Specific and Accurate Detection of the Citrus Greening Pathogen Candidatus liberibacter spp. Using Conventional PCR on Citrus Leaf Tissue Samples
Summary
Citrus Greening is a particularly destructive disease affecting citrus crops globally. Presented here is a simple method using PCR and genomic DNA extraction of citrus leaf tissue for the accurate and precise identification of the citrus greening pathogen, Candidatus liberibacter spp.
Abstract
Citrus greening, also known as huanglongbing, is a destructive citrus disease ravaging citrus farms globally. This disease causes asymmetrical yellow leaf mottling, vein yellowing, defoliation, root decay, and ultimately, the death of the citrus plant. When infected, the citrus plants have stunted growth and produce flowers out of season. These flowers rarely yield fruit, and those that do yield small, bitter, irregularly shaped citrus fruit that are not desirable. This disease is spread by the Asian citrus psyllid, Diaphorina citri, and by the grafting of infected citrus tissue. The pathogen has a long and variable incubation period within the citrus plant—sometimes years, before symptoms appear. Attempts to culture this pathogen in vitro have been unsuccessful, possibly due to the low and uneven concentration of the pathogen within infected citrus tissue, or because it is difficult to replicate the environmental conditions conducive to growth of the pathogen. It is very difficult to identify the disease before it has spread, due to its long incubation period and researchers' inability to culture the pathogen. As a result, the disease only becomes apparent after suddenly destroying a citrus farmer's entire yield. Presented here is a method for the accurate and specific detection of the citrus greening pathogen, Candidatus liberibacter spp. using a genomic DNA extraction kit and PCR. This method is simple, efficient, cost effective, and adaptable for quantitative analysis. This method can be adapted for use on any citrus tissue; however, it is potentially limited by the amount of pathogen present in the tissue. Nevertheless, this method will allow citrus farmers to identify infected citrus plants earlier, and curb the spread of this destructive disease before it can further spread.
Introduction
Citrus greening, also known as huanglongbing, has caused a significant loss of citrus trees. One representative case is Florida, where the disease is forecasted to cause a 70% reduction in the production of Florida orange boxes from 244 million boxes in the 1997 – 1998 season to 70 million boxes as of the 2016 – 2017 season1. Over 90% of citrus trees in Florida are infected2, and it is estimated that the Florida citrus industry loses about one billion dollars each year due to citrus diseases, wherein citrus greening plays a major role3. Citrus greening is spread primarily by the invasive Asian citrus psyllid Diaphorina citri, but can also be spread by grafting of infected tissue. The disease causes yellowing of veins, asymmetrical yellow leaf mottling, premature defoliation, twig dieback, root decay, and death of the plant. Importantly, the disease causes the citrus plant to produce flowers out of season, which rarely produce fruit. The fruit that is produced by infected citrus plants are immature, green, and bitter tasting4.
The purpose of this method is to accurately and precisely identify the motile bacterium Candidatus liberibacter spp., the causative agent of citrus greening, living within the phloem of infected citrus trees5. Genomic DNA is extracted from whole leaf tissue, which contains the live bacteria. This extracted genomic DNA is used as a template for conventional PCR, where oligonucleotides complementary to the bacterium's 16S rDNA sequence are used to amplify this sequence. Oligonucleotides complementary to a citrus FBOX gene are used as an internal amplification control. We chose to use this method, because it has been proven to be successful in previous research6.
This method has the clear advantage of being simple, relatively inexpensive, and able to be performed in any normally equipped biochemistry lab. In addition, PCR remains the most accurate and precise method for detecting this pathogen, due to the difficulty of culturing this pathogen7, and the pathogen's ability to survive and reproduce for years within an asymptomatic host8. Many different specialty PCR assays have been used to successfully detect this pathogen; however, conventional PCR remains the simplest assay for accurate and specific detection, especially within asymptomatic hosts8.
Protocol
1. Isolate Genomic DNA from Plant Tissue Using a Genomic Extraction Kit
- Obtain citrus leaf tissue by cutting a whole fresh leaf from a citrus tree using clean scissors, and place the leaf in a clean plastic sandwich bag.
NOTE: The bag containing the leaf tissue should be placed in a 4 °C refrigerator or on ice in an insulated container as soon as possible after collection to avoid spoilage. - Add a small amount of liquid nitrogen to chill a mortar and pestle. While they are chilling, use scissors to cut out a small piece of leaf tissue, approximately 1 square inch in size. Place the cut tissue in the mortar to instantly freeze it. Add more liquid nitrogen, if needed.
NOTE: Insulated gloves should always be worn while handling liquid nitrogen. - Quickly grind the plant tissue into a fine powder using the mortar. Continue grinding until the liquid nitrogen completely evaporates, and a fine green powder remains.
- Briefly chill a metal spatula in liquid nitrogen for 10 - 15 s, then scoop the ground tissue inside the mortar into a 1.5 mL microcentrifuge tube, using the spatula.
- Add 600 µL of Nuclei Lysis Solution (see Table of Materials) using a 1000 µL pipette and vortex the sample for 1 – 3 s, then incubate in a 65 °C water bath for 15 min.
- Add 3 µL of RNase Solution to the lysate using a 10µL pipette, invert the tube 2 – 5 times to mix it, and incubate at 37 °C in a cabinet incubator for 15 min.
NOTE: Allow the mixture to cool to room temperature before proceeding. - Add 200 µL of Protein Precipitation Solution, vortex at high speed for 20 s, then centrifuge for 3 min at 13,000 x g to form a firm pellet. While the sample is being centrifuged, add 600 µL of room temperature isopropanol to a fresh 1.5 mL microcentrifuge tube.
- Using a 1000 µL pipette, carefully remove the supernatant from the centrifuged sample, transferring it to the fresh 1.5 mL microcentrifuge tube containing isopropanol. The pellet may now be discarded.
NOTE: Avoid contaminating the supernatant with protein by leaving a miniscule amount of supernatant above the pellet. - Invert the tube until the DNA becomes visible as a mass of thread-like strands, then centrifuge at 13,000 x g for 1 min at room temperature.
- Decant the supernatant, add 600 µL of 70% ethanol to the pellet, invert the tube several times to wash the DNA, and centrifuge at 13,000 x g for 1 min.
- Carefully aspirate the ethanol, leaving the loose DNA pellet, then open and invert the tube, resting on absorbent paper. Air dry at room temperature for 15 min.
- Add 100 µL of DNA Rehydration Solution to the dried DNA pellet, then incubate the DNA in a 65 °C water bath for 1 h, while mixing periodically by tapping the tube.
- Place 1 µL of purified water on a microcuvette, and insert it into a spectrophotometer to be used as a blank. Calculate the concentration of DNA present by placing 1 µL of sample on the microcuvette, and inserting into the spectrophotometer. DNA concentration (ng/µL) can be calculated as (A260– A320) × dilution factor × 50 ng/µL.
NOTE: The protocol can be paused here. The sample can be stored at 4 °C.
2. Perform PCR on Genomic DNA Samples
- Combine 10 µL 2x PCR Master Mix (see Table of Materials), 1 µL forward primer (10 pM/µL), 1 µL reverse primer (10 pM/µL) (see Table 2 for primer sequences), 100 ng of genomic DNA extracted from the sample, and purified water (up to 20 µL final volume) in a 50 µL PCR tube, flick to mix, and briefly centrifuge.
- Insert the PCR tube into the thermocycler and run accordingly (See Table 1).
NOTE: The protocol can be paused here. If necessary, store sample at 4 °C after the reaction is complete.
3. Electrophorese the Amplified DNA in an Agarose Gel
- Weigh 0.4 g agarose powder and add it along with 1x TAE buffer to a clean 200 mL Erlenmeyer flask, filling up to 50 mL.
- Microwave on high for 1.5 min, shaking every 30 s, until agarose is fully dissolved.
NOTE: Insulated gloves should be worn while handling liquid agarose to prevent burns. - Add 2.5 µL of 10 mg/mL ethidium bromide to the liquid agarose and shake to mix, then pour the liquid agarose gel into a level gel mold. Quickly insert standard 12-tooth gel combs after pouring.
- Allow the gel to cool for 20 min at room temperature, then remove gel combs, and place the gel into a level gel box filled with 1x TAE buffer.
- Using a micropipette, load the total amplified DNA samples into wells. Add 5 µL of DNA ladder (see Table of Materials) in a separate well from your samples.
- Electrophorese the samples for 35 min at a constant 90 V, then remove the gel and place in a UV transilluminator.
- Photograph and analyze the gel banding pattern while using UV transillumination at 302 nm.
Representative Results
A positive result yields a distinct band corresponding to 500 bp for Candidatus Liberibacter asiaticus Las606/Lss6, and/or a distinct band corresponding to 700 bp for Laa2/Laj5, an insert in the 16S ribosomal β operon9. The internal amplification control band must also appear at 400 bp. This band corresponds to amplification of the citrus FBOX gene10, and must appear for a result to be considered valid (Figure 1 and Figure 2). Lack of both 500 bp and 700 bp bands while a 400 bp band is present represents a negative result. Performing PCR on healthy samples may yield some non-specific banding patterns around 700 bp and 400 bp; however, only a single bold band corresponding to these molecular weights indicates a positive result (Figure 1).
| Operation | Temp. | Time | Cycles |
| Initial Denaturation | 95 °C | 3 – 5 min | 1 |
| Denaturation | 95 °C | 30 sec | 35 |
| Annealing | 55 °C | 30 sec | |
| Elongation | 72 °C | 40 sec | |
| Final elongation | 72 °C | 10 min | 1 |
Table 1. Thermocycler settings used to amplify target 16S rDNA genes and citrus FBOX.
| Name of Primer | Sequence | Reference | |
| LAA2_F | 5'-TAT AAA GGT TGA CCT TTC GAG TTT-3' | Hocquellet, et al., 1999 | |
| LAJ5_R | 5'-ACA AAA GCA GAA ATA GCA CGA ACA A-3' | Hocquellet, et al., 1999 | |
| FBOX_F | 5'-TTG GAA ACT CTT TCG CCA CT-3' | This assay | |
| FBOX_R | 5'-AGC AGA CCT GGC TAT TAT ACG ACT G-3' | This assay | |
| LSS_F | 5'-GGA GAG GTG AGT GGA ATT CCG A-3' | Fujikawa, et al., 2012 | |
| LSS_R | 5'-ACC CAA CAT CTA GGT AAA AAC C-3' | Fujikawa, et al., 2012 | |
Table 2. Name of primers, their DNA sequence, and reference.
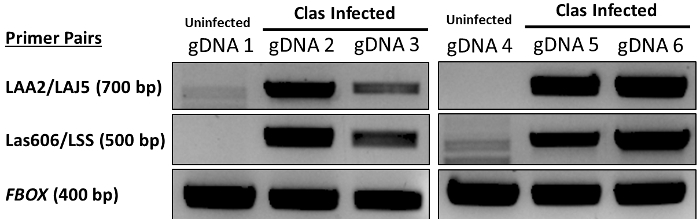
Figure 1. Analysis of conventional PCR results from four infected citrus leaf tissue samples and two heathy samples used as negative controls. 'gDNA' indicates a genomic DNA sample extracted from citrus leaf tissue. Samples gDNA 1 and gDNA 4 are from uninfected citrus leaf samples; gDNA 2, 3, 5, and 6 are Candidatus liberibacter asiaticum (Clas) infected samples. DNA was extracted as written in the protocol. PCR was performed using the extracted genomic DNA as a template, per the protocol (See Table 1). LAA2/LAJ5, LAS606/LSS, and FBOX refer to the primer sets used (See Table 2). Please click here to view a larger version of this figure.
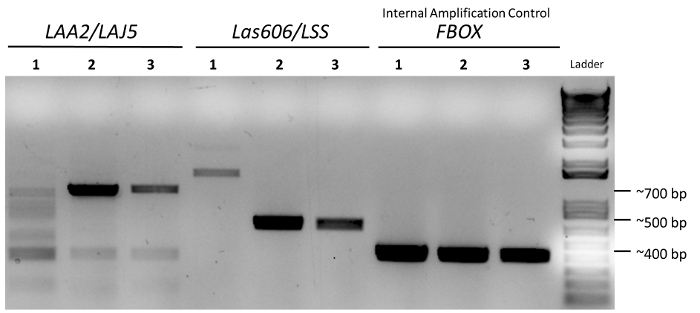
Figure 2. Complete gel photograph of three amplified gDNA samples. Sample 1 is healthy citrus tissue; samples 2 and 3 are Clas infected tissue samples. These samples correspond to gDNA 1, 2, and 3, as seen in Figure 1. Please click here to view a larger version of this figure.
Discussion
It is critical that all steps are followed exactly to achieve optimum experimental conditions. Whole leaf tissue was used during this demonstration; however, the protocol can be modified to theoretically include any type of plant tissue. Previously, researchers have extracted genomic DNA from leaf vein tissue, rather than whole leaf tissue, and achieved similar results11. If the band corresponding to the citrus FBOX gene fails to appear, the result is invalid, and there may be one or more of several technical errors in the operating procedure. Common errors include: using contaminated or improperly sterilized water, micropipette tips, or microcentrifuge tubes; failing to add one or more components necessary in the PCR mix or during genomic DNA extraction; failing to maintain the proper thermocycler conditions; using reagents or other ingredients that are past their expiration date; failing to add ethidium bromide or another DNA stain to the agarose gel; electrophoresing the amplified DNA at too high a voltage or for too long; and using old or improperly mixed TAE buffer, or simply using water instead of TAE buffer to fill the gel box.
Primers LAA2_F and LAJ5_R were chosen from previous research due to their success at identifying Candidatus Liberibacter asiaticum and africanum. These primers amplify ribosomal subunit genes rplA and rplJ, yielding an amplified 669 bp fragment from C. l. africanum or a 703 bp fragment from C. l. asiaticum9. Primers LSS_F and LSS_R were chosen for this assay from previous research due to their established low false negative rate, accuracy, and specificity for C. l. asiaticum. The LSS primer set amplifies a 500 bp sequence in all three 16S rDNA genes present in C. l. asiaticum6. The FBOX primers were constructed for use as an internal amplification control and amplify a 400 bp sequence found in citrus plants. Previous research shows that FBOX is stably expressed in citrus plants, and would make a great candidate for use as a reference gene when adapting this assay for RT-qPCR10.
This assay can be adapted to be used for any type of citrus tissue, and can be modified for use with additional PCR-based assays, such as RT-qPCR. The limitations of this assay are few, and most are inherent to conventional PCR. These limiting factors include: the presence of ethanol or other inhibitory chemicals in the PCR mixture, the lack of quantitation without performing a more advanced assay, the need for a thermocycler in smaller labs, and the presence of DNA contamination.
While the specificity of this assay has not been directly compared to the more specific RT-qPCR methods, this assay is useful for scientists performing research on Candidatus liberibacter, and farmers seeking to diagnose potentially infected citrus trees in their fields, especially if more specific methods are not available or cost-effective. Indeed, it is conceivable that a single citrus grower or researcher could quickly and cheaply assay several hundred samples in a few days, by using multichannel pipettes combined with several 96-well plates, without purchasing an expensive RT-qPCR thermocycler.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
Conceptualization, H.C. and Z. Q. F.; Investigation, H. C., J. C, and Z. Q. F; Resources, Z. Q. F.; Writing – Original Draft, I. A. P.; Writing – Review & Editing, I. A. P., J. C., H. C., S. L. T., and Z. Q. F.; Visualization, H. C.; Supervision, Z. Q. F.; Funding Acquisition, Z. Q. F and F. Q. L.
Project supported by a South Carolina Improving Teacher Quality Higher Education State Grant titled, Enhancing Middle Grades Science Teachers' Knowledge of Plant Processes, Structures, and Functions Through Engagement in Plant Sciences Research (Plant Science)
Funds awarded by the United Sates Department of Education (CFDA Number 84.367B)
The authors would like to thank Dr. Nian Wang of the University of Florida for providing genomic DNA samples from infected Citrus sinensis Valencia.
Materials
| Wizard Genomic DNA Purification Kit | Promega | A1120 | Source of Nuclei Lysis, RNase, Protein Precipitation, and DNA Rehydration solutions |
| Liquid nitrogen | Air Products | N/A | |
| Mastercycler pro with control panel | Eppendorf | 6321000019 | |
| 1250 W Microwave | Panasonic | N/A | |
| 5424 table top centrifuge | Eppendorf | 05-403-93 | |
| Isotemp 215 digital water bath | Fisher scientific | 15-462-15Q | |
| Metal spatula | Sigma-Aldrich | Z283274 | |
| Mortar and pestle | Sigma-Aldrich | Z247464 | |
| LSE Vortex Mixer | Corning | 6775 | |
| 2-propanol | Sigma-Aldrich | I9516 | |
| Sterile 10 μL tips | TipOne | 1161-3730 | |
| Sterile 200 μL tips | TipOne | 1163-1730 | |
| Sterile 1250 μL tips | TipOne | 1161-1750 | |
| Variable Volume Pipettor Kit | VWR | 75788-460 | |
| Ethidium bromide | Sigma-Aldrich | E1510 | |
| Agarose | IBI Scientific | IB70041 | |
| EDTA | Thermo Fisher Scientific | 17892 | |
| Acetic acid | Fisher Chemical | A38-212 | |
| Tris base | Sigma-Aldrich | 10708976001 | |
| μcuvette | Eppendorf | 6138000018 | |
| Stackable casting tray and combs | Carolina | 213655 | |
| PowerPac Basic Power Supply | Bio-Rad | 1645050 | |
| Mini-Sub Cell GT System | Bio-Rad | 1704487EDU | |
| Biospectrometer basic | Eppendorf | 6135000009 | |
| 0.2 mL PCR tubes | Fisher scientific | AB-0620 | |
| 1.5 mL microcentrifuge tubes | Thermo Fisher Scientific | 3439 | |
| 2X Taq Green PCR Master Mix | Promega | M7122 | |
| Forward Primer | Thermo Fisher Scientific | N/A | |
| Reverse Primer | Thermo Fisher Scientific | N/A | |
| Water, PCR grade | Sigma-Aldrich | 3315953001 | |
| Gel Doc XR+ | Bio-Rad | 1708195 | |
| 1 KB+ DNA ladder | Thermo Fisher Scientific | 10787018 |
Referenzen
- Field Office, F. l. o. r. i. d. a. . Citrus October Forecast Maturity Test Results and Fruit Size. , (2016).
- Singerman, A., Useche, P. Impact of citrus greening on citrus operations in Florida. Food and Resource Economics Department, UF/IFAS Extension. , (2015).
- Spreen, T. H., Hodges, A. . The economic impact of HLB on the florida citrus industry. , (2012).
- UICEP. . Pathology. , (2013).
- Jagoueix, S., Bove, J. M., Garnier, M. The phloem-limited bacterium of greening disease of citrus is a member of the alpha subdivision of the Proteobacteria. Int J Syst Bacteriol. 44 (3), 379-386 (1994).
- Fujikawa, T., Iwanami, T. Sensitive and robust detection of citrus greening (huanglongbing) bacterium "Candidatus Liberibacter asiaticus" by DNA amplification with new 16S rDNA-specific primers. Mol. Cell. Probes. 26 (5), 194-197 (2012).
- Davis, M. J., Mondal, S. N., Chen, H., Rogers, M. E., Brlansky, R. H. Co-cultivation of ‘Candidatus Liberibacter asiaticus’ with actinobacteria from citrus with huanglongbing. Plant Dis. 92 (11), 1547-1550 (2008).
- Department of Agriculture & National Agriculture Statistics Service. . Citrus Fruits 2015 Summary (September 2015). , (2015).
- Hocquellet, A., Toorawa, P., Bové, J. M., Garnier, M. Detection and identification of the two Candidatus Liberobacter species associated with citrus huanglongbing by PCR amplification of ribosomal protein genes of the β operon. Mol. Cell. Probes. 13 (5), 373-379 (1999).
- Mafra, V., et al. Reference genes for accurate transcript normalization in citrus genotypes under different experimental conditions. PLoS One. 7 (2), e31263 (2012).
- Li, W., Hartung, J. S., Levy, L. Quantitative real-time PCR for detection and identification of Candidatus Liberibacter species associated with citrus huanglongbing. J. Microbiol. Meth. 66 (1), 104-115 (2006).

