Generating Homo- and Heterografts Between Watermelon and Bottle Gourd for the Study of Cold-responsive MicroRNAs
Summary
Here we present a detailed protocol for efficiently making homo- and heterografts between watermelon and bottle gourd, in addition to methods of tissue sampling, data generation, and data analysis, for the investigation of cold-responsive microRNAs.
Abstract
MicroRNAs (miRNAs) are endogenous small non-coding RNAs of about 20 – 24 nt, known to play important roles in plant development and adaptation. There is an accumulating evidence showing that the expressions of certain miRNAs are altered when grafting, an agricultural practice commonly used by farmers to improve crop tolerance to biotic and abiotic stresses. Bottle gourd is an inherently climate-resilient crop compared to many other major cucurbits, including watermelon, rendering it one of the most widely used rootstocks for the latter. The recent advancement of high-throughput sequencing technologies has provided great opportunities to investigate cold-responsive miRNAs and their contributions to heterograft advantages; yet, adequate experimental procedures are a prerequisite for this purpose. Here, we present a detailed protocol for efficiently generating homo- and heterografts between the cold-susceptible watermelon and the cold-tolerant bottle gourd, in addition to methods of tissue sampling, data generation, and data analysis. The presented methods are also useful for other plant-grafting systems, to interrogate miRNA regulations under various environmental stresses, such as heat, drought, and salinity.
Introduction
Grafting has long been employed as an agricultural technique to improve crop production and tolerance to biotic and abiotic stresses1,2,3. In heterografting systems, elite rootstocks can enhance water and nutrients uptake of plants, strengthen resistance to soil pathogens, and limit the negative effects of metal toxicity4,5, which may confer the grafts an enhanced growth vigor and increased tolerance to environmental stresses. In many cases, heterografting can also impact fruit qualities in horticultural plants, leading to improved fruit flavor and increased content of health-related compounds6,7. It has been found that the long-distance transfer of phytohormones, RNAs, peptides, and proteins between the rootstock and the scion is a fundamental mechanism modulating the growth and development reprogramming of scion plants8,9,10. Grafting has been widely used in studies of long-distance signaling and transport in relation to environmental adaptation11. Grafting experiments are especially powerful for unambiguous detection of transmitted molecules in receiving tissue or vascular sap, and activation or suppression of molecular targets due to signal transmission12.
Non-coding RNAs, a big class of RNA that exert important regulatory functions in cells, have been reported to play a role in facilitating plant adaptation to abiotic stress13. miRNAs are endogenous small non-coding RNAs of about 20 – 24 nt. Studies have revealed the regulatory role of miRNAs in various aspects of plant activities, such as shoot growth, lateral root formation14,15,16, nutrient uptake, sulfate metabolism and homeostasis17, and responses to biotic and abiotic stress18. Recently, the expression of miRNAs and their target genes were related to salt stress tolerance in heterografted cucumber seedlings19. In the intervariety grafts of grape, the responses of miRNA expression to drought stress were found to be genotype-dependent20.
The rapid development and decreasing cost of high-throughput sequencing technology have provided a great opportunity for the study of miRNA regulations in agronomical plants. Watermelon (Citrullus lanatus [Thunb.] Mansf.), an important cucurbit crop grown throughout the world, is susceptible to low temperatures. Bottle gourd (Lagenaria siceraria [Molina] Standl.) is a more climate-resilient cucurbit commonly used by farmers to graft with watermelon. The primary goal of the current study is to establish a standard, efficient, and convenient method for making heterografts between watermelon (Citrullus lanatus [Thunb.] Mansf.) and bottle gourd (Lagenaria siceraria [Molina] Standl). This protocol also provides a detailed experimental scheme and analytical procedures for the study of the regulation of miRNA expressions following grafting, which is useful for revealing the mechanisms underlying heterografting advantages.
The plant materials used in this study include the watermelon cultivar and the bottle gourd landrace. Watermelon cultivar is a commercial cultivar with high yield but susceptible to low temperatures. Bottle gourd landrace is a popular rootstock for grafting with watermelon, cucumber, and bottle gourd, due to its excellent tolerance of low temperatures21.
Protocol
1. Seed Sterilization and Germination
- For surface sterilization, soak the bottle gourd seeds in a 500-mL beaker filled with water at 58 °C with occasional stirring, until the water temperature drops to 40 °C.
- Meanwhile, put 3 kg of peat soil into a nylon bag and, to sterilize it, autoclave it at 120 °C/0.5 MPa for 20 min.
- Keep soaking the bottle gourd seeds for 4 – 5 h more with no stirring.
- Once the water reaches room temperature, rinse the seeds 2x – 3x with distilled water.
- Drain the excess water and allow the seeds to sprout in a gauze bag at 28 °C in a growth chamber in the dark.
- After germination, sow the seeds into plastic pots (6 cm in diameter) filled with sterilized peat soil.
- When the bottle gourd seedlings have developed two flattened cotyledons, repeat steps 1.1 – 1.3 with watermelon seeds.
NOTE: This time management ensures that the sizes of the scion and rootstock match well for successful grafting.
2. Seedling Growth and Grafting
- Grow the seedlings in a growth chamber with a 16-h light /8-h dark cycle, keeping the temperature at 28 °C during the day (light) and at 22 °C during the night (dark). Irrigate the seedlings by adding water 1x a day in the late afternoon.
- Use the cut-grafting method22 to make heterografts when the seedlings of the bottle gourd (rootstock) are at the one true-leaf stage and the cotyledons of the watermelon (scion) have emerged (not yet flattened).
- Cut the hypocotyls of the watermelon seedlings at 2 – 3 cm below the cotyledons, and the top of the bottle gourd seedlings at the site immediately above the true leaves.
- Use a toothpick to make a hole in the top of the trimmed bottle gourd seedlings. Insert the trimmed watermelon seedlings into the holes of the bottle gourd seedlings to make heterografts.
- Use a similar method as presented in step 2.2 to make homografts.
NOTE: Homo- and heterografting combinations should always be made simultaneously (Figure 1), which, in this case, results in the following: watermelon/bottle gourd (WB, heterograft), watermelon/watermelon (WW, homograft) and bottle gourd /bottle gourd (BB, homograft).
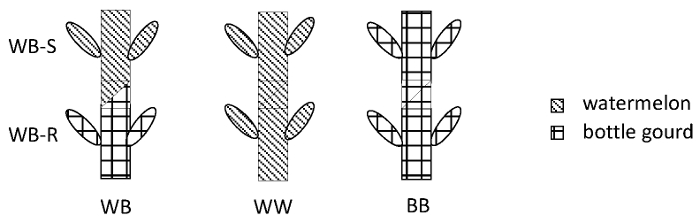
Figure 1: Illustration of the graft combinations and the grafted plant structures. WB = watermelon/bottle gourd heterografting; WW = watermelon/watermelon homografting; BB = bottle gourd/bottle gourd homo-grafting; WB-S = scion leaves of the watermelon/bottle gourd heterografts that were sampled; WB-R = rootstock leaves of the watermelon/bottle gourd heterografts that were sampled. Please click here to view a larger version of this figure.
3. Postgrafting Management, Cold Treatment, and Sampling
- Enwrap the grafted seedlings with transparent polyethylene bags to keep a relatively high humidity and maintain them for 7 d under environmental conditions of 16-h light/8-h dark cycles, keeping the temperature at 28 °C during the day (light) and at 22 °C during the night (dark).
- Uncover the transparent polyethylene bags on the 7th day. Let the plants grow for an additional 7 – 10 days under the same conditions.
- Divide the healthy uniform seedlings into two groups, one for cold treatment (stressed) and one for control (non-stressed). For the control group, leave the seedlings in the same growth chamber (at 28 °C) for an additional 48 h, while for the cold-stressed group, transfer the seedlings to a growth chamber with a constant temperature at 6 °C, with light/dark conditions as described in step 2.1.
- Sample the leaves of the scion and the rootstock from the grafts (Figure 1). Freeze the samples immediately in liquid nitrogen and store them at -70 °C until use.
4. Library Preparation and High-throughput Sequencing
- Transfer the frozen samples to a 2-mL microcentrifuge tube in liquid nitrogen.
- Add a stainless-steel bead (5 mm in diameter) to each tube containing the tissues.
- Homogenize the tissues to a fine powder using a bead mill homogenizer for 30 s.
- For each grafting combination, take equal amounts (0.1 g) of ground sample from ten seedlings and mix them in a 10-mL centrifuge tube. Add an appropriate amount of guanidium hydrochloride reagent (Table of Materials) based on the manufacturer's suggestions corresponding to the tissue weight.
- Remove genomic DNA contaminations by adding RNA-free DNase I to 150 U/mL at 37 °C for 1 h.
- Determine the total RNA quantity on a microcapillary electrophoresis system to ensure the RNA integrity number > 7.0.
NOTE: A RIN > 7.0 ensures a high integrity of the RNA samples. - Prepare small RNA libraries using a commercial kit (Table of Materials) according to the manufacturer's instructions. Use 1 µg of total RNA per sample to initiate.
- Thaw library normalization reagents and adapters according to the manufacturer's guidelines. Ligate small RNAs with the 5′ and 3′ adapters and elute and purify them. Then, reverse transcribe the 5′ and 3' ligated small RNAs following the manufacturer's guidelines.
- Perform PCR amplification according to the manufacturer's protocol. Assess the quality and quantity of the cDNA libraries using a microcapillary electrophoresis system.
- Load 1 µL of an RNA library on a microcapillary electrophoresis system to ensure the RIN > 7.0.
- Sequence the small RNA libraries on a high-throughput sequencing instrument as described elsewhere23.
5. miRNA and Target Gene Prediction
- For each grafting combination, use the open source UEA sRNA workbench 2.4-plant version24 to remove poor-quality sequences and to trim adaptor sequences from the raw reads. Discard sequences that are smaller than 18 nt or larger than 32 nt.
- Compare the high-quality "clean" sequences to the open source Rfam 11.0 database to recognize and remove reads of rRNA, tRNA, snoRNA, and other snRNAs.
- Align the remaining reads to the reference genomes using a short-read sequence alignment tool25. No mismatch is allowed in this step.
NOTE: The watermelon "97103" genome assembly V126 was used for alignment with the reads from the scion, and the bottle gourd "HZ" genome assembly V127 was used for reads from the rootstock. - Compare the remaining reads against known mature miRNAs in the open source miRBase 22.028. Reads that are homologous to known miRNAs are classified as conserved miRNAs.
- Compare the sequences that fail to match the known miRNA precursors with the genome sequence. Use the MIREAP29 algorithm to detect potential novel miRNAs under default settings.
6. Differential Expression and Gene Ontology Analysis
- Compare the expression levels of miRNAs based on their read counts. miRNAs with a P-value (Fisher's exact test) < 0.05 and an absolute log2 value > 2 are considered to be differentially expressed.
- Use an antisense oligonucleotide target site selection tool (TargetFinder)30 to predict potential complementary mRNAs (miRNA target genes) to the differentially expressed miRNAs under default parameters.
- Use the gene ontology (GO) enrichment analytical tool31 to reveal the miRNA target genes ontology (GO) patterns under a P-value threshold of 0.05 for statistical significance.
Representative Results

Figure 2: Phenotypes of various grafts at room temperature and cold-stressed conditions. (a) This panel shows homo- and heterografted seedlings at room temperature as the control. (b) This panel shows homo- and heterografted seedlings after 48 h of cold treatment. Please click here to view a larger version of this figure.
Using the described method, we obtained a high success (survival) rate of 98% for grafting. Phenotypes of various grafts at room temperature and cold-stressed conditions are shown in Figure 2. After 48 h of cold treatment, the homografted watermelon plants showed obvious growth retardation with wilted young leaves, while the homografted bottle gourd plants and the watermelon/bottle gourd heterografts exhibited much more vigorous growth. No symptoms of damage were observed in the leaves of the heterografts, which even outperformed the homografted bottle gourd plants, where the lowest true leaves were damaged. These results clearly demonstrate the advantage of heterografts in conferring cold tolerance.
Small RNA sequencing of the eight libraries yielded a total of 258 million raw reads. After the quality control (QC), a total of 146 million reads corresponding to approximately 30 million unique sequences were retained (Table 1). Based on this set of clean sRNA sequences, 323 miRNAs, including 10 known and 313 novel miRNAs, were predicted from the bottle gourd, and 20 known and 802 novel miRNAs were predicted from the watermelon.sRNAs of 24 nt made up the biggest class of sRNAs in all grafting combinations, regardless of room temperature or cold-stressed conditions (Figure 3).
| Treatment | Code | No. reads | sRNA | |
| Total | Unique | |||
| WW-CK | Raw | 30612962 | ||
| Clean | 19727501 | 3858868 | ||
| Mapped to genomic | 19059359 | 3777952 | ||
| BB-CK | Raw | 30845546 | ||
| CK | Clean | 16832061 | 3787866 | |
| Mapped to genomic | 16375142 | 3694388 | ||
| WB-CK-S | Raw | 39492123 | ||
| Clean | 26783053 | 6319473 | ||
| Mapped to genomic | 25919944 | 6132389 | ||
| WB-CK-R | Raw | 23763619 | ||
| Clean | 10187791 | 1784447 | ||
| Mapped to genomic | 8946929 | 1537867 | ||
| WW-CL | Raw | 27557577 | ||
| Clean | 17879038 | 3336242 | ||
| Mapped to genomic | 17153763 | 3259960 | ||
| BB-CL | Raw | 29780991 | ||
| Clean | 13342206 | 3235570 | ||
| Cold | Mapped to genomic | 12949972 | 3164329 | |
| WB-CL-S | Raw | 45708415 | ||
| Clean | 23071845 | 4310276 | ||
| Mapped to genomic | 22363113 | 4224166 | ||
| WB-CL-R | Raw | 30585408 | ||
| Clean | 19029266 | 3541729 | ||
| Mapped to genomic | 17364239 | 3196106 | ||
Table 1: Statistics of small RNAs in various grafts at room temperature or under cold treatment.
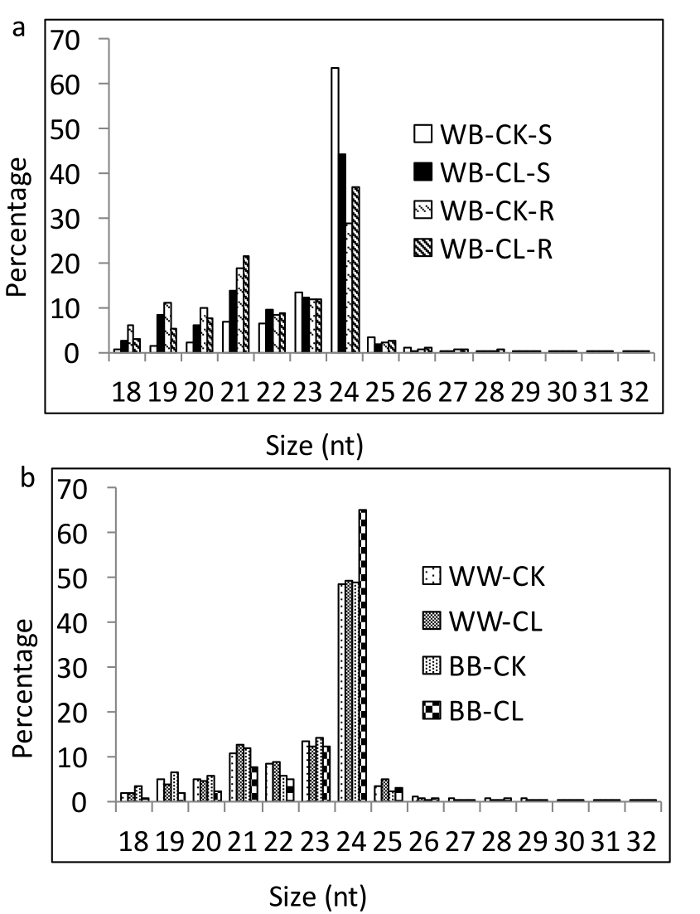
Figure 3: Size distribution of the sRNA reads in various grafts. (a) This panel shows the size distribution of the sRNA reads in heterografts under control or cold conditions. (b) This panel shows the size distribution of the sRNA reads in homografts under control or cold conditions. Please click here to view a larger version of this figure.
Upon a 48-h of cold treatment, 30 and 268 miRNAs were up- and downregulated, respectively, in the leaves of the scion in the heterografts. This was in sharp contrast to the results in the leaves of rootstock, where 31 and only 12 miRNAs were up- and downregulated, respectively (Figure 4). In the watermelon/watermelon homografts, 64 and 83 miRNAs were up- and downregulated, respectively. In the bottle gourd/bottle gourd homografts, these numbers were 30 and 28. Apparently, heterografting caused a profound reprogramming of the miRNA expressions. GO-enrichment analyses of the putative target genes of the differentially expressed miRNAs identified 78 enriched GO terms in the scion of the heterografts, with 40 classified into biological processes, 2 into cellular components, and 36 into molecular functions (Figure 5). We found that several known GO terms/pathways related to abiotic/biotic stress resistance and signal transduction, for instance, the chitin catabolic process (GO: 0006030, GO: 0006032), ethylene-activated signaling pathway (GO: 0009873), polyamine biosynthetic process (GO:0006596), and signal transduction by protein phosphorylation (GO: 0009755), were involved. Combined, our results suggest that the downregulation of miRNAs,by tuning the abundance of the transcripts of their target genes, may represent an important mechanism underlying enhanced cold tolerance. In the watermelon/bottle gourd grafts, the heterograft per se has a significant impact on the miRNA patterns that form the graft advantages.
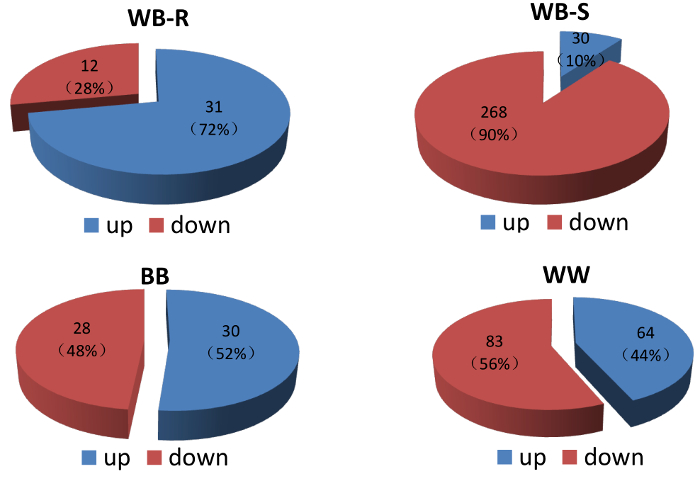
Figure 4: Comparison of the patterns of up- and downregulated miRNAs in response to cold stress in various grafts. WB-S = scion leaves of the watermelon/bottle gourd heterografts that were sampled; WB-R = rootstock leaves of the watermelon/bottle gourd heterografts that were sampled; WW = the watermelon/watermelon homografts; BB = the bottle gourd/bottle gourd homografts. Please click here to view a larger version of this figure.
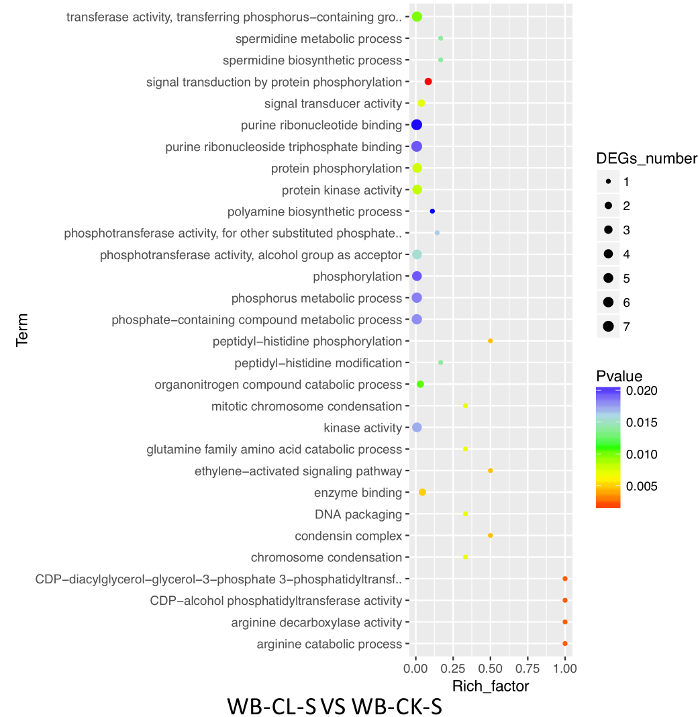
Figure 5: GO enrichment analyses of the putative target genes of differentially expressed miRNAs in the scion leaves of heterografts upon cold stress. WB-CL-S = scion leaves of the watermelon/bottle gourd heterografts under cold treatment; WB-CK-S = scion leaves of the watermelon/bottle gourd heterografts at room temperature. Please click here to view a larger version of this figure.
Discussion
In this protocol, we described in detail a highly efficient and reproducible method to make homo- and heterografts between watermelon and bottle gourd. This method, requiring no specific equipment, is very easy to operate and typically has a very high survival rate of grafting. The method can also be used to make grafts for other cucurbits, such as between watermelon, cucumber, and pumpkin.
It is worth noting that the relative size (age) of the rootstock and scion is critical to making a successful graft (step 2.2 of the Protocol). We observed that, if the rootstock used was too large compared to the scion, the graft union was more difficult to form because the stem of the scion was somewhat hollowed. Based on our previous proteomic data31, the inclusion of self-grafted scion and self-grafted rootstock as controls is strongly recommended (step 2.3 of the Protocol), because then, the impact of grafting injuries can be largely eliminated.
This protocol also provides a detailed experimental scheme and specific experimental procedures for investigating the abundances of miRNAs in the heterografting system. This method will also be useful for studies in other plant-grafting systems to reveal the mechanisms of local and long-distance miRNA regulation. In the Representative Results, we report the expression changes of only local miRNAs in the scion or rootstock in response to a low temperature. Accumulating reports have highlighted the involvement of long-distance small RNA transmission in grafting-related phenotypic changes. The protocol presented here, which combines the methods for grafting and high-throughput data analysis, can also be used for miRNA transmission analysis between the scion and the rootstock. The principle for differentiating transmitted miRNAs from local miRNAs is based on their sequence similarity to the reference genomes (i.e., a miRNA in the scion that is more like the rootstock genome is considered to be transferred from the rootstock, and vice versa).
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31772291), the Research Project for Public Interest in Zhejiang Province (2017C32027), the Key Science Project of Plant Breeding in Zhejiang (2016C02051), and the National Program for the Support of Top-notch Young Professionals (to P.X.).
Materials
| TRIzol Reagent | Invitrogen | 15596026 | |
| RNA-free DNase I | Takara | D2270A | |
| Truseq Small RNA sample prep Kit | Illumina | RS-200-0012 | |
| 2100 Bionalyser | Agilent | 5067 | |
| DNA Polymerase | Thermo Fisher Scientific | F530S | |
| UEA sRNA workbench 2.4-plant version (software) | NA | NA | http://srna-workbench.cmp.uea.ac.uk/ |
| Rfam 11.0 database (website) | NA | NA | http://rfam.janelia.org |
| miRBase 22.0 (website) | NA | NA | http://www.mirbase.org/ |
| MIREAP(software) | NA | NA | https://sourceforge.net/projects/mireap/ |
| TargetFinder (software) | NA | NA | http://targetfinder.org/ |
Referenzen
- Schwarz, D., Rouphael, Y., Colla, G., Venema, J. H. Grafting as a tool to improve tolerance of vegetables to abiotic stresses: Thermal stress, water stress and organic pollutants. Scientia Horticulturae. 127, 162-171 (2010).
- Li, Y., et al. Mechanisms of tolerance differences in cucumber seedlings grafted on rootstocks with different tolerance to low temperature and weak light stresses. Turkish Journal of Botany. 39 (4), 606-614 (2015).
- Li, C. H., Li, Y. S., Bai, L. Q., He, C. X., Yu, X. C. Dynamic Expression of miRNAs and Their Targets in the Response to Drought Stress of Grafted Cucumber Seedlings. Horticultural Plant Journal. 2 (1), 41-49 (2016).
- Rouphael, Y., Cardarelli, M., Colla, G., Rea, E. Yield, mineral composition, water relations, and water use efficiency of grafted mini-watermelon plants under deficit irrigation. HortScience. 43 (3), 730-736 (2008).
- Savvas, D., et al. Interactive effects of grafting and manganese supply on growth, yield, and nutrient uptake by tomato. HortScience. 44 (7), 1978-1982 (2009).
- Aloni, B., Cohen, R., Karni, L., Aktas, H., Edelstein, M. Hormonal signaling in rootstock-scion interactions. Scientia Horticulturae. 127, 119-126 (2010).
- Rouphael, Y., Caradrelli, M., Rea, E., Colla, G. Improving melon and cucumber photosynthetic activity, mineral composition, and growth performance under salinity stress by grafting onto Cucurbita hybrid rootstocks. Photosynthetica. 50 (2), 180-188 (2012).
- Louws, F. J., Rivard, C. L., Kubota, C. Grafting fruiting vegetables to manage soilborne pathogens, foliar pathogens, arthropods and weeds. Scientia Horticulturae. 127 (2), 127-146 (2010).
- Asins, M. J., et al. Genetic analysis of rootstock-mediated nitrogen (N) uptake and root-to-shoot signalling at contrasting N availabilities in tomato. Plant Science. 263, 94-106 (2017).
- Yin, L. K., et al. Role of protective enzymes in tomato rootstocks to resist root knot nematodes. Acta Horticulturae. 1086 (1086), 213-218 (2015).
- Gaion, L. A., Carvalho, R. F. Long-Distance Signaling: what grafting has revealed?. Journal of Plant Growth Regulation. 37 (2), 694-704 (2018).
- Turnbull, C. G., Hennig, L., Köhler, C. Grafting as a research tool. Plant Developmental Biology. , 11-26 (2010).
- Li, C., et al. Grafting-responsive miRNAs in cucumber and pumpkin seedlings identified by high-throughput sequencing at whole genome level. Physiologia Plantarum. 151 (4), 406-422 (2014).
- Lakhotia, N., et al. Identification and characterization of miRNAome in root, stem, leaf and tuber developmental stages of potato (Solanum tuberosum L.) by high-throughput sequencing. BMC Plant Biology. 14 (1), 6 (2014).
- Jones-Rhoades, M. W., Bartel, D. P., Bartel, B. MicroRNAs and their regulatory roles in plants. Annual Review of Plant Biology. 57, 19-53 (2006).
- Puzey, J. R., Kramer, E. M. Identification of conserved Aquilegia coerulea microRNAs and their targets. Genetic. 448 (1), 46-56 (2009).
- Matthewman, C. A., et al. miR395 is a general component of the sulfate assimilation regulatory network in Arabidopsis. FEBS Letters. 586 (19), 3242-3248 (2012).
- Ali, E. M., et al. Transmission of RNA silencing signal through grafting confers virus resistance from transgenically silenced tobacco rootstocks to non-transgenic tomato and tobacco scions. Journal of Plant Biochemistry and Biotechnology. 25 (3), 245-252 (2016).
- Li, Y. S., Li, C. H., Bai, L. Q., He, C. X., Yu, X. C. MicroRNA and target gene responses to salt stress in grafted cucumber seedlings. Acta Physiologiae Plantarum. 38 (2), 1-12 (2016).
- Pagliarani, C., et al. The accumulation of miRNAs differentially modulated by drought stress is affected by grafting in grapevine. Plant Physiology. 173 (4), 2180-2195 (2017).
- Liu, N., Yang, J. H., Guo, S. G., Xu, Y., Zhang, M. F. Genome-wide identification and comparative analysis of conserved and novel microRNAs in grafted watermelon by high-throughput sequencing. PLoS One. 8 (2), e57359 (2013).
- Song, G. Development of 2JC-350 automatic grafting machine with cut grafting method for vegetable seedling. Transactions of the Chinese Society of Agricultural Engineering. 22 (12), 103-106 (2006).
- Kumar, D., et al. Uncovering leaf rust responsive miRNAs in wheat (triticum aestivum l.) using high-throughput sequencing and prediction of their targets through degradome analysis. Planta. 245 (1), 1-22 (2016).
- Kohli, D., et al. Identification and characterization of wilt and salt stress-responsive microRNAs in chickpea through high-throughput sequencing. PLoS One. 9 (10), e108851 (2014).
- Salzberg, S. L. Computational challenges in next-generation genomics. International Conference on Scientific and Statistical Database Management. ACM. 2, (2013).
- Guo, S. G., et al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nature Genetics. 45, 51-58 (2013).
- Wang, Y., et al. Gourdbase: a genome-centered multi-omics database for the bottle gourd (lagenaria siceraria), an economically important cucurbit crop. Scientific Reports. 8 (1), 306 (2018).
- Wang, X. F., Liu, X. S. Systematic Curation of miRBase Annotation Using Integrated Small RNA High-Throughput Sequencing Data for C. elegans and Drosophila. Frontiers in Genetics. 2, 25 (2011).
- Bo, X. C., Wang, S. Q. TargetFinder: a software for antisense oligonucleotide target site selection based on MAST and secondary structures of target mRNA. Bioinformatics. 21 (8), 1401-1402 (2005).
- . GOATOOLS: Tools for Gene Ontology Available from: https://doi.org/10.5281/zenodo.31628 (2015)
- Wang, L. P., Li, G. J., Wu, X. H., Xu, P. Comparative proteomic analyses provide novel insights into the effects of grafting wound and hetero-grafting per se on bottle gourd. Scientia Horticulturae. 200 (8), 1-6 (2016).

