example data and data calculation. (Sheet 1) An example data for a male pronucleus is shown. No. indicates the consecutive number of the image. The row intensities for each region (ROI, Ref, and BG) were indicated. (Sheet 2) An example for data calculation is shown. Protocol number indicates the corresponding places in the protocol section. Notes explain the meaning of each score. The numerical formula can be referred by clicking the cells. Examples of recovery curve and mobile fraction bar graph are shown. Please click here to download this file.
| Day -3 | Day -2 | Day -1 | Day of FRAP | Day +1 | Day +3 | Day +19 | |
| mRNA preparation | Cutting template plasmid (over night) |
1). Purification template plasmid | |||||
| 2). In vitro transcription | |||||||
| 3). In vitro poly A tailing | |||||||
| 4). mRNA quality check by electrophoresis | |||||||
| FRAP | 1). Preparation of manipulation pipet and chamber | ||||||
| 2). mRNA injection | |||||||
| 3). zFRAP analysis | |||||||
| 4). Calculation of recovery curve and mobile fraction*2 | |||||||
| IVF and analysis of pre-implantation development | eCG injection | 1). hCG injection | 1). Sperm capacitation | ||||
| 2). Pre-incubation of media (HTF) | 2). In vitro fertilization | ||||||
| Preparation of the media (HTF, CZB, H-CZB and PVP-CZB) | 3). Evaluation preimplantation development | ||||||
| Embryo transfer | Preparation of pseudopregnant female (mating with vasectomized male*1) |
Transfer of 2-cell stage embryos to pseudopregnant female | Evaluation of birth rate | ||||
| *1: Vasectomized male mouse should be prepared at least 2 weeks before mating | |||||||
| *2: Calculation can be prolonged into the next day (Day +1) | |||||||
Table 1: Time table of the experiments. The day of the FRAP analysis is regarded as day 0.The experiments that should be performed are indicated on the respective days for each item.
| Categories | No. of injected zygotes | No. of recovered zygotes after mRNA injection (%)* | No. of zygotes analyzed | No. of transferred | No. of recipients | No. of pups (%)*** | Weight of pups (g) | No. of transgenic pups |
| 2-cell stage embryos (%)** | ||||||||
| Intact | – | – | 99 | 98 (99.0) | 9 | 61 (62.2)a | 1.64±0.02 | n.d |
| No FRAP | 420 | 398 (94.8) | 82 | 78 (95.1) | 7 | 41 (52.6)a, b | 1.66±0.03 | n.d |
| FRAP | 79 | 78 (98.7) | 7 | 32 (41.0)b | 1.69±0.03 | 0 | ||
| *: calculated by dividing with no. of injected zygotes; **: calculated by dividing with no. of zygotes analyzed; ***: calculated by dividing with no. of transferred 2-cell stage embryos. a,b: superscripts indicate significant difference (P<0.05). n.d: not determined. This table has been modified from [Ooga et al 2017]7. | ||||||||
Table 2: The birth rate of analyzed embryos: The developmental potential of the embryos, which had been FRAP-analyzed, to full-term was examined. The obtained birth rate of these embryos is shown. As controls, intact and no FRAP-analyzed embryos were also examined. The weights of pups derived from these transferred embryos are shown as well.
Discussion
As revealed in this study, the zFRAP analysis does not cause critical damage to full-term development, suggesting this method is a very useful tool to reveal the association between molecular events and the embryonic developmental potential. During reprogramming, into the two-cell like state of ES cells, by which the differential potential of those cells becomes as high as that of two-cell stage embryos, chromatin looseness changes into that comparable with two-cell stage embryos5. Accordingly, it is possible that zygotic chromatin looseness is important for embryonic developmental potential. Somatic cell nuclei transferred into the cytoplasm of oocytes showed lower chromatin looseness compared with not only paternal pronuclei but also maternal pronuclei, suggesting the lack of acquisition open chromatin in somatic cell nuclear transfer zygotes is involved in their poor developmental potential. Collectively, the zFRAP analysis seems to be useful for elucidating genome reprogramming mechanisms.
The zFRAP system may make it possible to select zygotes with high developmental potential. In the preliminary study, in addition to SCNT, it was revealed that round spermatid injection (ROSI)-derived zygotes harbor abnormal chromatin looseness. Besides, it was found that some of them showed chromatin looseness at the level comparable to IVF-derived zygotes as well, suggesting that these zygotes have high developmental potential. Importantly, zygotes of which chromatin looseness was evaluated by zFRAP could develop to term. In the future, therefore, it may be possible to distinguish the SCNT- and ROSI-derived zygotes with higher developmental potential from lower ones by the evaluation of chromatin looseness by zFRAP.
To date, previous studies have shown the utility of live imaging method for the analysis of epigenetic state in preimplantation embryos. Some of the live imaging methods can reveal each epigenetic modification (e.g. DNA/histone modification) in preimplantation embryos13,14, but using zFRAP, it is possible to evaluate the zygotic chromatin looseness without killing the cells. Chromatin looseness seems to be affected by epigenetic modifications. For example, heterochromatin in which repressive epigenetic modification such as H3K9me3 and H3K27me3 are enriched showed lower histone mobility than euchromatic region3,15. Indeed, treatment with a chemical compound, which converts epigenetic state, caused the alteration of chromatin looseness in zygotes. Therefore, using zFRAP, it is possible to reveal the summative epigenetic state of chromatin structure. It was thought that this would be an advantage for evaluating the developmental potential of the zygotes.
The injection of mRNA encoding eGFP-H2B for zFRAP has a demerit but increases the versatility of zFRAP. The demerit of the mRNA injection is the damage caused by microinjection. To avoid this, it is very effective to utilize the eGFP-H2B transgenic mouse. If eGFP-H2B transgenic mouse line is used, the offspring rate may be increased compared to the case of using mRNA microinjection. Conversely, it is the merit of mRNA injection that zFRAP allows the use of any kind of strain of wild-type mice. The researchers who want to use zFRAP with their interest of mouse strain do not need to prepare the desired mouse strain with eGFP-H2B transgene. This merit becomes more prominent in the analysis of transgenic (e.g. overexpression/knockdown) or knockout mouse line. The researchers who want to utilize zFRAP for their research topics with the transgenic/knockout line do not have to prepare the eGFP-H2B transgenic line from a limited number of their interest ones. This indicates an advantage for the progression of research.
Live imaging analysis with preimplantation embryos requires specialized knowledge and very expensive and finely tuned specialized devices. Therefore, an introduction of such an experimental system is not an easy decision. The experimental system that is established in this study needs only a thermo-heater aside from a confocal laser scanning microscope. In addition, learning the method is easy so that a beginner in a laboratory can learn the method of zFRAP within 1 month. However, there are some issues that still need consideration. First is focus drift. During observation, it is possible that the pronuclei or the zygotes deviate from the position where they presented before observation starts. If the focus drift occurs, ROIs also deviate from the correct position. As a result, quantitative data becomes worthless. To avoid this issue, the researcher has to use the lid of the glass bottom dishes for protection against the window from the air conditioner and wait for the oil expansion over the objective lens. If focus drift occur, data from the deviating zygotes should be discarded and a 2nd FRAP analysis with the same zygotes is not recommended. In this study, shortening the observation time from 150 s3 to about 25 s7 was very helpful. Second is longer incubation outside the incubator. Since longer exposure to the environment outside the incubator is definitely harmful for the embryonic development, it was recommended to finish the FRAP-analysis within 1 h per batch. The number of the zygotes FRAP-analyzed in a HEPES-buffered media on the glass bottom dishes should be 6 – 10. In this experimental condition, the maximum number is around 30 for 4 h but if more zygotes are needed, 20 zygotes per h can be analyzed by postponing the assessment of fluorescent intensity in the reference and background region. Third is "time-related deterioration of the laser of the confocal laser microscopy". If the bleaching laser is insufficient, the correct quantitative data cannot be obtained because of insufficient bleaching. Indeed, the parental asymmetry of zygotic chromatin looseness cannot be acquired with a weak bleaching laser. To avoid this, it is recommended to check whether the laser power has weakened as the occasion arises. In this study, 110-µW bleaching was enough for the acquisition of parental asymmetry.
zFRAP analysis can be used for other kinds of proteins, in addition to core histones. Since core histones show prominently low mobility, longer total observation time (about 25 s in this study) is needed in the zFRAP analysis. Therefore, to analyze a considerable number of zygotes, culturing in HEPES-buffered media on the heater for a long time is unavoidable. On the other hand, for the zFRAP analysis of high mobility proteins, the total observation time can be set short, enabling the time of culturing in HEPES-buffered media on the heater to be short. Therefore, since longer exposure to the environment outside the incubator is highly detrimental for the embryonic development, zFRAP analysis for proteins other than core histones, which have high mobility by nature, seem to show lower toxicity than core histones. It is hoped that this experimental system will help further the investigation of the relationships between various molecular events and embryonic development.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
We thank Satoshi Kishigami, Sayaka Wakayama, Hiroaki Nagatomo, Satoshi Kamimura, and Kana Kishida for providing critical comments and technical support. This work was partially funded by the Ministry of Education, Culture, Sports, Science and Technology program for promoting the reform of national universities to M.O.; the Japan Society for the Promotion of Science (16H02593), Asada Science Foundation, and the Takeda Science Foundation to T.W. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Materials
| Confocal laser scaning microscope | Olympus | FV1200 | FV1000 can be also used for FRAP analysis (reference #7) |
| Thermo plate | Tokai Hit | TP-110RH26 | |
| Inverted microscope | Olympus | IX71 | |
| Micro manupilator | Narishige | MMO-202ND | |
| Pieze Micro Micromanipulator | Prime tech | PMAS-CT150 | |
| 35 mm culture dish | Falcom | 351008 | 35 x 100 mm style; for IVF |
| 60 mm culture dish | Falcom | 351007 | 60 x 15 mm style; for embryo culture |
| 50 mm culture dish | Falcom | 351006 | 50 x 9 mm style; for manipulation on the stage |
| 50 mm glass bottom dish | Matsunami | D910400 | 50 mm dish, 27 mm φ hole size; for FRAP analysis |
| Mineral oil | Organic spceiality chemicals | 625071 | For FRAP analysis on the glass bottom dishes |
| Mineral oil | Sigma | M8410-1L | For IVF and embryo culture |
| glass capillary | Drummond | 1-000-1000 | For handling mouse zygotes |
| Borosilicate glass | Prime tech | B100-75-10-PT | For microinjection of mRNA |
| Micropipett puller | Sutter instrument | P-97/IVF | For preparation of the injection or holding neadle (out side diameter: 80 µm, inner diameter: 10 µm) |
| Microforge | Narishige | MF-900 | |
| mMESSAGE MACHINE SP6 Transcription kit | Thermo Fisher scientific |
AM1340 | |
| Poly (A) tailing kit | Thermo Fisher scientific |
AM1350 | |
| PVP solution 10% (PVP-HTF) | IrvineSceientific | 99311 | HEPES buffered HTF containing 10% PVP |
| Sodium HEPES | Sigma | H3784 | |
| pTOPO eGFP-H2B | Template plasmid for eGFP-H2B mRNA (reference #6) | ||
| NorthernMax Gly Sample loading Dye | Thermo Fisher scientific |
AM8551 | For electrophoresis of in vitro transcribed mRNA |
| Phenol chloroform isamyl alcohol | nacalai tesque | 25970-14 | |
| Chloroform | nacalai tesque | 08401-65 | |
| Not I | TOYOBO | NOT-111X | |
| Ethachinmate | WAKO | 318-01793 | |
| 3664 Otical power meter | Hioki | 3664 | Power meter for laser power |
| Stereomicmicroscope | Olympus | SZX16 |
Referenzen
- Burton, A., Torres-Padilla, M. E. Epigenetic reprogramming and development: a unique heterochromatin organization in the preimplantation mouse embryo. Brief Funct Genomics. 9, 444-454 (2010).
- Okada, Y., Yamaguchi, K. Epigenetic modifications and reprogramming in paternal pronucleus: sperm, preimplantation embryo, and beyond. Cell Mol Life Sci. 74, 1957-1967 (2017).
- Ooga, M., Fulka, H., Hashimoto, S., Suzuki, M. G., Aoki, F. Analysis of chromatin structure in mouse preimplantation embryos by fluorescent recovery after photobleaching. Epigenetics. 11, 85-94 (2016).
- Meshorer, E., et al. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 10, 105-116 (2006).
- Boskovic, A., et al. Higher chromatin mobility supports totipotency and precedes pluripotency in vivo. Genes Dev. 28, 1042-1047 (2014).
- Yamagata, K., Ueda, J. Long-term live-cell imaging of mammalian preimplantation development and derivation process of pluripotent stem cells from the embryos. Dev Growth Differ. 55, 378-389 (2013).
- Ooga, M., Wakayama, T. FRAP analysis of chromatin looseness in mouse zygotes that allows full-term development. PLoS One. 12, e0178255 (2017).
- Quinn, P., Begley, A. Effect of human seminal plasma and mouse accessory gland extracts on mouse fertilization in vitro. Australian journal of biological sciences. 37, 147-152 (1984).
- Chatot, C. L., Lewis, J. L., Torres, I., Ziomek, C. A. Development of 1-cell embryos from different strains of mice in CZB medium. Biology of reproduction. 42, 432-440 (1990).
- Bae, J., Sung, B. H., Cho, I. H., Song, W. K. F-actin-dependent regulation of NESH dynamics in rat hippocampal neurons. PLoS One. 7, e34514 (2012).
- Dieteren, C. E., et al. Defective mitochondrial translation differently affects the live cell dynamics of complex I subunits. Biochim Biophys Acta. 1807, 1624-1633 (2011).
- Subramanian, V., et al. H2A.Z acidic patch couples chromatin dynamics to regulation of gene expression programs during ESC differentiation. PLoS Genet. 9, e1003725 (2013).
- Hayashi-Takanaka, Y., et al. Tracking epigenetic histone modifications in single cells using Fab-based live endogenous modification labeling. Nucleic Acids Res. 39, 6475-6488 (2011).
- Yamazaki, T., Yamagata, K., Baba, T. Time-lapse and retrospective analysis of DNA methylation in mouse preimplantation embryos by live cell imaging. Dev Biol. 304, 409-419 (2007).
- Puschendorf, M., et al. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat Genet. 40, 411-420 (2008).

