Hybrid Clear/Blue Native Electrophoresis for the Separation and Analysis of Mitochondrial Respiratory Chain Supercomplexes
Summary
Here we present a protocol to extract, resolve and identify mitochondrial supercomplexes which minimizes exposure to detergents and Coomassie Blue. This protocol offers an optimal balance between resolution, and preservation of enzyme activities, while minimizing the risk of losing labile protein-protein interactions.
Abstract
Complexes of the oxidative phosphorylation machinery form supramolecular protein arrangements named supercomplexes (SCs), which are believed to confer structural and functional advantages to mitochondria. SCs have been identified in many species, from yeast to mammal, and an increasing number of studies report disruption of their organization in genetic and acquired human diseases. As a result, an increasing number of laboratories are interested in analyzing SCs, which can be methodologically challenging. This article presents an optimized protocol that combines the advantages of Blue- and Clear-Native PAGE methods to resolve and analyze SCs in a time-effective manner. With this hybrid CN/BN-PAGE method, mitochondrial SCs extracted with optimal amounts of the mild detergent digitonin are exposed briefly to the anionic dye Coomassie Blue (CB) at the beginning of the electrophoresis, without exposure to other detergents. This short exposure to CB allows to separate and resolve SCs as effectively as with traditional BN-PAGE methods, while avoiding the negative impact of high CB levels on in-gel activity assays, and labile protein-protein interactions within SCs. With this protocol it is thus possible to combine precise and rapid in gel activity measurements with analytical techniques involving 2D electrophoresis, immuno-detection, and/or proteomics for advanced analysis of SCs.
Introduction
Mitochondria produce energy through oxidative phosphorylation, where respiratory complexes I-II-III-IV oxidize substrates and transfer electrons to oxygen, generating a gradient that allows phosphorylation of ADP by the ATP synthase (CV). In the past years, extensive studies have shown that respiratory chain complexes are not solely incorporated in a linear way in the inner mitochondrial membrane, but are also organised into supercomplexes (SCs) arrangements1,2. In mammalian mitochondria, SCs exist in varying stoichiometries: CI/CIII2/CIV1-4 (which is named the respirasome, and which is capable of NADH:O2 oxidoreduction in vitro)2, CI/CIII2, and CIII2/CIV1-2 3,4. Furthermore, respiratory complexes are distributed under different ratios between their free form and SCs arrangements. Therefore, it is estimated that 85%–100% of CI, 55%–65% of CIII, and 15%–25% of CIV are found in SCs4. These supramolecular structures are thought to decrease ROS production, stabilize or assist in the assembly of individual complexes, regulate respiratory chain activity, and prevent protein aggregation in the protein rich inner mitochondrial membrane5,6,7,8. Their remodelling ability upon variation in energy demand and their importance in the pathogenesis of diseases is being investigated in several labs3,7,9,10,11,12,13,14. Studies have demonstrated that pathological changes in SCs assembly are present in a variety of disorders, including, but not limited to, genetic defect in cardiolipin synthesis15, heart failure16, ischemia-reperfusion17, diabetes12, and aging18.
Native electrophoresis and immunodetection are widely used in SCs studies to resolve OXPHOS complexes quaternary arrangements2,19,20,21. Native electrophoresis can further be combined with specific in gel activity assays or 2D-SDS PAGE to enable precise molecular determination of the various SCs assemblies1,19. The ability to study SCs is critically dependent on the extraction conditions, including type and concentration of detergent used, ionic strength and pH, as well as on electrophoretic migration conditions, which comprise buffer composition, presence of CB, gel size, and acrylamide percentage2.
Protocols and SCs band resolution vary greatly among papers, making comparison between studies difficult and adaptation of methods challenging22. Therefore, this paper proposes a robust and optimal protocol to extract SCs from isolated mitochondria of different sources with the non-ionic detergent digitonin, and to resolve high molecular weight SCs bands. The optimized detergent concentration, composition of the extraction buffer, and the absence of Coomassie Blue in sample preparation minimize disruption of protein complexes. This protocol (see Figure 1 for an overview) combines CN-PAGE and BN-PAGE for optimal SCs assemblies resolution on large gel, and is compatible with in-gel activity assays allowing better visualization of reactive bands, along with the use of immunodetection for a detailed analysis SCs arrangements and composition.
Protocol
1. SC Extraction
- Prepare 100 mL of extraction buffer (see Table 2) by dissolving EDTA in water. Increase pH with KOH until completely dissolved, then adjust pH to 7.5 with HCl. Add remaining components to the solution, complete to final volume with water, and keep on ice. In a tube, dissolve digitonin in extraction buffer to make a 10% stock solution, vortex thoroughly until completely dissolved, and keep on ice.
NOTE: Extraction buffer can be prepared in advance and kept at 4 °C for 2 months maximum. If wavy bands start to appear at the bottom of the gel, it means extraction buffer is too old. When preparing the 10% digitonin solution, prepare a stock volume counting 500 µL per sample if using mouse liver mitochondria. Digitonin solubility varies upon provenance and product lot (see Table of Materials). - Mitochondria isolated from animal tissue (mouse heart, muscle, liver; rat heart) or cells (human fibroblast) using standard protocols23,24,25 can be used for the extraction of SCs. Once mitochondria are obtained, quantify protein content using the bicinchoninic acid assay kit according to the manufacturer’s recommendations. Supplement isolated mitochondria with proteases and phosphatases inhibitors at this step as needed.
NOTE: SCs can be extracted on either fresh or thawed mitochondria. It is recommended to extract SCs from all samples at the same time, to insure they are treated with the same batches of solutions and under the same conditions. - Based on the mitochondrial protein concentration obtained, and the final digitonin/protein ratio desired, calculate the volume of stock digitonin solution and extraction buffer required as per Table 1. For SC extraction, add 1 µL of extraction buffer (Table 2) containing digitonin for each 10 µg of mitochondrial protein. The digitonin/protein ratio can vary from 2 to 8 g/g. A digitonin titration should always be performed for each new type of sample used (See Figure 2 for an example).
- Pellet mitochondria, in a 1.5 mL tube by centrifugation at 16,000 x g for 10 min at 4 °C.
- Discard supernatant and re-suspend the mitochondrial pellet in the calculated volume of ice-cold extraction buffer containing digitonin. Place tubes on a mini tube rotator and incubate for 30 min at 4 °C at a medium rotation speed. Make sure that samples are getting properly mixed.
- Centrifuge samples at 20,400 x g for 45 min at 4 °C to remove insolubilized fragments.
- Transfer supernatant in a new tube on ice and quantify proteins. This fraction represents the respiratory supercomplexes extract. If electrophoresis is not performed on the same day, store samples at -80 °C.
NOTE: 1) Avoid freeze/thaw cycles of the extract, as this disrupts higher molecular arrangements of SCs. Aliquot sample before the first freeze/thaw cycle if needed. 2) To perform a standard BN-PAGE experiment, CB should be added to the SCs extract at this step. CB should be added in a 1g/8g ratio relative to the amount of detergent used.
2. Gradient gel Casting and Electrophoresis
- Prepare 3x gradient buffer and acrylamide stocks for making the gradient gel, aliquot, and store at -20 °C (see Table 3).
- Prepare anode and cathode buffers and keep at 4 °C (see Table 5).
- Open the casting chamber and place an outer glass plate (20 cm x 22 cm) in the chamber. Position one set of spacers (1.5 mm) using the alignment card to ensure they are seated firmly against the side and corners of the chamber. Place an inner glass plate (20 cm x 20 cm) on top of the spacers (this forms the gel sandwich), and put a plastic separation sheet on top of the glass plate.
- Repeat step 2.3 until the desired number of gels to cast is reached. For this protocol, 4 gels are casted. The casting chamber system we use (see Table of Materials) allows the casting of a maximum of 10 gels at a time. Take-up the remaining space in the chamber by first adding as many acrylic blocks as needed, and then glass plates if needed.
NOTE: The montage has to be tightly sealed; there should be no space between the gel sandwiches in the chamber. - Place a strip of parafilm in the groove before seating the gasket firmly in the gasket notch. Place the sealing plate on the chamber and tighten all 6 screws. Stand the casting chamber.
- Place gradient former on a stir plate with a magnetic stirrer in the “light” mixing chamber. Connect the tubing of casting chamber to the gradient former, secure the tubing in the cassette of the peristaltic pump, and make sure the stopcock of the gradient former is closed.
- To cast 4 gels, prepare 60 mL of 4% and 60 mL of 12% gel solutions (see Table 4) in an Erlenmeyer flask and swirl thoroughly to mix. Pour 60 mL of 4% gel solution in the “light” mixing chamber, and 60 mL of 12% in the “heavy” reservoir chamber of the gradient former. Set stir speed of the stir plate at 350 rpm. Open the stopcock and turn on the pump at 35 rpm.
- Once the light fraction is lower than the heavy fraction, pause the pump and open the valve stem between “light” and “heavy” reservoirs, let fractions volume equilibrate, and restart the pump.
NOTE: It is important that no bubbles enter the system and get trapped between glass plates. If this happens, undo montage, wash, and redo. - Once the gradient gel is completely poured, stop the pump, and overlay water (about 1 mL) on each gel sandwich to prevent drying of the gel. Let polymerize for 2 h.
- Prepare 25 mL stacking gel in Erlenmeyer and swirl to mix thoroughly. Remove water and insert 15 well combs in each gel sandwich. Pour stacking gel and let polymerize for 2 h.
NOTE: Gels can be casted and kept at 4 °C for 1 week. - Insert gel in sandwich clamps and remove comb. With the short glass plate facing down, insert the gel sandwich in the cooling core. Repeat on the other side, and place core in the electrophoresis tank.
- Pour 300 mL of Blue Cathode Buffer in inner chamber of the electrophoresis tank. Pour 2 L of Anode Buffer in outer chamber of electrophoresis tank.
NOTE: The electrode has to be submerged in cathode buffer, which requires approximately 300 mL. - Load between 75 μg and 175 μg of protein per well. Run gel at 150 V for 1.5 h (or until samples have all entered the gradient gel) in cold room (4 °C).
NOTE 1: 1) A minimum of 75 μg per well is required for good resolution of in-gel activity bands. Loading over 175 μg of protein will lead to a loss of clear bands due to excessive enzyme activity. 2) Replicates of the sample should be loaded in separate wells to enable parallel determination of in-gel activities and immunoblot analysis of OXPHOS complexes. Parallel determination of IGA for CI, CII, CIV and CV requires a minimum of 300 µg. Parallel immunoblot analysis of CI, CII, CII, CIV and CV requires a minimum of 375 µg. - Remove Blue Cathode Buffer with pipet or vacuum, replace by 300 mL Coomassie Blue-free Cathode Buffer, and run gel at 200 V overnight (16–20 h) in cold room (4 °C). Proceed to step 3 or 4 for in-gel activity measurement or immunoblotting.
3. In-gel Activity for Complexes I, II, IV and CV
- Before the end of electrophoresis, prepare in-gel activity buffers according to Table 6, and keep in the dark at RT. 20 mL of in-gel activity buffer is sufficient for 3 sample lanes.
NOTE: This CV in-gel activity assay is based on the reverse activity of ATPsynthase (i.e., ATP hydrolysis), and uses calcium as a co-factor, which precipitates in the gel. Calcium is less harmful than the lead used in other protocols. Furthermore, the use of this protocol does not require a pre-activation/conditioning of the gel23. - Stop electrophoresis and retrieve gel. Cut lanes, if necessary, and transfer gel lanes in plastic bags (3 sides cut, and plastic bag opened like a book). Seal 2 of the 3 sides with a heat sealer.
NOTE: To compare the composition of SC bands between experimental groups, it is recommended to run the same samples in replicates on the same gel. Cut lanes to incubate each replicate in different in-gel activity buffers (CI, II, IV, V). To confirm the specificity of the assays, additional replicates can be prepared to run in gel activities in presence of specific respiratory chain inhibitors. - For 3 experimental samples (i.e. 3 wells), add 20 mL of in-gel activity buffer, remove bubbles, and seal 4th side of plastic bag.
NOTE: Add inhibitors in negative control experiments if performed: CI: Rotenone 1 µM; CII: Sodium Malonate 10 mM; CIII: Antimycin-A 8 µM; CIV: KCN 0.6 mM; CV: Oligomycin 0.5 µM. - Incubate gel lanes at 37 °C in the dark and check every 15 min. Incubation time varies depending on the amount of protein and complexes. CI will react faster than CIV or CV. Optimal staining usually occurs after 2 h for CI, 4 h for CIV and 6 h for CII and CV.
- Rinse gel lanes in water to stop reaction, and image on a white background for CI, CII, CIV, or black background for CV.
NOTE: Gels can be kept in plastic bags at RT or 4 °C for several months.
4. Immunoblotting
- Prepare transfer buffer according to Table 7, and keep at RT. Prepare TBST and keep at RT.
- Place the entire gel, or selected lanes, in a container and add transfer buffer supplemented with SDS (0.25% final in transfer buffer). Place container on rocker and incubate for 1 h.
- Cut PVDF membrane (size corresponding to the size of the gel) and activate in 20 mL of methanol under agitation for 2 min. Replace by 20 mL transfer buffer and place under agitation for 2 min.
- Prepare transfer sandwich, from bottom to top, making sure there is no bubble between gel and activated PVDF membrane: clear side of cassette / black sponge / blotting paper / membrane / gel / blotting paper / black sponge / black side of cassette. Close and lock the cassette.
- Place transfer sandwich in transfer tank, with clear side of the sandwich facing the red side of the electrode, and pour transfer buffer to immerge the gel. Connect cooling system to the transfer tank and set at 4 °C. Connect to the power supply, set at 40 mA, and run for 24 h.
- Retrieve membranes, block for 1 h in 5% BSA in TBST, and incubate in primary antibody solution prepared in 5% BSA in TBST overnight at 4 °C.
NOTE: See Table 8 for antibodies used. - Rinse membranes in TBST 3x for 10 min each.
- Incubate membranes in secondary antibody solutions prepared in 5% BSA in TBST for 2 h at room temperature.
- Rinse membranes in TBST 3x for 10 min each.
- Add chemiluminescent solution to membranes and image.
5. Analysis
- In-gel activity assay images or immunoblots can be used to analyze SCs. To analyze composition of bands, align replicates and validate which complex reacted positively for each given band.
- To analyze the distribution of complexes, in various supramolecular assemblies, open images in ImageJ and use the Gel analysis tool (see Figure 5 for an example).
- Select lanes with the rectangle tool, and plot lanes.
- Draw lines to close the area under the curve of each bands of interest and click on each area with the wand tool to generate a Table containing the area under the curve values.
- To calculate the distribution of the complex, report the values for each band relative to that of the monomer.
Representative Results
Figure 2 shows results from a digitonin titration experiment aimed to identify the proper amount of digitonin required for the extraction of SCs. This amount will vary depending on the tissue/cell type and whether the sample was frozen or not. For this experiment, a CIV in-gel activity was performed to visualize SCs isolated from fresh mouse liver mitochondria. Ratios from 2/1 to 10/1 g digitonin/g of protein were tested. The optimal amount of digitonin for this sample is 4 g/g, as it provides a good resolution of monomeric CIV, and high molecular weight SCs. At a lower ratio, bands are not clear and resolve into a smear during electrophoresis, whereas the use of higher ratio of digitonin leads to disruption of high molecular weight SC.
Figure 3 and Figure 4 show the results of a complete experiment performed on a preparation of mouse liver mitochondria extracted with 4 g digitonin /g protein. Proteins were separated using hybrid BN/CN-PAGE, standard BN-PAGE, or CN-PAGE. All three gels were casted at the same time and lanes were loaded with replicates of the same sample. Following electrophoresis, individual lanes were cut and processed for in gel activity measurement (CI, CII, CIV and CV on Figure 3) and immunoblotting (CI, CII, CIII, CIV, CV on Figure 4).
Addition of CB either momentarily in cathode buffer (i.e. hybrid CN/BN-PAGE) or in sample and cathode buffer throughout electrophoresis (i.e. BN-PAGE), considerably improves the mobility and resolution of SC bands, and individual respiratory complexes compared to CN-PAGE (Figure 3). Bands are easily distinguishable with the hybrid technique or BN-PAGE after in-gel activity for CIV, whereas in the same sample resolved by CN-PAGE, SCs and monomeric CIV reactive bands cannot be identified.
Figure 3 and Figure 4 show that the resolution and banding pattern of OXPHOS monomers and supramolecular assemblies is qualitatively comparable between hybrid CN/BN-PAGE and BN-PAGE. However, notable differences exist. First, the electrophoretic mobility of OXPHOS complexes is slightly reduced when proteins are separated using hybrid CN/BN-PAGE conditions vs standard BN-PAGE, due to reduced amount of CB. This mobility shift is greater for CIV monomers, followed by CV monomers, and CI (Figure 3 and Figure 4). Second, the blue background is lower in the hybrid CN/BN-PAGE compared to BN-PAGE (Figure 3, left lanes). As a result, high background levels following BN-PAGE completely masks the in-gel activity staining for CII, and enhances the background noise associated with the activity of CIV dimers (Figure 3). Third, the activity of CV is higher when samples are run under hybrid CN/BN-PAGE conditions compared to BN-PAGE (Figure 3), due to the reduced amount of CB, which is known to interfere with CV catalytic activity.26 CN/BN-PAGE also allows better preservation of CV supramolecular assemblies, as shown by a greater proportion of total CV activity being associated with CV dimers (Figure 3). Moreover, CV oligomers are visible under CN/BN-PAGE, while they are completely dissociated under BN-PAGE conditions. Interestingly, distinct bands displaying CV activity are also observed between CV monomers and dimers, when samples are run under CN/BN-PAGE (Figure 2).
Figure 5 shows a representative analysis of OXPHOS complex distribution in supramolecular assemblies. The image shows CI in gel activity of samples obtained from 4 distinct healthy mouse liver mitochondria preparations. Densitometry analysis allows to measure the area under the curve of CI-reactive bands, and to present the relative distribution of C1 activity in the monomeric (I1) and supramolecular forms (I1III2, I1III2IV1, I1III2IVn). Similar analysis can be performed following immunoblot.
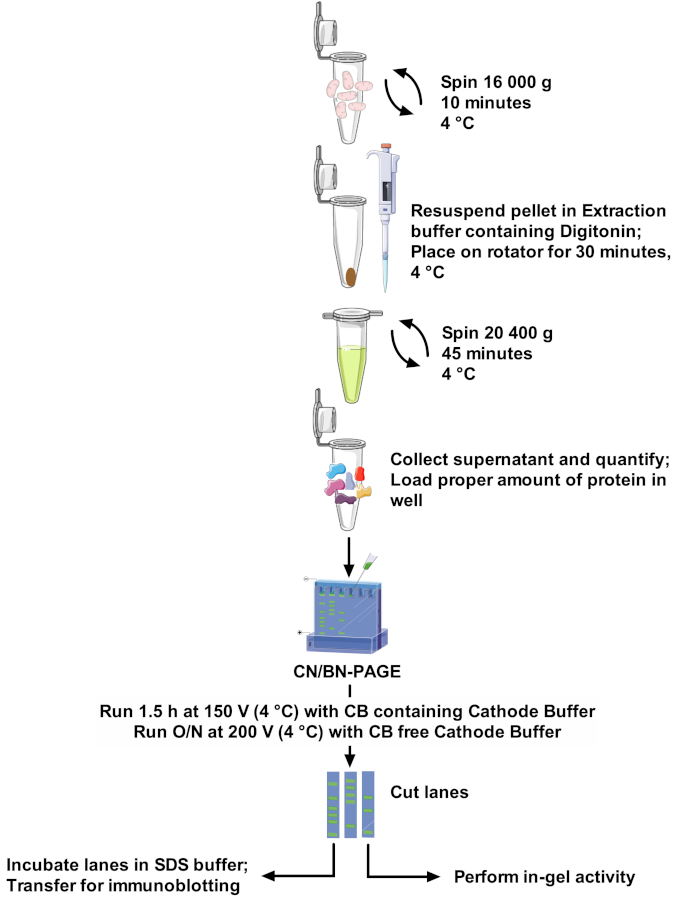
Figure 1: Assay workflow. Please click here to view a larger version of this figure.
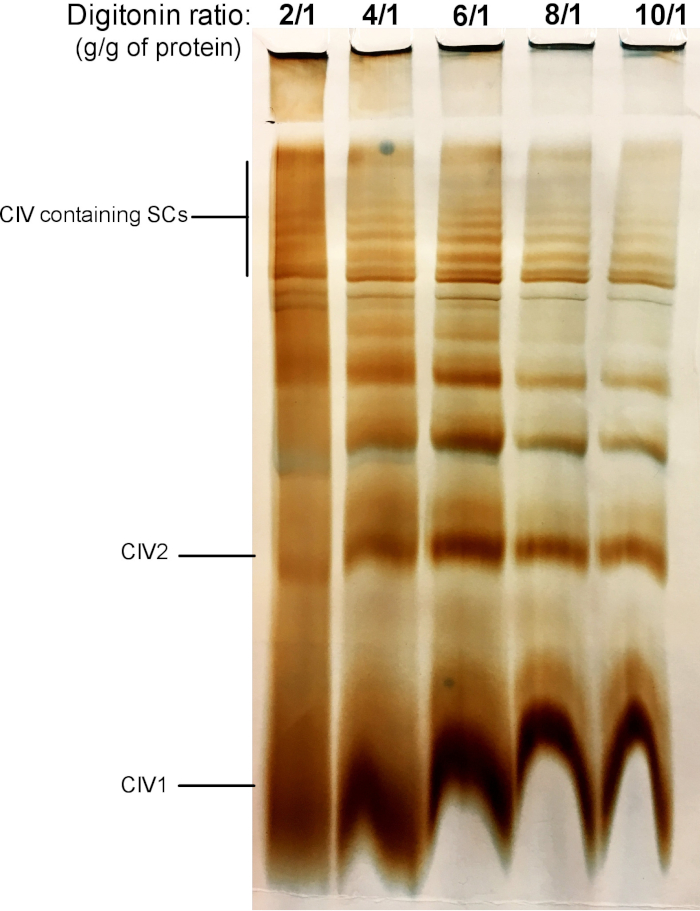
Figure 2: Digitonin titration to extract supercomplexes from fresh mouse liver mitochondria. This example shows aliquots of mouse liver mitochondria, isolated from one animal that was treated with increasing amounts of digitonin to extract respiratory supercomplexes. Samples were then resolved by hybrid CN/BN PAGE, and in-gel activity of CIV was determined. CIV1: complex IV monomers; CIV2: Complex IV dimers; SC: supercomplexes. Please click here to view a larger version of this figure.
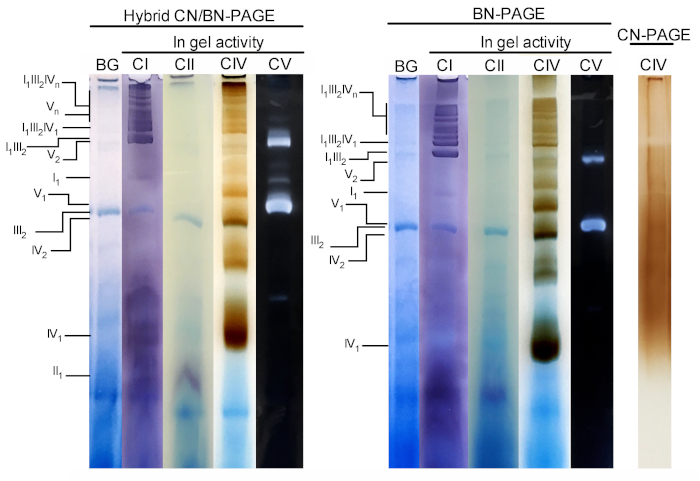
Figure 3: In-gel activity of OXPHOS complexes following hybrid CN/BN-PAGE, BN-PAGE or CN-PAGE. Liver mitochondria isolated from one mouse were treated with digitonin (4 g/g ratio digotonin/protein) to extract respiratory supercomplexes. Aliquots of this sample were then loaded on multiple wells in three distinct gels and submitted to CN/BN-PAGE, BN-PAGE or CN-PAGE. Each replicate lane within each gel was then cut and immediately used for in-gel activity assays (labeled CI, CII, CIV and CV). One lane was used as control to show background (labeled BG) staining with Coomassie Blue. OXPHOS complexes and supramolecular assemblies are identified using the standard nomenclature, with numbers in indices indicating the molecular stoichiometry of each OXPHOS complex. It should be noted that the position of CIII-containing supramolecular assemblies is based on immunodetection since in-gel activity for CIII was not performed in this particular experiment. Please click here to view a larger version of this figure.
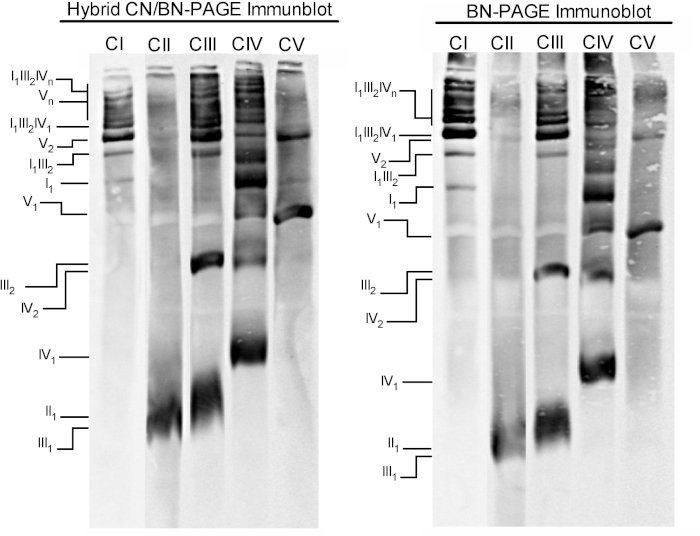
Figure 4: Immunoblot analysis of OXPHOS complexes following hybrid CN/BN-PAGE or BN-PAGE. Replicates from the experiments described in the Figure 3 legend were electro-transferred on a single membrane. After transfer, individual lanes were cut and incubated with specific antibodies recognizing CI, CII, CIII, CIV, and CV. OXPHOS complexes and supramolecular assemblies are identified using the standard nomenclature, with numbers in indices indicating the molecular stoichiometry of each OXPHOS complex. Please click here to view a larger version of this figure.
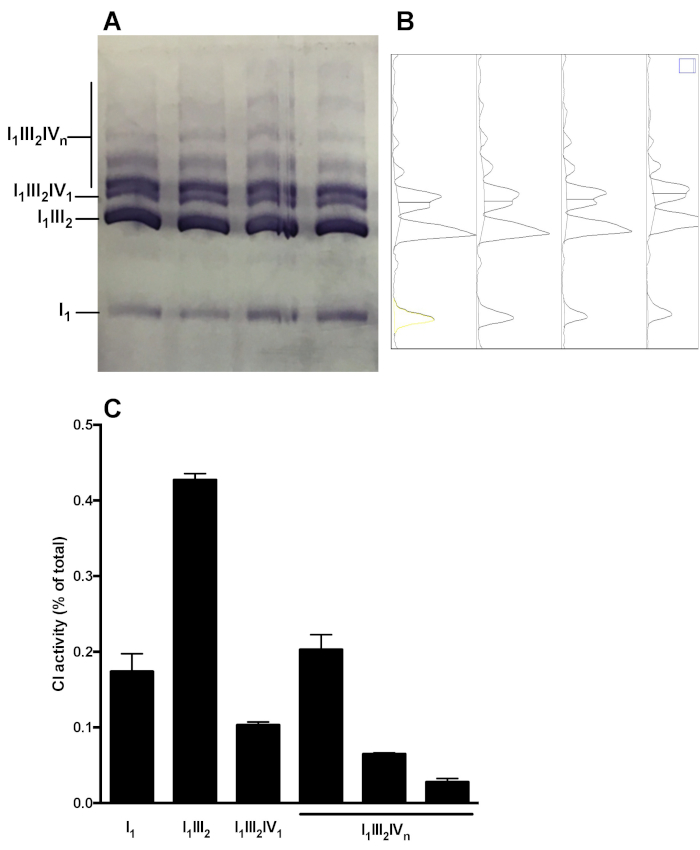
Figure 5: Quantification of CI distribution in monomeric and supramolecular assemblies. (A) CI in-gel activity determined following Hybrid CN/BN-PAGE of in liver mitochondria SC extracts obtained from 4 mice. (B) Densitograms obtained using ImageJ’s Gel Analysis Tool showing distinct peaks corresponding to CI monomers (I1) and various CI-containing supramolecular complexes (I1III2, I1III2IV1, and I1III2IVn). (C) quantification of the relative distribution of C1 activity. The data represent mean and SEM of the 4 mice. Please click here to view a larger version of this figure.
| Digitonin/protein ratio (g/g) | 2 g/g | 4 g/g | 6 g/g | 8 g/g |
| Volume of extraction buffer (µL) | 400 | 300 | 200 | 100 |
| Volume of 10 % stock digitonin (µL) | 100 | 200 | 300 | 400 |
| Total extraction buffer volume (µL) | 500 | 500 | 500 | 500 |
Table 1: Volumes required to extract SCs from 5 mg of mitochondrial proteins using various digitonin/protein ratios.
| Compound | Final Concentration |
| EDTA, pH 7.5 | 1 mM |
| HEPES | 30 mM |
| Potassium Acetate | 150 mM |
| Glycerol | 12% |
| 6-aminocaproic Acid | 2 mM |
Table 2: SC extraction buffer (final concentrations). Keep at 4 °C for a maximum of 3 months.
| Compound | Final Concentration |
| 3X Gel Buffer: Aliquot and keep at -20 °C, pH 7.5 | |
| Imidazole/HCl pH-7.0 | 75 mM |
| 6-aminocaproic Acid | 1.5 M |
| Acrylamide Buffer: Aliquot and keep at -20 °C | |
| Acrylamide | 99.5% |
| Bis-Acrylamide | 3% |
Table 3: Gel stock buffers.
| For 2 gels: | 4% (60 mL) | 12% (60 mL) | Stacking (4%) (25 mL) |
| 3X Gel Buffer | 19.8 mL | 19.8 mL | 8.25 mL |
| Acrylamide Buffer | 4.8 mL | 14.4 mL | 2 mL |
| H2O | 35 mL | 13.1 mL | 14.6 mL |
| Glycerol | – | 12 mL | – |
| APS 10% | 360 μL | 60 μL | 150 μL |
| TEMED | 24 μL | 12 μL | 10 μL |
Table 4: 4%–12% gradient gel.
| Compound | Final Concentration |
| Anode Buffer: Keep at 4 °C, pH 7.5 | |
| Imidazole | 25 mM |
| Cathode Buffer: Keep at 4 °C, pH 7.5 | |
| Tricine | 50 mM |
| Imidazole | 7.5 mM |
| With or without Coomassie Blue (G250) | 0.022% |
Table 5: Electrophoresis buffers.
| Compound | Final Concentration |
| Complex I activity Buffer: prepare fresh in 5 mM TRIS-HCl pH 7.4 | |
| Nitrotetrazolium blue | 3 mM |
| NADH | 14 mM |
| Complex II activity Buffer: prepare fresh in 5 mM TRIS-HCl pH 7.4 | |
| Succinate | 20 mM |
| PMSF | 0.2 mM |
| Nitrotetrazolium blue | 3 mM |
| Complex IV activity Buffer: prepare fresh in 50 mM Na-Phosphate pH 7.2 | |
| Cytochrome C | 0.05 mM |
| Diaminobenzidine | 2.3 mM |
| ATPsynthase activity Buffer: prepare fresh in water, adjust pH to 8 with KOH | |
| Glycine | 50 mM |
| MgCl2 | 5 mM |
| HEPES | 50 mM |
| CaCl2 | 30 mM |
| ATP | 5 mM |
Table 6: In-gel activity assay buffers.
| Compound | Final Concentration |
| Transfer Buffer | |
| Tris Base | 25 mM |
| Glycine | 192 mM |
| SDS | 4% |
| Methanol | 20% |
| TBST | |
| Tris Base | 20 mM |
| NaCl | 137 mM |
| Tween 20 | 0.1% |
Table 7: Immunoblotting buffers.
| Complex | Subunit | Clone |
| I | NDUFA9 | 20C11B11B11 |
| II | SDHA | 2E3GC12FB2AE2 |
| III | UQCRC2 | 13G12AF12BB11 |
| IV | COX4 | 1D6E1A8 |
| V | ATPB | 3D5 |
Table 8: Antibodies used for immunoblotting to detect respiratory chain SC. See Table of Materials for companies and lot numbers.
Discussion
Mitochondrial supercomplexes are being actively studied to elucidate their physiological role, and their importance in the pathogenesis of numerous human diseases, whether they are acquired or genetic mitochondrial diseases3,7,9,10,11,12,13,14. In order to obtain reliable results, several aspects need to be considered. This protocol has been tested with mouse liver mitochondria, mouse skeletal muscle mitochondria (results not shown), rat heart mitochondria, and human fibroblast mitochondria (results not shown), but could certainly be adapted to other sources of isolated mitochondria. The method optimally combines various aspects of BN and CN-PAGE protocols, which allow to reduce exposure to detergents and anionic compounds to a minimum compared to published protocols20,27,28.
Sample preparation
Sample preparation represents a crucial step for successful separation of SCs. Buffer composition should be carefully selected in order to achieve proper solubilization of proteins and proteins assemblies, while preserving as much as possible their functional and structural integrity. Ionic strength and pH of the extraction buffer are two important factors to consider. Salt concentrations that are too low (< 50 mM K-Acetate or NaCl) will result in poor solubilization of proteins in presence of non-ionic detergents, while salt concentrations above 500 mM will promote protein stacking/aggregation, and precipitation of CB and proteins29. SCs should therefore be extracted using buffers at near physiological ionic strength. With regards to pH, the use of a near physiological pH is recommended.
Detergent type and detergent/protein ratio are also critical for optimal SC extraction. For maximal preservation of native SCs, digitonin is preferred26. As shown in the present protocol and other published methods23,30,31,32, this mild detergent preserves the supramolecular composition of multiple SC assemblies, and the dimeric and oligomeric structure of ATPsynthase (Figure 3 and Figure 4). Titration of the samples of interest with various amounts of digitonin is critical in order to identify the conditions that allow optimal solubilization, while preserving enzyme activity and physiological protein interactions. Titration should be performed with ratios ranging between 2 and 8 g/g26. Optimal results for liver, skeletal muscle and cardiac mitochondria are respectively obtained with 4, 5, and 6 g digitonin/g protein. It should be noted that digitonin can be replaced by Triton X-100, which under optimal conditions results in similar migration and SC composition as those observed with digitonin2. However, this detergent should be used with caution, since relatively small increase in the detergent/protein ratio (e.g., from 1 to 1.5 g/g) can result in a complete dissociation of SCs assemblies2, which can result in experimental inconsistencies. After extraction, samples are traditionally supplemented with Coomassie Blue to give proteins a charge when applied to the gel, except for traditional CN-PAGE20,26. In order to minimize protein exposure to Coomassie blue and potential dissociation of labile proteins, samples are not supplemented with Coomassie blue in this protocol.
Electrophoresis
Both CN-PAGE and BN-PAGE have been used to study mitochondrial OXPHOS complexes, each of them having distinct advantages and limitations. The milder conditions used under CN-PAGE (mainly the absence of CB, which has a detergent-like effect), allows better preservation of ATP synthase in-gel activity, and limits the dissociation of labile proteins in high molecular weight SCs and ATP synthase assemblies26. However, the absence of the anionic dye CB in the protein extract and electrophoresis buffers causes the proteins to migrate based on their intrinsic charge and isoelectric point, which reduces the electrophoretic mobility of proteins within the gel26. Moreover, in the absence of CB, proteins with insufficient negative charge tend to aggregate, thus reducing the resolution of protein complexes in the gel20,26. To circumvent these limitations, the so-called high-resolution CN-PAGE has been developed by Wittig and Schragger20. In this protocol, sodium deoxycholate (DOC) and various mild non-ionic detergents (DDM, Triton X100) are added to the cathode buffer to keep membrane proteins solubilized and impose a negative charge shift on proteins, which results in a considerable improvement of resolution20.
A distinctive feature of the present hybrid CN/BN protocol is that a comparable resolution can be reached without these detergents. Momentary addition of CB to the cathode buffer at the beginning of the electrophoresis is sufficient to limit protein aggregation and enhance mobility in the gel (Figure 3 and Figure 4). As a result, this hybrid technique enables excellent resolution of distinct SC assemblies and very low or no exposure to detergents. The presence of low amounts of CB also allows better preservation of CV activity, improved preservation of dimeric and oligomeric CV assemblies (Figure 3 and Wittig and Schägger 200526), and a reduction of the blue background noise that can hinder the quantification of in gel activities, particularly for CII and CIV (Figure 2). Moreover, the absence of CB in the protein extract limits the disruption of labile protein interactions within SCs. For example, physical association of the ATP synthase with ANT to form the synthasome 33 or with Cyclophilin-D to regulate PTP opening 34 are better seen in absence of CB. Momentary exposure to CB during electrophoresis only may therefore be useful to reveal novel protein interactions within SCs. Overall, this hybrid CN/BN-PAGE protocol thus allows to combine precise and rapid in gel activity measurements with analytical techniques involving 2D electrophoresis, immuno-detection and/or proteomics for advanced analysis of SCs. It should be noted that with the growing interest for SCs, an increasing number of studies use small 10 x 10 cm gels for native PAGE. While this approach may be sufficient to identify gross changes in the abundance SC assemblies, the lower separation capacity of small gels is likely limited to resolve subtle rearrangements or to cut distinct bands for proteomic analysis. Moreover, several studies using smaller gels have reported that the respirasome migrates at the same size as the ATPsynthase dimer, making it difficult to dissociate them22. Therefore, the use of large gels should be favored.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank Jenna Rossi for technical assistance, and Dr. Mireille Khacho, Dr. David Patten and Dr. Ujval Anil Kumar for helpful discussion while developing this method. This work was funded by the Canadian Institutes of Health Research (CIHR) and the National Sciences and Engineering Council of Canada (NSERC). AC is a recipient of Doctoral Award – Frederick Banting and Charles Best Canada Graduate Scholarships (CIHR).
Materials
| 3,3'-Diaminobenzidine tetra-hydrochloride hydrate | Sigma | D5637 | |
| 6-amino caproic acid | sigma | A2504 | |
| Acrylamide | Sigma | A3553 | |
| Adenosine 5'-triphosphate disodium salt hydrate | sigma | A3377 | |
| Anti-ATPB antibody [3D5] – Mitochondrial Marker | Abcam | ab14730 | Lot number GR3174539-12, RRID AB_301438 |
| Anti-SDHA antibody [2E3GC12FB2AE2] | Abcam | ab14715 | Lot number GR3235943-1, RRID AB_301433 |
| Anti-UQCRC2 antibody [13G12AF12BB11] | Abcam | ab14745 | Lot number GR304308-3, RRID AB_2213640 |
| Bis N,N'-Methylene-Bis-Acrylamide | Biorad | 1610201 | |
| Brilliant Blue G | Sigma | 27815 | |
| COX4 Monoclonal Antibody (1D6E1A8) | Invitrogen | 459600 | Lot number TI2637158, RRID AB_2532240 |
| Cytochrome c from equine heart | sigma | C7752 | |
| Digitonin | Sigma | D141 | |
| Fisherbrand FH100M Multichannel Peristaltic Pumps | Thermo Fisher | 13-310-660 | |
| Imidazole | Sigma | I0250 | |
| Inner Glass Plates. Pkg of 2, 20 x 20 cm, glass plates for 20 cm PROTEAN II xi and PROTEAN II XL electrophoresis cells, use with outer plate | Biorad | 1651823 | |
| Model 485 Gradient Former. 40-175 ml acrylamide gradient former, includes valve stem and tubing kit, for use with Mini-PROTEAN multi-casting chamber systems | Biorad | 1654120 | |
| NDUFA9 Monoclonal Antibody (20C11B11B11) | Invitrogen | 459100 | Lot number TD2536591, RRID AB_2532223 |
| Nitrotetrazolium Blue chloride | Sigma | N6639 | |
| Outer Glass Plates. Pkg of 2, 22.3 x 20 cm, glass plates for 20 cm PROTEAN II xi and PROTEAN II XL electrophoresis cells, use with inner plate | Biorad | 1651824 | |
| Phenylmethanesulfonyl floride | Sigma | P7626 | |
| Powerpack 1000 | Biorad | Serial Number 286BR 07171 | |
| PROTEAN II Sandwich Clamps. Pkg of 2, clamps for running gels, for 20 cm PROTEAN II xi electrophoresis cell, 1 left and 1 right | Biorad | 1651902 | |
| PROTEAN II xi Cell. Large format vertical electrophoresis cell, 16 x 20 cm gel size, 4 gel capacity, spacers and combs | Biorad | 1651811 | |
| PROTEAN II xi Comb. Pkg of 1, 15-well, 1.5 mm, comb for PROTEAN II xi electrophoresis cell | Biorad | 1651873 | |
| PROTEAN II xi Multi-Gel Casting Chamber. Multi-gel casting chamber, 20 x 20 cm gel size, for up to ten 1.5 mm thick gels, includes sealing plate, gasket, separation sheets, acrylic blocks, PROTEAN II XL alignment cards | Biorad | 1652025 | |
| PROTEAN II xi Spacers. Pkg of 4, 1.5 mm, spacers for 20 cm PROTEAN II xi electrophoresis cell | Biorad | 1651849 | |
| SCIENCEWARE Utility Bags (10 x 12") 4 mil, Bel-Art, Box of 100 | VWR | 11215-388 | |
| Thick Blot Paper. Pkg of 25 sheets, 15 x 20 cm, absorbent filter paper, for use with Trans-Blot cassette | Biorad | 1703956 | |
| Trans-Blot Cell With Plate Electrodes and Super Cooling Coil. Transfer cell and cooling coil (#170-3912), includes 2 gel holder cassettes, buffer tank, lid with cables, fiber pads, 1 pack blot paper | Biorad | 1703939 | |
Referenzen
- Shagger, H. Respiratory Chain Supercomplexes. IUBMB Life. 52 (3-5), 119-128 (2001).
- Acin-Perez, R., Fernandez-Silva, P., Peleato, M. L., Perez-Martos, A., Enriquez, J. A. Respiratory active mitochondrial supercomplexes. Molecular Cell. 32 (4), 529-539 (2008).
- Greggio, C., et al. Enhanced Respiratory Chain Supercomplex Formation in Response to Exercise in Human Skeletal Muscle. Cell Metabolism. 25 (2), 301-311 (2017).
- Schagger, H., Pfeiffer, K. The ratio of oxidative phosphorylation complexes I-V in bovine heart mitochondria and the composition of respiratory chain supercomplexes. Journal of Biological Chemistry. 276 (41), 37861-37867 (2001).
- Milenkovic, D., Blaza, J. N., Larsson, N. G., Hirst, J. The Enigma of the Respiratory Chain Supercomplex. Cell Metabolism. 25 (4), 765-776 (2017).
- Acin-Perez, R., Enriquez, J. A. The function of the respiratory supercomplexes: the plasticity model. Biochimica et Biophysica Acta. 1837 (4), 444-450 (2014).
- Vartak, R., Porras, C. A., Bai, Y. Respiratory supercomplexes: structure, function and assembly. Protein Cell. 4 (8), 582-590 (2013).
- Lapuente-Brun, E., et al. Supercomplex Assembly Determines Electron Flux in the Mitochondrial Electron Transport Chain. Science. 340 (6140), 1567-1570 (2013).
- Lazarou, M., Smith, S. M., Thorburn, D. R., Ryan, M. T., McKenzie, M. Assembly of nuclear DNA-encoded subunits into mitochondrial complex IV, and their preferential integration into supercomplex forms in patient mitochondria. The FEBS Journal. 276 (22), 6701-6713 (2009).
- Sun, D., Li, B., Qiu, R., Fang, H., Lyu, J. Cell Type-Specific Modulation of Respiratory Chain Supercomplex Organization. International Journal of Molecular Sciences. 17 (6), (2016).
- D’Aurelio, M., Gajewski, C. D., Lenaz, G., Manfredi, G. Respiratory chain supercomplexes set the threshold for respiration defects in human mtDNA mutant cybrids. Human Molecular Genetics. 15 (13), 2157-2169 (2006).
- Antoun, G., et al. Impaired mitochondrial oxidative phosphorylation and supercomplex assembly in rectus abdominis muscle of diabetic obese individuals. Diabetologia. 58 (12), 2861-2866 (2015).
- Kanaan, G. N., Patten, D. A., Redpath, C. J., Harper, M. -. E. Atrial Fibrillation Is Associated With Impaired Atrial Mitochondrial Energetics and Supercomplex Formation in Adults With Type 2 Diabetes. Canadian Journal of Diabetes. , (2018).
- Kuter, K., et al. Adaptation within mitochondrial oxidative phosphorylation supercomplexes and membrane viscosity during degeneration of dopaminergic neurons in an animal model of early Parkinson’s disease. Biochimica et Biophysica Acta. 1862 (4), 741-753 (2016).
- McKenzie, M., Lazarou, M., Thorburn, D. R., Ryan, M. T. Mitochondrial Respiratory Chain Supercomplexes Are Destabilized in Barth Syndrome Patients. Journal of Molecular Biology. 361 (3), 462-469 (2006).
- Rosca, M. G., et al. Cardiac mitochondria in heart failure: decrease in respirasomes and oxidative phosphorylation. Cardiovascular Research. 80 (1), 30-39 (2008).
- Jang, S., et al. Elucidating Mitochondrial Electron Transport Chain Supercomplexes in the Heart During Ischemia-Reperfusion. Antioxidants & Redox Signaling. 27 (1), 57-69 (2017).
- Frenzel, M., Rommelspacher, H., Sugawa, M. D., Dencher, N. A. Ageing alters the supramolecular architecture of OxPhos complexes in rat brain cortex. Experimental Gerontology. 45 (7), 563-572 (2010).
- Krause, F. Detection and analysis of protein-protein interactions in organellar and prokaryotic proteomes by native gel electrophoresis: (Membrane) protein complexes and supercomplexes. Electrophoresis. 27 (13), 2759-2781 (2006).
- Wittig, I., Karas, M., Schagger, H. High Resolution Clear Native Electrophoresis for In-gel Functional Assays and Fluorescence Studies of Membrane Protein Complexes. Molecular Cell Proteomics. 6, 1215-1225 (2007).
- Wittig, I., Braun, H. P., Schagger, H. Blue native PAGE. Nat Protoc. 1 (1), 418-428 (2006).
- Jang, S., Javadov, S. Current Challenges in Elucidating Respiratory Supercomplexes in Mitochondria: Methodological Obstacles. Frontiers in Physiology. 9, 238-238 (2018).
- Cuillerier, A., et al. Loss of hepatic LRPPRC alters mitochondrial bioenergetics, regulation of permeability transition and trans-membrane ROS diffusion. Human Molecular Genetics. 26 (16), 3186-3201 (2017).
- Pallotti, F., Lenaz, G. . Methods in Cell Biology. 80, 3-44 (2007).
- Wieckowski, M. R., Giorgi, C., Lebiedzinska, M., Duszynski, J., Pinton, P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nature Protocols. 4, 1582 (2009).
- Wittig, I., Schägger, H. Advantages and limitations of clear-native PAGE. PROTEOMICS. 5 (17), 4338-4346 (2005).
- Jha, P., Wang, X., Auwerx, J. Analysis of Mitochondrial Respiratory Chain Supercomplexes Using Blue Native Polyacrylamide Gel Electrophoresis (BN-PAGE). Current Protocols in Mouse Biology. 6 (1), 1-14 (2016).
- Beutner, G., Porter, G. A. Analyzing Supercomplexes of the Mitochondrial Electron Transport Chain with Native Electrophoresis, In-gel Assays, and Electroelution. Journal of Visualized Experiments. (124), e55738 (2017).
- Von Hagen, J. . Proteomics Sample Preperation. , 485 (2008).
- Couoh-Cardel, S. J., Uribe-Carvajal, S., Wilkens, S., García-Trejo, J. J. Structure of dimeric F1F0-ATP synthase. The Journal of Biological Chemistry. 285 (47), 36447-36455 (2010).
- Strauss, M., Hofhaus, G., Schröder, R. R., Kühlbrandt, W. Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. The EMBO Journal. 27 (7), 1154-1160 (2008).
- Schägger, H., Pfeiffer, K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. The EMBO Journal. 19 (8), 1777-1783 (2000).
- Wittig, I., Schägger, H. Structural organization of mitochondrial ATP synthase. Biochimica et Biophysica Acta – Bioenergetics. 1777 (7), 592-598 (2008).
- Giorgio, V., et al. Dimers of mitochondrial ATP synthase form the permeability transition pore. Proceedings of the National Academy of Sciences of the United Stated of America. 110 (15), 5887 (2013).

