A 3-dimensional (3D)-printed Template for High Throughput Zebrafish Embryo Arraying
Summary
Here, we present a protocol to design and fabricate a zebrafish embryo arraying template, followed by a detailed procedure on the use of such template for high throughput zebrafish embryo arraying into a 96-well plate.
Abstract
The zebrafish is a globally recognized fresh water organism frequently used in developmental biology, environmental toxicology, and human disease related research fields. Thanks to its unique features, including large fecundity, embryo translucency, rapid and simultaneous development, etc., zebrafish embryos are often used for large scale toxicity assessment of chemicals and drug/compound screening. A typical screening procedure involves adult zebrafish spawning, embryos selection, and arraying the embryos into multi-well plates. From there, embryos are subjected to exposure and the toxicity of chemical, or the effectiveness of the drugs/compounds can be evaluated relatively quickly based on phenotypic observations. Among these processes, embryos arraying is one of the most time-consuming and labor-intensive steps that limits the throughput level. In this protocol, we present an innovative approach that makes use of a 3D-printed arraying template coupled with vacuum manipulation to speed up this laborious step. The protocol herein describes the overall design of the arraying template, a detailed experimental setup and step-by-step procedure, followed by representative results. When implemented, this approach should prove beneficial in a variety of research applications using zebrafish embryos as testing subjects.
Introduction
As a popular model organism, zebrafish is widely used in the fields of medicine and toxicology1,2,3,4. Compared to in vitro platforms, zebrafish offer much greater biological complexities that one or two cell types could not offer. Besides being a whole organism model, the zebrafish's large fecundity, rapid and simultaneous embryonic development, and high organ translucency have given this model unique advantages to be used for large scale toxicity or drugs/compound screening5. The hundreds of embryos produced by one pair of adult zebrafish each week surpass any other whole animal models and have made it suitable for high throughput screening.
A typical screening procedure using zebrafish involves a significant amount of manual work, such as adult zebrafish spawning, embryo selection, and arraying embryos into suitable containers where they are subjected to exposure through water immersion. The development of the embryos is monitored and observable endpoints such as mortality, hatchability and abnormality are often evaluated manually and used as the preliminary identifications of the toxicity of chemicals or indications of the effectiveness of drugs or compounds. To speed up the screening procedure, approaches such as automated imaging and computer-assisted image analysis have been explored previously. For example, microscopes with high content imaging capabilities have been adapted to perform automated bright-field or fluorescence imaging on zebrafish embryos at various developmental stages from 96/384 well plates6. Microfluidic devices coupled with microscopes were used to position zebrafish larvae through current manipulation for imaging of brain neurons7. These approaches could significantly improve the efficiency of image acquisitions compared to traditional manual operation. Moreover, with large number of images being generated, image analysis tools have also been developed to speed up the data processing, as demonstrated by Liu et al. and Tu et al.8,9.
As the throughput level of imaging and image analysis increases, it became clear that the rate-limiting step for screening lies in the process of preparing zebrafish embryos for exposure, which typically means arraying them into 96- or 384-well plates. To solve this bottleneck step, vision-guided robotics were developed by Mandrell et al.10 and us11 previously to replace manual handling but the instruments were rather sophisticated and there is a deep learning curve to implement such techniques. Therefore, to provide an easy-to-use approach becomes one important factor to further improve the throughput level of zebrafish screening and is the main objective of this work.
In this work, we designed and fabricated an embryo arraying template by 3D printing. Such an arraying template was designed to entrap zebrafish embryo into wells that fit with a standard 96-well plate. Instead of selecting embryos and arraying them into individual well one by one, one could perform embryo entrapment and array all 96 embryos into a multiwall plate at once. Using this template and the following protocol, one could significantly increase the efficiency of arraying embryos into multiwall plates, which would in term boost the screening capacity at least tenfold, compared to manual operation. The protocol described below includes an overall design for the arraying template, zebrafish spawning, embryo collection, and arraying. Figure 1 shows the overall design of the arraying template. Figure 2 shows an overview of the step-by-step protocol on using the template described in Parts 3 and 4.
Protocol
1. Design and Fabrication of a Zebrafish Embryos Arraying Template
- Design the arraying template with a 12 by 8, 96-well layout that fits a standard 96-well plate. Use the dimensions listed in Figure 1A for the upper embryo entrapment chamber (see also the Supplemental File).
- Use the dimensions shown in Figure 1B and 1D for the entrapment well.
- Use the dimensions in Figure 1C for the bottom vacuum chamber.
- Use the dimensions in Figure 1B for the air in/outlet.
- Use a 3D-printer (with 0.1 mm precision) to print the template; see Table of Materials for recommended resin to be used for printing.
NOTE: 3D printers with a 0.1 mm precision are recommended for fabrication of the arraying template (see Table of Materials). The suggested color for the surface of the template is dark grey or black.
2. Zebrafish Embryo Spawning
- Place two pairs of male and female fish per mating box one day prior to spawning. Separate males and females by a clear plastic divider.
- Take off the dividers in the morning to mix male and female fish.
- Remove the male and female fish and collect zebrafish embryos using a fine-mesh strainer. Wash the embryos with 250 mL egg water (see Table of Materials).
- Transfer the collected embryos to petri dishes (90 mm in diameter) with Holtfreter's solution (see Table of Materials) and remove dead and unfertilized embryos using a stereomicroscope.
- Place the embryos in a 28.5 °C incubator. At 4 h post fertilization (hpf), observe the embryos and remove any dead and unhealthy embryos. The embryos are now ready for the next step.
3. Preparation of Arraying Template
- Wash the template 2–3 times with 500 mL deionized water and put it into the drying oven (45 °C) for 5 min.
- Tape the bottom chamber with a piece of sealing film (Figure 2, Step 1).
- Connect a vacuum pump through the air outlet at the bottom of the template.
NOTE: The recommended max vacuum for the vacuum pump is 0.1 Mpa. Be aware of the strength of the vacuum used. If the negative pressure is too strong, cut a cross-shaped hole on the sealing film to lower the pressure.
4. Arraying Zebrafish Embryos into a 96-well plate
- Using a plastic transfer pipette, place approximately 150 embryos into the template, as demonstrated in Figure 2, Step 2.
- Connect the vacuum pump to the air outlet to generate negative pressure in the chamber sealed by the sealing film in step 3.3.
- Shake the entire template horizontally until each well has one embryo entrapped (Figure 2, Step 3).
NOTE: If the Holtfreter's Solution dries up before the embryos are trapped in each well, add additional Holtfreter's Solution in the entrapment chamber and repeat this step. - Discard extra Holtfreter's Solution and embryos that are not entrapped in the wells (Figure 2, Step 4).
- Turn off and disconnect the vacuum pump.
- Place a standard 96-well plate upside down against the template (Figure 2, Step 5) and rotate both at the same time (Figure 2, Step 6).
- Tap the bottom of the template or connect the air outlet to a compressed gas dusting can to transfer all trapped embryos from the template to the 96-well plate (Figure 2, Step 6).
- Repeat step 4.1 to 4.8 to prepare additional multi-well plates.
- Remove the sealing film and wash the template 3 times from top to bottom with 500 mL deionized water for future use.
NOTE: Do not use any organic solvents, like ethanol, to clean the template.
Representative Results
Figure 3 shows a typical 3D-printed arraying template. This template uses photosensitive resin as raw material and was made by a 3D printer; a layer of black paint was applied to provide a better contrast to the color of embryos. The position of 96 wells (12 by 8) was designed to fit with a standard 96-well plate. Similarly, a 384 (24 by 16) well template could also be designed and fabricated using the same method. The upside chamber was slightly bigger than a standard 96-well plate to provide a better fitting. Grooves were also designed to hold the extra embryos during arraying.
At a modest rate, 20 plates could be prepared using the 3D-printed arraying template within 30 min, while only two to three plates could be prepared manually. Table 1 shows a comparison between manual, robotic, and arraying template operations. Figure 4 shows a comparison between two plates, one arrayed by the 3D-printed arraying template, one prepared manually. Using the arraying template, there was a negligible amount of liquid transferred with the embryos, which also made it convenient for further exposure experiments.
Figure 5 shows that there were no significant effects on the overall health status of the embryos after being plated by manual and arraying template methods. To make sure the embryos were not affected by such an arraying process, we followed embryo development after arraying them into the 96-well plates for 3 days. The hatching rate, survival rate, and malformation rate of zebrafish embryos at 24, 48, and 72 hpf were all comparable with the embryos arrayed manually.
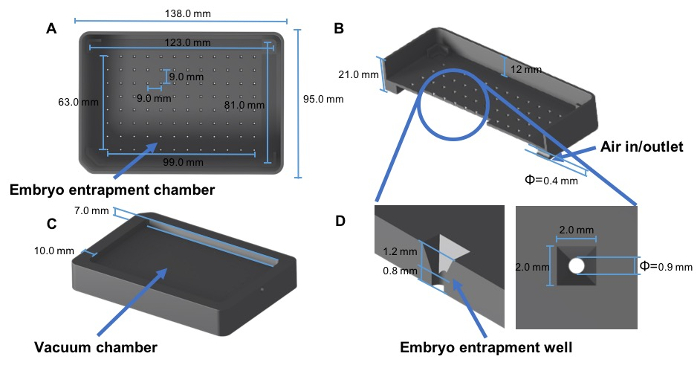
Figure 1: Design of a zebrafish embryo arraying template. (A and C) 3-d Max drawing showing the overview of the template. (B) Cross-section view of the template. (D) A partial view of the template. Please click here to view a larger version of this figure.
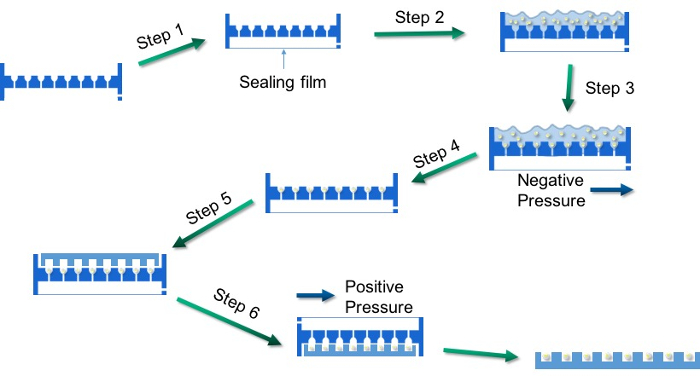
Figure 2: Typical steps when using the arraying template. Please click here to view a larger version of this figure.
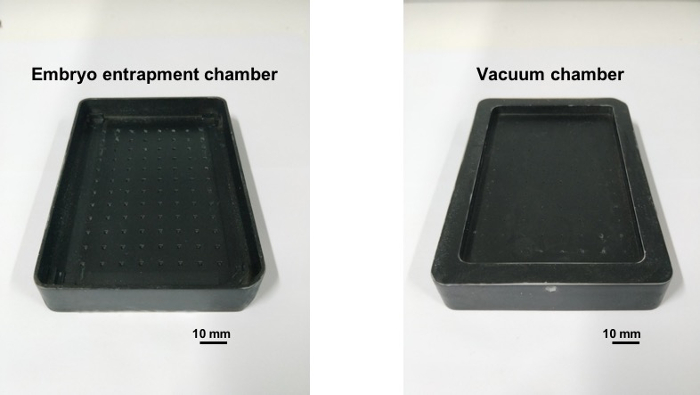
Figure 3: Photographs of a 3D-printed arraying template. Please click here to view a larger version of this figure.
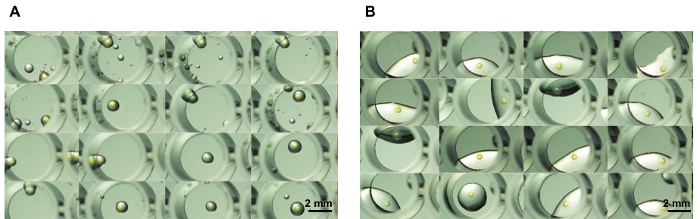
Figure 4: Microscopic images of embryos after transferred to 96-well plate. (A) Partial image of a 96-well plate arrayed by the template. (B) Partial image of a 96-well plate arrayed manually. Please click here to view a larger version of this figure.
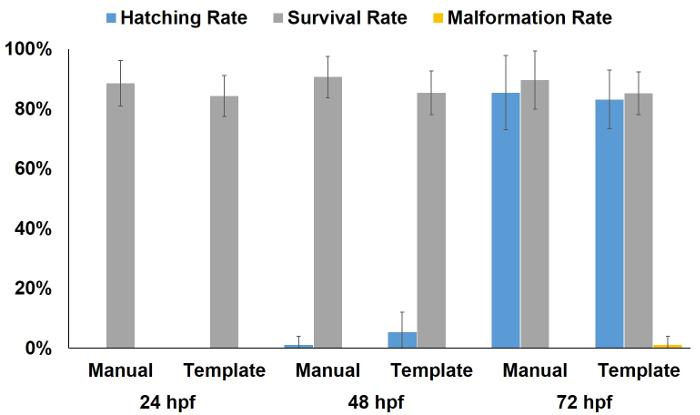
Figure 5: The overall health status of the embryos after being plated by manual and arraying template methods. Based on the hatching, abnormal, and survival rates of the embryos after being plated using either manual or the arraying template methods, no significant effects on the overall health of the embryos were observed in either case. Error bars are standard deviations. Please click here to view a larger version of this figure.
| Manual | Robotic Platform | Arraying Template | |
| Plating Time | 10–30 min/plate | 5–10 min/plate | 1–2 min/plate |
| Cost | Low | High | Low |
| Required Training | Minimum | High | Minumum |
| Working Area | Small | Large | Small |
| Effect On The Embryos | Little to none | Little to none | Little to none |
| Amount of liquid added per well | Random | 5–10 µL | Minimum |
Table 1: Comparison of manual, robotic, and arraying template operations.
Discussion
There are two critical steps in this protocol that require close attention for a successful implementation of 3D-printed template for arraying zebrafish embryos.
The most important factor on the design of the arraying template is the entrapment well. To makes sure there is only one embryo trapped in each well, one should pay close attention to the diameter and the depth of the entrapment well, and the diameter of the through hole. The recommended diameter is within 1.5 to 2 times of the diameter of a typical embryo (including the chorion). The depth of the entrapment well should be within 2 times of the diameter of a typical embryo (including the chorion) to avoid stacking of embryos in the same well. The diameter of the through hole should be approximately half of the diameter of a typical embryo (including the chorion) to avoid embryos being squeezed through the hole by the negative pressure. Due to the complexity of the design, 3D printing technology is recommended for fabrication of the arraying template.
The essence of the arraying template is to entrap individual embryos at designated locations, i.e. positions that fit with a 96- or 384-well plate. To create a proper negative pressure to hold the individual embryos in place without damaging them, one must adjust the negative pressure generated by the vacuum device. Too much pressure may destroy the embryos, while too little pressure may not trap them in the wells. Therefore, it is highly recommended to observe the embryos entrapped in the template under a stereomicroscope before transferring them into the multiwell plates. Furthermore, during entrapment, the embryos might be exposed to air during this process. Should this happen, add additional Holtfreter's medium in the entrapment chamber according to step 4.3. Also, because of the possibility of embryos being exposed to air, the current protocol does not work on dechorionated zebrafish embryos.
The arraying template presented in this work provides an innovative approach for establishing a low-cost and high-throughput screening platform using zebrafish embryos. Compared to the previously established robotic platforms or commercially available flow cytometry-based instrument, this method is easy to use and does not require sophisticated training. Taking advantages of the 3D printing technology, one could easily change the format of the arraying template to fit different purposes.
The template and protocol in its current form still require some manual work. Streamlining the procedure could further improve the throughput level and increase the amount of multiwell plates prepared within a short period of time.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
This work was supported by the "1000plan Youth" program, the Startup Funds from Tongji University, and NSFC Grant# 21607115 and 21777116 (Lin).
Materials
| Zebrafish Facility | Shanghai Haisheng Biotech Co., Ltd. | Z-A-S5 | |
| Mating box | Shanghai Haisheng Biotech Co., Ltd. | ||
| Wash Bottle, 500 ml | Sangon Biotech | F505001-0001 | |
| Sodium chloride | Vetec | V900058-500G | |
| Potassium Chloride | Sinopharm Chemical Reagent Co.,Ltd | 10016318 | |
| Calcium chloride | Sinopharm Chemical Reagent Co.,Ltd | 20011160 | |
| Sodium bicarbonate | Vetec | v900182-500G | |
| Methylene Blue Hydrate | TCI | M0501 | |
| Hydrochloric acid | Sinopharm Chemical Reagent Co.,Ltd | 10011008 | |
| Sea Salts | Instant Ocean | SS15-10 | |
| Pipetter | Fisherbrand | 13-675M | |
| Controlled Drop Pasteur Pipet | Fisherbrand | 13-678-30 | |
| Microscope | OLYMPUS | SZ61 | |
| Biochemical incubator | Shanghai Yiheng Scientific Instrument Co., Ltd. | LRH-250 | |
| 3D printer | UnionTech | Lite600 | |
| Photosensitive resin | UnionTech | UTR9000 | |
| Vacuum pump | Shanghai Yukang Scientific Instrument Co., Ltd. | SHB-IIIA | |
| Adhesive PCR Plate Seals | Solarbio | YA0245 | |
| 96 well plate | Costar | 3599 | |
| Multi 8-channel pipette 30 – 300 μl | Eppendorf | 3122000.051 | |
| Compressed Gas Duster | Shanghai Zhantu Chemical Co., Ltd. | ST1005 | |
| DI Water | Thermo | GenPure Pro UV/UF | |
| Drying oven | Shanghai Yiheng Scientific Instrument Co., Ltd. | BPG-9106A | |
| System water | Water out of the facility’s water system | ||
| Egg water | Dilute 60mg “Instant Ocean” sea salts and 0.25 mg/L methylene blue in 1 L DI water | ||
| Holtfreter’s solution | Dissolve 7.0 g Sodium chloride (NaCl), 0.4 g Sodium bicarbonate (NaHCO3), 0.1 g Potassium Chloride (KCl), 0.235 g Calcium chloride (CaCl2.2H2O) in 1.9 L DI water. Adjust pH to 7 using HCl and adjust volume to 2 L using Di water |
Referenzen
- Howe, K., et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 496 (7446), 498-503 (2013).
- Leslie, M. Zebrafish larvae could help to personalize cancer treatments. Science. 357 (6353), 745-745 (2017).
- Lin, S., et al. Understanding the Transformation, Speciation, and Hazard Potential of Copper Particles in a Model Septic Tank System Using Zebrafish to Monitor the Effluent. ACS Nano. 9 (2), 2038-2048 (2015).
- Lin, S., et al. Aspect ratio plays a role in the hazard potential of ceo2 nanoparticles in mouse lung and zebrafish gastrointestinal tract. ACS Nano. 8 (5), 4450-4464 (2014).
- Baraban, S. C., Dinday, M. T., Hortopan, G. A. Drug screening in Scn1a zebrafish mutant identifies clemizole as a potential Dravet syndrome treatment. Nature Communications. 4, (2013).
- Lin, S., et al. High content screening in zebrafish speeds up hazard ranking of transition metal oxide nanoparticles. ACS Nano. 5 (9), 7284-7295 (2011).
- Kuipers, J., Kalicharan, R. D., Wolters, A. H. G., van Ham, T. J., Giepmans, B. N. G. Large-scale Scanning Transmission electron microscopy (nanotomy) of healthy and injured zebrafish brain. Journal of Visualized Experiments. (111), (2016).
- Liu, R., et al. Automated Phenotype Recognition for Zebrafish Embryo Based In vivo High Throughput Toxicity Screening of Engineered Nano-Materials. PLoS One. 7 (4), (2012).
- Tu, X., et al. Automatic Categorization and Scoring of Solid, Part-Solid and Non-Solid Pulmonary Nodules. in CT Images with Convolutional Neural Network. Scientific Reports. 7, 8533 (2017).
- Mandrell, D., et al. Automated zebrafish chorion removal and single embryo placement: optimizing throughput of zebrafish developmental toxicity screens. Journal of Laboratory Automation. 17 (1), 66-74 (2012).
- Lin, S., Zhao, Y., Nel, A. E., Lin, S. Zebrafish: An in vivo model for nano EHS studies. Small. 9 (9-10), 1608-1618 (2013).

