Modeling Tuberculosis in Mycobacterium marinum Infected Adult Zebrafish
Summary
Here, we present a protocol to model human tuberculosis in an adult zebrafish using its natural pathogen Mycobacterium marinum. Extracted DNA and RNA from the internal organs of infected zebrafish can be used to reveal the total mycobacterial loads in the fish and the host's immune responses with qPCR.
Abstract
Mycobacterium tuberculosis is currently the deadliest human pathogen causing 1.7 million deaths and 10.4 million infections every year. Exposure to this bacterium causes a wide disease spectrum in humans ranging from a sterilized infection to an actively progressing deadly disease. The most common form is the latent tuberculosis, which is asymptomatic, but has the potential to reactivate into a fulminant disease. Adult zebrafish and its natural pathogen Mycobacterium marinum have recently proven to be an applicable model to study the wide disease spectrum of tuberculosis. Importantly, spontaneous latency and reactivation as well as adaptive immune responses in the context of mycobacterial infection can be studied in this model. In this article, we describe methods for the experimental infection of adult zebrafish, the collection of internal organs for the extraction of nucleic acids for the measurement of mycobacterial loads and host immune responses by quantitative PCR. The in-house-developed, M. marinum-specific qPCR assay is more sensitive than the traditional plating methods as it also detects DNA from non-dividing, dormant or recently dead mycobacteria. As both DNA and RNA are extracted from the same individual, it is possible to study the relationships between the diseased state, and the host and pathogen gene-expression. The adult zebrafish model for tuberculosis thus presents itself as a highly applicable, non-mammalian in vivo system to study host-pathogen interactions.
Introduction
Zebrafish (Danio rerio) is a widely used animal model in biomedical research and it is an accepted model for common vertebrate biology. The zebrafish has been adapted to many fields of research modeling human diseases and disorders ranging from cancer1 and cardiac disease2 to infection and immunological studies of several bacterial 3 and viral infections4,5. In addition, the ex utero development of zebrafish embryos has made the zebrafish a popular model in developmental biology6 and toxicology7,8.
In many fields of research, including infection biology, the optically transparent zebrafish larvae are commonly used. The first immune cells appear within 24 h post fertilization (hpf), when primitive macrophages are detected9. Neutrophils are the next immune cells to appear around 33 hpf10. Zebrafish larvae are thus feasible for studying the early stages of infection and the role of innate immunity in the absence of adaptive immune cells11. However, the adult zebrafish with its fully functional adaptive immune system provides an additional layer of complexity for infection experiments. T cells can be detected around 3 days post fertilization12, and B cells are able to produce functional antibodies by 4 weeks post fertilization13. The adult zebrafish has all the main counterparts of the mammalian innate and adaptive immune system. The main differences between the immune systems of fish and humans are found in antibody isotypes as well as in the anatomy of lymphoid tissues. The zebrafish has only three antibody classes14, whereas humans have five15. In the absence of bone marrow and lymph nodes, the primary lymphoid organs in the fish are the kidney and the thymus16 and the spleen, the kidney and the gut serve as secondary lymphoid organs17. Despite these differences, with its full immune arsenal of innate and adaptive cells, the adult zebrafish is a highly applicable, easy-to-use, non-mammalian model for host-pathogen interaction studies.
The zebrafish has lately been established as a feasible model to study tuberculosis18,19,20,21,22. Tuberculosis is an airborne disease caused by Mycobacterium tuberculosis. According to the World Health Organization, tuberculosis caused1.7 million deaths in 2016 and is the leading cause of death by a single pathogen worldwide23. Mice24,25, rabbits26 and non-human primates27 are the best-known animal models in tuberculosis research but each face their limitations. The non-human primate model of M. tuberculosis infection resembles the human disease most closely, but using this model is limited due to serious ethical considerations. Other animal models are hindered by the host-specificity of M. tuberculosis that affects the disease pathology. Probably the biggest issue in modeling tuberculosis is the wide spectrum of infection and disease outcomes in the human disease: tuberculosis is a very heterogeneous disease ranging from sterilizing immunity to latent, active and reactivated infection28, which can be hard to reproduce and model experimentally.
Mycobacterium marinum is a close relative of M. tuberculosis with ~3,000 orthologous proteins with 85% amino acid identity29. M. marinum naturally infects zebrafish producing granulomas, the hallmarks of tuberculosis, in its internal organs19,30. Unlike other animal models used in tuberculosis research, zebrafish produces many offspring, it requires only a limited space and importantly, it is neurophysiologically the least developed vertebrate tuberculosis model available. Additionally, the M. marinum infection causes latent infection, active disease or even sterilization of mycobacterial infection in adult zebrafish closely mimicking the spectrum of disease outcomes of human tuberculosis19,31,32. Here, we describe methods for the experimental tuberculosis model of adult zebrafish by injecting M. marinum into the abdominal cavity and using quantitative PCR to measure the mycobacterial loads and immune responses from zebrafish tissue samples.
Protocol
All zebrafish experiments have been approved by the Animal Experiment Board in Finland (ESAVI/8245/04.10.07/2015). Methods are performed according to the act (497/2013) and the government decree (564/2013) on the protection of animals used for scientific or educational purposes in Finland.
1. Culturing of Mycobacterium marinum
NOTE: Since Mycobacterium marinum is a pathogen capable of causing superficial infections in humans, find out the local guidelines for personal safety and biohazard waste disposal before starting to work with this bacterium.
- Culture M. marinum on a 7H10 plate supplemented with 10% OADC (oleic acid, albumin, dextrose, catalase) enrichment and 0.5% v/v of glycerol at 29 °C for at least 5 days. Maintain M. marinum cultures by taking fresh bacteria from the freezer every two weeks and transferring onto a new plate every other week.
NOTE: M. marinum is a natural fish pathogen infecting aquatic species and it is important to take precautions not to contaminate zebrafish stocks with the bacteria. Infected zebrafish and items contaminated with bacteria have to be kept separate from the breeding facilities. - Use a sterile 1-µL inoculation loop to aseptically transfer a loopful of M. marinum bacterial mass into a cell culture flask containing 10 mL of 7H9 medium with 10% ADC (albumin, dextrose, catalase) enrichment, 0.2% v/v polysorbate 80 and 0.2% v/v of glycerol. Culture for 3–4 days to approximately an OD600 (optical density) of 0.7 at 29 °C in the dark without shaking. Leave the cap loose, or use a filter cap, to allow for the sufficient exchange of gases.
NOTE: Polysorbate 80 is added to the medium to prevent the aggregation of bacteria. In addition, exposure to light leads to phenotypic changes in bacterial colonies (e.g., the color changes from white to yellow). To avoid this, keep the cultures in the dark. - Measure the OD600value with a spectrophotometer. Dilute the liquid culture to an OD600 of 0.07–0.09 and continue culturing for 2 days at 29 °C in the dark without shaking. Leave the cap loose.
NOTE: During these two days, the bacterial suspension will reach an OD600of approximately 0.5 corresponding to an early log-phase.
2. Preparation of Bacterial Solution for Infecting Adult Zebrafish
- Transfer the bacterial suspension into a big sterile cuvette or a 15 mL tube and place it in the dark at room temperature for 15 min to allow the biggest clumps to settle.
- Transfer the top 5–7 mL of the suspension into a clean tube or a cuvette and measure the OD600. Use this top phase of the suspension for the infections.
- Collect 1 mL of M. marinum culture into a fresh tube and centrifuge for 3 min at 10,000 x g. Remove the supernatant and resuspend the pellet in 1 mL of sterile 1x PBS.
- Dilute to reach the desired bacterial concentration by using sterile 1x PBS with 0.3 mg/mL phenol red as a tracer. Divide the diluted suspension into three aliquots.
NOTE: Use a predetermined OD600 vs. CFU (colony-forming units)/µL curve to estimate the dilution required to get the wanted number of bacteria in the 5 µL injection volume34. The correlation between OD600 and concentration of the bacterial suspension needs to be validated before starting the actual infection experiments. Reserve two weeks for collecting this validation data. - Using a 1 mL syringe, slowly pull the suspension through a 27 G needle 3x. For each aliquot, perform this step just before use.
NOTE: Do not use the same bacterial solution for more than 2 h.
3. Experimental M. marinum Infection with Intraperitoneal Injection
- Pipette a 5 µL droplet of the diluted bacterial solution onto a piece of parafilm film and pull the droplet into a 30 G insulin needle.
- Use 5–8 month-old wild-type fish and rag1−/−hu1999 mutant fish for the experiment. Anesthetize adult zebrafish in the tank water with 0.02% 3-aminobenzoic acid ethyl ester (pH 7.0). Position the fish ventral side up into a slit on a moist foamed plastic.
NOTE: Rag-/- mutant fish are not able to undergo somatic recombination and produce functional T and B cells. - Inject the needle between the pelvic fins at an approximately 45° angle. Keep the needle opening upwards to observe that the entire opening is inside the abdominal cavity. Slowly inject the bacterial suspension and carefully remove the needle.
NOTE: In case the red tracer is leaking out of the fish upon injection, exclude the fish from the experiment. - Immediately after injection, transfer the fish into a recovery tank with fresh tank water.
- Take samples of the bacterial suspension on 7H10 plates every 15 min from the bacterial aliquot in use and incubate the bacteria at 29 °C for 5 days and verify the infection dose by counting the colonies on the plates.
- Check the well-being of the fish regularly and euthanize any fish with symptoms of the infection with over 0.02% concentration of 3-aminobenzoic acid ethyl ester (pH 7.0).
NOTE: Approximately, 7% of the adult zebrafish infected with 34 ± 15 CFU and 30% of zebrafish infected with 2029 ± 709 CFU will have had symptoms by 8 weeks19. The symptoms may include abnormal swimming, lack of response to touch, gasping, edema or observable wasting. - Maintain the zebrafish according to the common standards35.
4. Collection of Internal Organs
- Euthanize the zebrafish with an overdose of 3-aminobenzoic acid ethyl ester (over 0.02% concentration, pH 7.0) in the tank water.
- Insert one pin posterior to the branchiostegal rays and another through the tail to tack the fish onto the platform.
- Open the whole abdominal cavity with a scalpel and collect the internal organs by using a small spoon and sharp-ended tweezers. Start from the heart and work along the spine towards the tail to detach all internal organs in one block.
NOTE: Be sure to collect all kidney tissue by scraping along the spine with the spoon. - Finally, use tweezers to detach the gut next to the cloaca and transfer the organs into a 1.5 mL homogenization tube with six 2.8 mm ceramic beads. Immediately place on dry-ice to freeze the sample. The sample can be stored at -80 °C until homogenized.
- Rinse the instruments with 70% ethanol between individuals.
5. Homogenization and RNA extraction from an organ block.
NOTE: The method is modified from the Stanford University protocols36.
- Add guanidine thiocyanate-phenol solution used for nucleic acid extraction (Table of Materials) on the top of the sample to a total volume of 1,500 µL. Ensure that the sample covers a maximum of 10% of the total volume.
CAUTION: The expected volume of the harvested tissue is 100 µL. Guanidine thiocyanate-phenol solution contains toxic and irritating compounds and requires protective clothing, nitrile gloves and working in a fume hood. Do not combine with bleach as this will cause formation of toxic gases. Read the material safety data sheet (MSDS) before use. - Homogenize samples using a bead-beating homogenizer 3 times for 40 s at 3,200 rpm. Cool on ice for 30 s between the cycles. Sonicate the homogenized samples in a water bath for 9 min.
- Centrifuge the samples at 12,000 x g for 10 min at 4 °C and move 1,000 µL of the cleared homogenate into a fresh microcentrifuge tube.
- Add 200 µL of chloroform, immediately mix by vortexing for 15 s and incubate for 2 min at room temperature.
CAUTION: Chloroform is a toxic and irritant compound if inhaled, swallowed or contacted with skin or eyes. Use necessary safety equipment for personal protection and work in a fume hood. Read the material safety data sheet (MSDS) before use. - Centrifuge at 12,000 x g for 15 min at 4 °C to separate the aqueous and organic phases.
- Carefully, transfer 500 µL of the top phase to a fresh tube to avoid contaminating the RNA. Remove and discard the rest of the aqueous phase (~100 µL) and store the interphase and organic phase at 4 °C for DNA extraction.
NOTE: The top phase contains the RNA. - Add 500 µL of 2-propanol and immediately mix by vortexing for 15 s. Incubate for 10 min at room temperature to precipitate the RNA.
- Centrifuge at 12,000 x g for 10 min at 4 °C to pellet the RNA. Remove the supernatant by pipetting.
- Add 1 mL of 75% ethanol and vortex for 10 s.
NOTE: The protocol can be paused here, and the samples kept overnight at 4 °C. - Centrifuge at 7,500 x g for 5 min at 4 °C. Remove the supernatant by pipetting.
- Repeat the wash steps 5.9–5.10. Remove the supernatant carefully by pipetting and let the pellet air-dry in a fume hood.
- Dissolve the RNA pellet in 500 µL of nuclease-free water and keep the samples on ice. Measure the concentrations with a microvolume spectrophotometer or with equivalent equipment. Store the RNA at -80 °C.
6. Purification of Co-extracted Zebrafish and Mycobacterial DNA
- Prepare a back-extraction buffer (BEB) by dissolving 118.2 g of guanidine thiocyanate (final concentration 4 M), 3.68 g of sodium citrate (final concentration 50 mM) and 30.29 g of Tris free base (final concentration 1 M) in 120 mL of nuclease-free water (this may require stirring overnight). Add nuclease-free water to a final total volume of 250 mL and filter to sterilize the solution.
NOTE: This buffer can be stored at room temperature for up to 6 months. Do not combine BEB with bleach as they react to produce toxic gases such as hydrogen chloride and hydrogen cyanide. - Use the interphase and organic phase of the sample to extract mycobacterial DNA. Add 500 μL of BEB to each tube. Mix extensively for 10 min by inversion at room temperature.
- Centrifuge the tubes at 12,000 x g for 30 min at room temperature and carefully transfer 500 µL of the upper aqueous phase containing the DNA to a new tube.
- Add 400 μL of 2-propanol. Mix by inverting and incubate for 10 min at room temperature.
- Centrifuge the samples at 12,000 x g for 15 min at 4 °C. A pellet containing the DNA should be visible at this point. Carefully remove the supernatant by pipetting.
- Add 800 μL of 70% ethanol. Wash the pellet by inversion. Do not vortex the samples at this point, as genomic DNA breaks down easily.
- Centrifuge the samples at 12,000 x g for 15 min at 4°C and remove the supernatant by pipetting. Repeat the ethanol wash (steps 6.6 and 6.7).
- Remove the ethanol by carefully pipetting. Let the samples air-dry for 5–10 min. Dissolve the pellet in 200 μL of nuclease-free water.
- Measure DNA concentrations with a microvolume spectrophotometer or with equivalent equipment. DNA can be stored at 4 °C or at -20 °C for long-term storage.
7. Quantitative PCR for Measuring Mycobacterial Loads
- Prepare qPCR reaction mixes with no-ROX (carboxy-X-rhodamine) against the M. marinum internal transcribed spacer (ITS) between 16S-23S ITS according to the manufacturer's instructions with MMITS1 primers (Table 1). Pipette the reaction mix and sample dilutions as duplicates on a 96-well plate suitable for qPCR. Include a DNA standard dilution series of a known amount of bacteria in each run.
NOTE: The expected M. marinum load per fish can range from 0 CFU to 1,000,000 CFU at 4 wpi. The qPCR assay can be also performed with other qPCR kits but the annealing temperature for the primers has to be re-optimized. - Seal the plate with an optically transparent film and centrifuge the plate at 2,000 x g for 2 min at 4 °C.
- Run the qPCR program shown in the Table 2.
- Using the standard curve, calculate the number of bacteria in the entire fish sample.
8. DNase Treatment of the RNA Samples
- To remove any possible remaining traces of genomic DNA from the RNA, carry out DNase I treatment. Thaw the RNA samples on ice.
NOTE: Be sure to use only RNase-free equipment and solutions and wipe the working surface and pipettes with a decontamination reagent eliminating RNases (Table of Materials) before starting to work. Wear a long-sleeved lab coat and gloves to protect your samples. - Prepare 10 μL DNase I reaction mixes on ice according to the manufacturer's instructions. Mix 1 μL of DNase I, 1 μL of 10x DNase buffer and 8 μL of RNA sample including a maximum of 1 μg of RNA.
- Gently mix the reactions (no vortexing) and incubate for 30 min at 37 °C.
- Prior to heat-inactivation, add 1 µL of 50 mM EDTA to each 10 µL sample. If EDTA is not added, the RNA will undergo chemical degradation when heated.
- Incubate for 10 min at 65 °C to heat-inactivate DNase I. Continue directly to cDNA synthesis or store the DNase-treated RNA at -80 °C.
9. cDNA Synthesis
- Keep all reagents and samples on ice and prepare the reaction mixes according to the manufacturer's instructions. For a 5 μL reaction mix, include 1 μL of Reverse Transcription Master Mix, 3 μL of nuclease-free water and 1 μL of DNase treated RNA.
- Gently mix the reverse transcription reactions and briefly spin the tube, if needed.
- Place samples in a PCR machine and use the program shown in Table 3.
- Dilute the cDNA in nuclease-free water for qPCR to a maximum concentration of 2.5 ng/μL, if needed. cDNA can be stored at -20 °C.
10. Measuring Zebrafish Gene Expression by Quantitative PCR
- Prepare a qPCR master mix on ice according to the manufacturer's instructions and protect from light. Use the primers introduced in Table 1.
NOTE: To calculate the fold of induction for each gene, measure the expression also from a pooled baseline sample extracted from 6 healthy zebrafish. - Prepare replicates of each sample and pipette the reaction mixes onto a qPCR plate. Seal the plate with an optically transparent film and centrifuge the plate at 2,000 x g for 2 min at 4 °C before starting the run.
- Run the qPCR program shown in the Table 4 with the annealing temperature depending on the primer pair used (Table 1).
- Analyze the gene expression ratio compared to a house-keeping gene (loopern437) with the ΔCt method using the equation:
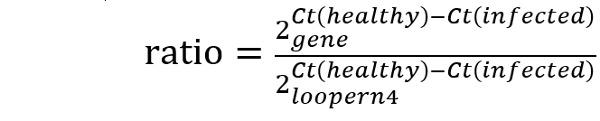
Representative Results
The natural fish pathogen Mycobacterium marinum infects the internal organs of the zebrafish and produces a systemic infection with histologically visible granulomas19. Adult zebrafish are infected with M. marinum by an intraperitoneal injection. The DNA and RNA are extracted, and the mycobacterial load is measured by quantitative polymerase chain reaction (qPCR) using DNA as the template. The outline of the method is shown in Figure 1.
The initial number of mycobacteria used for infecting the fish is a critical determinant for the outcome of infection. A high infection dose of M. marinum (~2,000 CFU) leads to a progressive disease in which the mycobacterial loads continue to increase until the average bacterial load reaches around five million bacteria (Figure 2A) ultimately killing the fish. A low dose (~20–90 CFU) of M. marinum leads to the development of a disease spectrum similar to that seen in human tuberculosis (Figure 2B). The bacterial load continues to increase until around 4–7 weeks (Figure 2A and Figure 3A), after which in the majority of the fish the disease reaches a steady-state. Figure 2B shows an example of the distribution of disease outcomes with a low dose infection: About 7% of the infected zebrafish were unable to restrict the bacterial growth. These individuals developed a primary progressive disease and they died within two months after the infection. Around 10% of the individuals cleared the mycobacterial infection by 4 weeks. The remaining 65% of the fish population developed a latent mycobacterial infection with steady bacterial burdens. However, between 8 and 32 weeks of infection, in 18%, the latent infection spontaneously reactivated leading to the progression of the disease.
By using rag-/- mutant fish, it is possible to study the role of adaptive immune responses in the adult fish. Rag-/- mutant fish cannot sufficiently limit the growth of mycobacteria leading to higher bacterial loads (Figure 3A) and increased morbidity (Figure 3B), clearly demonstrating the importance of adaptive immunity in controlling mycobacterial infection. Also, the importance of adaptive responses in evoking certain cytokine responses in mycobacterial infection can be studied in this model. Here, we show that the adaptive response is required for the efficient induction of interleukin 4 (IL4) (Figure 3C) but is dispensable for the induction of interferon-γ (IFNγ) at 4 wpi (Figure 3D). Interferon-γ is a cytokine driving the response against intracellular pathogens whereas interleukin 4 is a common mediator in the adaptive immune response against extracellular pathogens. The significantly higher expression levels of il4 in the wild-type group compared to rag-/- mutant fish refers to important adaptive humoral responses in the mycobacterial infection (Figure 3C).
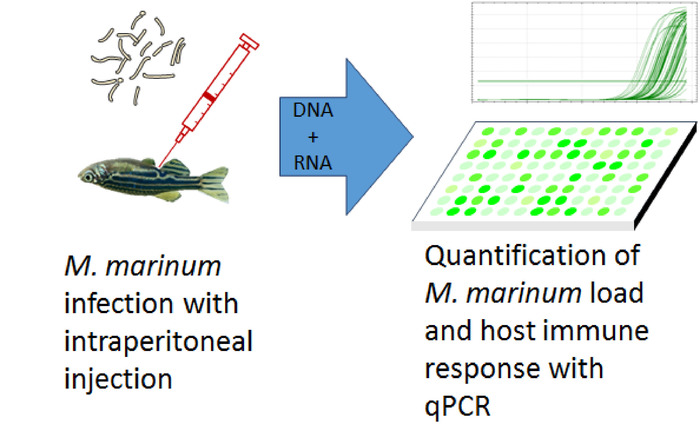
Figure 1: Workflow of studying the development of mycobacterial loads in the adult zebrafish. Adult zebrafish are infected with an intraperitoneal injection of M. marinum. DNA and RNA are extracted from the internal organs of the fish and the M. marinum load and host's immune responses are analyzed with quantitative polymerase chain reaction (qPCR). Please click here to view a larger version of this figure.
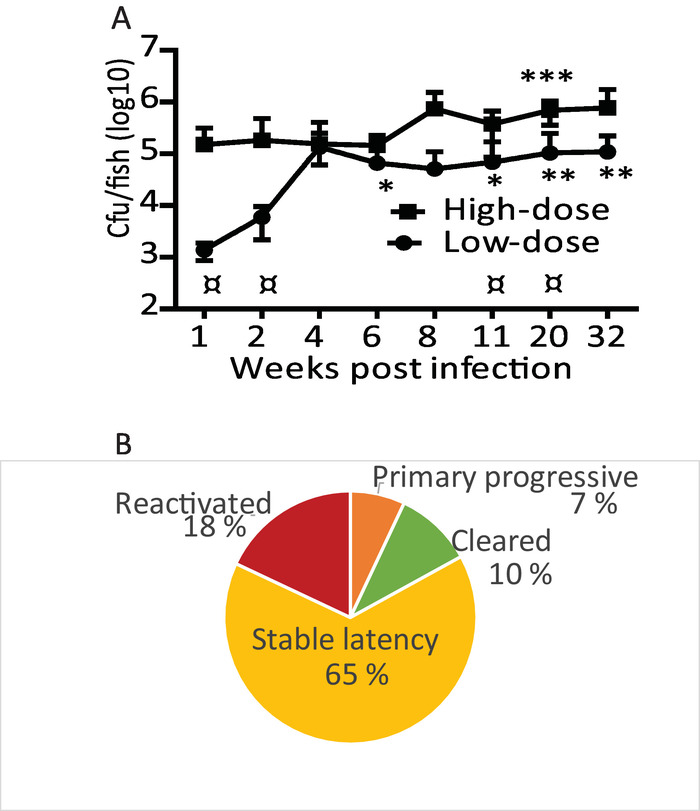
Figure 2: Injection of M. marinum into adult zebrafish causes a spectrum of disease states. (A) Zebrafish were injected with a low (34 ±15 CFU) or a high dose (2029 ±709 CFU) of M. marinum. Average loads for 5 fish (except 32 weeks high dose, n = 2) are shown with SD. Low-dose statistics: * p <0.05 compared with 1 week, ** p <0.05 compared with 1 and 2 week. High-dose statistics: *** p <0.05 compared with 1, 2, 8, 11 and 20 wk. ¤ low dose vs. high dose p<0.05. Modified from Parikka et al. 201219. (B) Typical distribution of disease outcomes within a wild-type zebrafish population infected with a low-dose of M. marinum. Please click here to view a larger version of this figure.
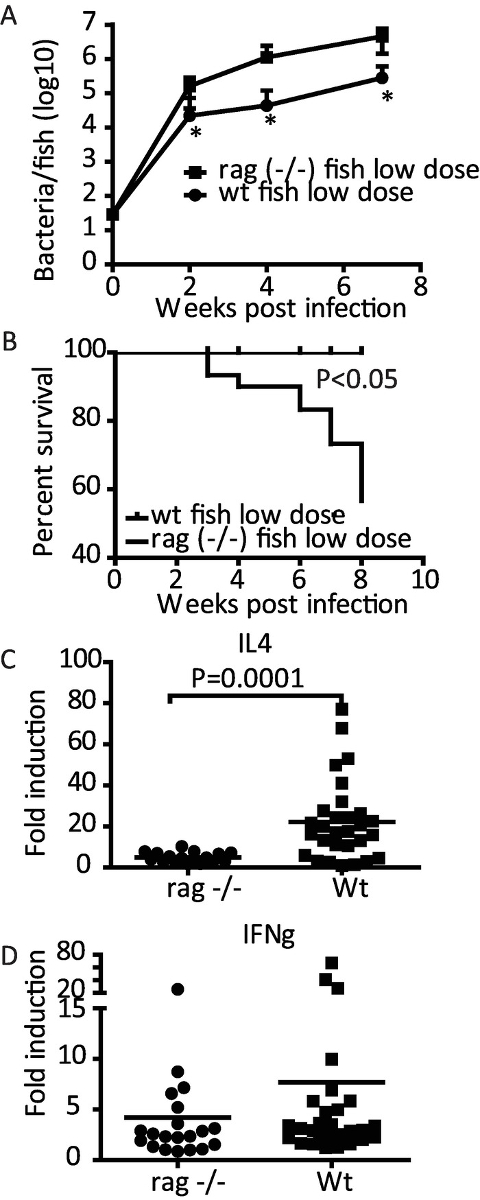
Figure 3: Adaptive immunity affects the course of mycobacterial infection in the adult zebrafish. Adult wild-type (wt) and rag1 (−/−) zebrafish were infected with a low dose (n = 30) of M. marinum. (A) The average mycobacterial loads were measured by qPCR at 2, 4, and 7 weeks post infection (wpi) (n = 10) *P<0.05. (B) The fish were euthanized upon the development of symptoms of the disease and survival plots were created. (C). The expression levels of il4 were measured at 4 wpi. (D) The expression levels of IFNγ were measured at 4 wpi. (A and B)modified from Parikka et al. 201219. (C and D) Modified from Hammarén et al. 201438. Please click here to view a larger version of this figure.
| Gene | Primer sequence | Annealing temperature |
| MMITS1 | F: CACCACGAGAAACACTCCAA | 65 |
| 16S–23SITS Locus AB548718 for M. marinum quantification | R: ACATCCCGAAACCAACAGAG | |
| loopern4 | F: TGAGCTGAAACTTTACAGACACAT | 61 |
| Expressed repetitive elements | R: AGACTTTGGTGTCTCCAGAATG | |
| il4 | GCAGGAATGGCTTTGAAGGG | 59.5 |
| ZDB-GENE-100204-1 | GCAGTTTCCAGTCCCGGTAT | |
| ifnγ1-2 | F: GGGCGATCAAGGAAAACGACCC, | 61 |
| ZDB-GENE-040629-1 | R: TAGCCTGCCGTCTCTTGCGT |
Table 1: Primer sequences and annealing temperatures. The sequences of the primers used and their optimized annealing temperatures. The primers for the M. marinum 16S-23S rRNA transcript have been optimized for a No-ROX qPCR kit and the other primers for a ROX including qPCR kit.
| Step | Time | Temperature |
| 1 | 3 min | 95 °C |
| 2 | 5 s | 95 °C |
| 3 | 10 s | 65 °C |
| 4 | 5 s | 72 °C (fluorescence detection) |
| 5 | Go to step 2. 39 times | |
| 6 | Melting curve analysis 55-95°C with 0.5°C intervals | |
| 7 | Forever | 4 °C |
Table 2: qPCR program for measuring M. marinum DNA. A qPCR protocol designed according to the manufacturer's instructions and optimized for measuring M. marinum DNA from zebrafish samples.
| Time | Temperature |
| 5 min | 25 °C |
| 30 min | 42 °C |
| 5 min | 85 °C |
| forever | 4 °C |
Table 3: cDNA synthesis program. Protocol for synthesizing cDNA from the extracted RNA of an infected zebrafish according to the manufacturer's instructions.
| Step | Time | Temperature |
| 1 | 30 s | 95 °C |
| 2 | 12 s | 95 °C |
| 3 | 30 s | Annealing °C |
| 4 | Go to step 2. for 39 times | |
| 5 | Melting curve analysis 65–95 °C with 0.5 °C intervals | |
| 6 | Forever | 4 °C |
Table 4: qPCR program for measuring host's gene expression. A qPCR protocol designed according to the manufacturer's instructions and optimized for measuring the expression of different zebrafish genes.
Discussion
Here we describe a qPCR-based application to measure mycobacterial loads from DNA extracted from experimentally infected adult zebrafish tissues. This application is based on primers designed around the 16S-23S rRNA internal transcribed spacer sequence40. The total mycobacterial load in a fish sample is estimated using a standard curve prepared from DNA extracted from a known number of cultured mycobacteria and assuming that one bacterium has one copy of its genome at any given moment. The detection limit of the M. marinum –qPCR is approximately 100 colony forming units18. A clear advantage of the method compared to traditional plating is that both active and non-dividing dormant bacteria can be detected. In addition, a common problem of contaminating growth on culture plates from zebrafish tissues is circumvented by this approach. However, as DNA is used as the template, it is possible that some of the copies measured can be derived from the DNA of bacteria that have died very recently. A significant advantage of the nucleic acid-based protocol is that as both DNA and RNA (as well as proteins, not described here) can be extracted from the same individual, the mycobacterial load of the individual can be combined with gene expression data of both the host and the bacteria.
The dosing of M. marinum is a critical determinant of the outcome of infection. The low M. marinum (~20–90 CFU) infection dose produces a spectrum of disease states with latency as the most common form. If the infection dose is in the order of thousands, a more progressive disease develops in the majority of individuals. As the natural infectious dose is known to be low in human tuberculosis41, using a low dose of M. marinum in the zebrafish model is likely to produce a more natural infection. To reach the correct dose, make sure to validate the relation between the OD600 and colony-forming units prior to starting any experiments. The normal variation of the plated infection doses is approximately 30% and is not considered a problem. However, it is important to verify that the entire 5 µL volume of the bacterial suspension remains inside the fish. Leaking of the injection solution will cause extra variation in the infection dose. In addition to the infection dose, the strain of the bacteria can affect the disease progression. It has been shown that the virulence of the different M. marinum strains can alter between the strains. The two most commonly used strains are ATCC927 (fish isolated strain used in these experiments) and the M strain. However, these strains differ greatly in their virulence. The human-isolated M strain develops a more progressive disease whereas the fish-isolated strain produces a milder disease well-suitable for studying the latent form of the mycobacterial infection33. The virulence of the bacteria within a strain may also alter if the bacteria is serially transferred from one culture to another, which can be prevented by taking fresh mycobacteria from a freezer stock often enough.
In vitro culturing of slowly dividing M. marinum without antibiotics is prone to contaminations. Therefore, the handling of the bacteria requires strict aseptic approach carried out in a laminar flow hood. Possible contamination in the culture can be detected as too high an OD600 value, odd-looking bacterial suspension or colonies. M. marinum colonies are normally fuzzy-edged, flat and matt white in color. M. marinum cultures are sensitive to light and they start producing yellow pigment when exposed to light. This yellow pigment can be used to distinguish M. marinum colonies from other bacteria after the infections. Contaminants can cause the fish to die soon after infection. Usually, the first symptoms of a low-dose infection do not appear before 3 weeks post infection and any mortalities before this time point are likely due to contamination in the bacterial suspension or trauma induced during injection.
Adult zebrafish are very sensitive to 3-aminobenzoic acid ethyl ester and do not survive if the exposure time is too long or the concentration of the anesthetic is too high. Therefore, when infecting, the zebrafish should be exposed to the anesthetic only for the minimum of time to achieve good anesthesia (~1–2 min). After infection, adult zebrafish are kept in groups at water temperature of 26–28 °C. If the temperature is higher or lower than this, it might affect the M. marinum growth rate and kinetics of the infection. Also, the tank water quality (microbiological quality, salt concentration, pH, oxygen saturation) is an important part ensuring a successful experiment. Health monitoring of the fish needs to be carried out daily and fish that show symptoms of infection need to be removed from the group and euthanized. If the fish die in the tank, the other fish may become re-infected through the gut, which will affect the progression of infection.
The commonly used zebrafish larval model of tuberculosis is applicable to study innate immunity, which directs the experiments to a limited subset of host-microbe interactions. The use of adult zebrafish makes it possible to study both innate and adaptive immune responses in a versatile model18,32,39. Adult zebrafish presents itself as a convenient vertebrate model to study the entire spectrum of tuberculosis. With a low-dose exposure of M. marinum, 7% of the fish population develop primary progressive disease, 10% are able to sterilize the infection, 65% develop latent disease and in 18% spontaneous reactivation occurs (Figure 2). The disease spectrum closely resembles that seen in human tuberculosis, in which the vast majority develop a latent disease, approximately 4–14% produce primary active infection within the first five years after the infection42, 10–20% of heavily exposed persons seem to be able to sterilize the infection43 and 5–10% of the latent infections reactivate44. Differences in hosts' genetics affects the susceptibility to tuberculosis and the disease progression45. This is also seen in the zebrafish population that is genetically very heterogeneous unlike many other laboratory animals46,47. The natural genetic variation makes it a highly applicable model in the search for optimal immune responses in this multifactorial disease.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
This work has been supported by the Finnish Cultural Foundation (H.L.), Tampere Tuberculosis Foundation (H.L., L.-M.V., M.M.H., M.P.), Foundation of the Finnish Anti-Tuberculosis Association (Suomen Tuberkuloosin Vastustamisyhdistyksen Säätiö) (H.L., M.M.H., M.P.), Sigrid Jusélius Foundation (M.P.), Emil Aaltonen Foundation (M.M.H.), Jane and Aatos Erkko Foundation (M.P.) and Academy of Finland (M.P.). Leena Mäkinen, Hanna-Leena Piippo and Jenna Ilomäki are acknowledged for their technical assistance. The authors acknowledge the Tampere Zebrafish Laboratory for their service.
Materials
| Mycobacterium marinum | American Type Culture Collection | ATCC 927 | |
| Middlebrock 7H10 agar | BD, Thermo Fisher Scientific | 11799042 | |
| Middlebrock OADC enrichment | BD, Thermo Fisher Scientific | 11718173 | |
| Middlebrock 7H9 medium | BD, Thermo Fisher Scientific | 11753473 | |
| Middlebrock ADC enrichment | BD, Thermo Fisher Scientific | 11718173 | |
| Tween 80 | Sigma-Aldrich | P1754 | |
| Glycerol | Sigma-Aldrich | G5516-500ML | |
| GENESYS20 Spectrophotometer | Thermo Fisher Scientific | ||
| Phosphate buffered saline tablets (PBS) | Sigma-Aldrich | P4417-50TAB | |
| Phenol red | Sigma-Aldrich | P3532 | |
| 27G needle | Henke Sass Wolf | 4710004020 | |
| 1 ml syringe | Henke Sass Wolf | 4010.200V0 | |
| Omnican 100 30G insulin needle | Braun | 9151133 | |
| 3-aminobenzoic acid ethyl ester (pH 7.0) | Sigma-Aldrich | A5040 | |
| 1.5 ml homogenization tube | Qiagen | 13119-1000 | |
| 2.8 mm ceramic beads | Qiagen | 13114-325 | |
| Ethanol, ETAX Aa | Altia | ||
| 2-propanol | Sigma-Aldrich | 278475 | |
| Chloroform | VWR | 22711.290 | |
| Guanidine thiocyanate | Sigma-Aldrich | G9277 | FW 118.2 g/mol |
| Sodium citrate | Sigma-Aldrich | 1613859 | FW 294.1 g/mol |
| Tris (free base) | Sigma-Aldrich | TRIS-RO | FW 121.14 g/mol |
| TRI reagent | Molecular Research Center | TR118 | Guanidine thiocyanate-phenol solution |
| PowerLyzer24 homogenizator | Qiagen | ||
| Sonicator m08 | Finnsonic | ||
| Nanodrop 2000 | Thermo Fisher Scientific | ||
| SENSIFAST No-ROX SYBR, Green Master Mix | Bioline | BIO-98005 | |
| qPCR 96-well plate | BioRad | HSP9601 | |
| Optically transparent film | BioRad | MSB1001 | |
| C1000 Thermal cycler with CFX96 real-time system | BioRad | ||
| RNase AWAY | Thermo Fisher Scientific | 10666421 | decontamination reagent eliminating RNases |
| DNase I | Thermo Fisher Scientific | EN0525 | |
| Reverse Transcription Master Mix | Fluidigm | 100-6298 | |
| SsoFast Eva Green master mix | BioRad | 172-5211 |
Referenzen
- Zhao, S., Huang, J., Ye, J. A fresh look at zebrafish from the perspective of cancer research. Journal of Experimental & Clinical Cancer Research. 34, 80 (2015).
- Bournele, D., Beis, D. Zebrafish models of cardiovascular disease. Heart failure reviews. 21 (6), 803-813 (2016).
- Torraca, V., Mostowy, S. Zebrafish Infection: From Pathogenesis to Cell Biology. Trends in cell biology. 28 (2), 143-156 (2018).
- Varela, M., Figueras, A., Novoa, B. Modelling viral infections using zebrafish: Innate immune response and antiviral research. Antiviral Research. 139, 59-68 (2017).
- Goody, M. F., Sullivan, C., Kim, C. H. Studying the immune response to human viral infections using zebrafish. Developmental and comparative immunology. 46 (1), 84-95 (2014).
- Thisse, C., Zon, L. I. Organogenesis–heart and blood formation from the zebrafish point of view. Science. 295 (5554), 457-462 (2002).
- Eimon, P. M., Rubinstein, A. L. The use of in vivo zebrafish assays in drug toxicity screening. Expert Opinion on Drug Metabolism & Toxicology. 5 (4), 393-401 (2009).
- Sukardi, H., Chng, H. T., Chan, E. C. Y., Gong, Z., Lam, S. H. Zebrafish for drug toxicity screening: bridging the in vitro cell-based models and in vivo mammalian models. Expert Opinion on Drug Metabolism & Toxicology. 7 (5), 579-589 (2011).
- Wittamer, V., Bertrand, J. Y., Gutschow, P. W., Traver, D. Characterization of the mononuclear phagocyte system in zebrafish. Blood. 117 (26), 7126-7135 (2011).
- Harvie, E. A., Huttenlocher, A. Neutrophils in host defense: new insights from zebrafish. Journal of leukocyte biology. 98 (4), 523-537 (2015).
- Yoshida, N., Frickel, E., Mostowy, S. Macrophage-Microbe interactions: Lessons from the Zebrafish Model. Frontiers in Immunology. 8, 1703 (2017).
- Langenau, D. M., et al. In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish. Proceedings of the National Academy of Sciences of the United States of America. 101 (19), 7369-7374 (2004).
- Lewis, K. L., Del Cid, N., Traver, D. Perspectives on antigen presenting cells in zebrafish. Developmental and comparative immunology. 46 (1), 63-73 (2014).
- Hu, Y., Xiang, L., Shao, J. Identification and characterization of a novel immunoglobulin Z isotype in zebrafish: Implications for a distinct B cell receptor in lower vertebrates. Molecular immunology. 47 (4), 738-746 (2010).
- Danilova, N., Bussmann, J., Jekosch, K., Steiner, L. A. The immunoglobulin heavy-chain locus in zebrafish: identification and expression of a previously unknown isotype, immunoglobulin Z. Nature immunology. 6 (3), 295-302 (2005).
- Zapata, A., Diez, B., Cejalvo, T., Frias, C. G., Cortes, A. Ontogeny of the immune system of fish. Fish & shellfish. 20 (2), 126-136 (2006).
- Traver, D., Paw, B. H., Poss, K. D., Penberthy, W. T., Lin, S., Zon, L. I. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nature immunology. 4 (12), 1238-1246 (2003).
- Hammaren, M. M., et al. Adequate Th2-Type Response Associates with Restricted Bacterial Growth in Latent Mycobacterial Infection of Zebrafish. Plos Pathogens. 10 (6), e1004190 (2014).
- Parikka, M., et al. Mycobacterium marinum Causes a Latent Infection that Can Be Reactivated by Gamma Irradiation in Adult Zebrafish. PLoS Pathog. 8 (9), 1-14 (2012).
- Tobin, D. M., et al. Host Genotype-Specific Therapies Can Optimize the Inflammatory Response to Mycobacterial Infections. Cell. 148 (3), 434-446 (2012).
- Lesley, R., Ramakrishnan, L. Insights into early mycobacterial pathogenesis from the zebrafish. Current opinion in microbiology. 11 (3), 277-283 (2008).
- Berg, R. D., Ramakrishnan, L. Insights into tuberculosis from the zebrafish model. Trends in molecular medicine. 18 (12), 689-690 (2012).
- Ordonez, A. A., et al. Mouse model of pulmonary cavitary tuberculosis and expression of matrix metalloproteinase-9. Disease Models & Mechanisms. 9 (7), 779-788 (2016).
- Kramnik, I., Beamer, G. Mouse models of human TB pathology: roles in the analysis of necrosis and the development of host-directed therapies. Seminars in Immunopathology. 38 (2), 221-237 (2016).
- Manabe, Y. C., et al. The aerosol rabbit model of TB latency, reactivation and immune reconstitution inflammatory syndrome. Tuberculosis. 88 (3), 187-196 (2008).
- Pena, J. C., Ho, W. Monkey Models of Tuberculosis: Lessons Learned. Infection and immunity. 83 (3), 852-862 (2015).
- Cadena, A. M., Fortune, S. M., Flynn, J. L. Heterogeneity in tuberculosis. Nature Reviews Immunology. 17 (11), 691-702 (2017).
- Stinear, T. P., et al. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome research. 18 (5), 729-741 (2008).
- Swaim, L. E., Connolly, L. E., Volkman, H. E., Humbert, O., Born, D. E., Ramakrishnan, L. Mycobacterium marinum infection of adult zebrafish causes caseating granulomatous tuberculosis and is moderated by adaptive immunity. Infection and immunity. 74 (11), 6108-6117 (2006).
- Myllymaki, H., Bauerlein, C. A., Ramet, M. The Zebrafish Breathes new Life into the Study of Tuberculosis. Frontiers in Immunology. 7, 196 (2016).
- Luukinen, H., et al. Priming of Innate Antimycobacterial Immunity by Heat-killed Listeria monocytogenes Induces Sterilizing Response in Adult Zebrafish Tuberculosis Model. Disease Models and Mechanisms. 11, (2018).
- Sar, A. M., Abdallah, A. M., Sparrius, M., Reinders, E., Vandenbroucke-Grauls, C., Bitter, W. Mycobacterium marinum strains can be divided into two distinct types based on genetic diversity and virulence. Infection and immunity. 72 (11), 6306-6312 (2004).
- Madigan, M., Martinko, J. . Brock Biology of Microorganisms. , (2016).
- Nüsslein-Volhard, C., Dahm, R. . Zebrafish:a practical approach. , (2002).
- Vanhauwaert, S., et al. Expressed Repeat Elements Improve RT-qPCR Normalization across a Wide Range of Zebrafish Gene Expression Studies. Plos One. 9 (10), e109091 (2014).
- Hammaren, M. M., et al. Adequate Th2-Type Response Associates with Restricted Bacterial Growth in Latent Mycobacterial Infection of Zebrafish. Plos Pathogens. 10 (6), e1004190 (2014).
- Oksanen, K. E., et al. An adult zebrafish model for preclinical tuberculosis vaccine development. Vaccine. 31 (45), 5202-5209 (2013).
- Roth, A., Fischer, M., Hamid, M. E., Michalke, S., Ludwig, W., Mauch, H. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. Journal of clinical microbiology. 36 (1), 139-147 (1998).
- Rajararna, M. V. S., Ni, B., Dodd, C. E., Schlesinger, L. S. Macrophage immunoregulatory pathways in tuberculosis. Seminars in immunology. 26 (6), 471-485 (2014).
- Vynnycky, E., Fine, P. The natural history of tuberculosis: the implications of age-dependent risks of disease and the role of reinfection. Epidemiology and infection. 119 (2), 183-201 (1997).
- Cobat, A., et al. Two loci control tuberculin skin test reactivity in an area hyperendemic for tuberculosis. Journal of Experimental Medicine. 206 (12), 2583-2591 (2009).
- Delogu, G., Goletti, D. The Spectrum of Tuberculosis Infection: New Perspectives in the Era of Biologics. Journal of Rheumatology. 41, 11-16 (2014).
- Abel, L., et al. Genetics of human susceptibility to active and latent tuberculosis: present knowledge and future perspectives. Lancet Infectious Diseases. 18 (3), E75 (2018).
- Guryev, V., et al. Genetic variation in the zebrafish. Genome research. 16 (4), 491-497 (2006).
- Brown, K. H., et al. Extensive genetic diversity and substructuring among zebrafish strains revealed through copy number variant analysis. Proceedings of the National Academy of Sciences of the United States of America. 109 (2), 529-534 (2012).

