Combining Non-reducing SDS-PAGE Analysis and Chemical Crosslinking to Detect Multimeric Complexes Stabilized by Disulfide Linkages in Mammalian Cells in Culture
Summary
Disulfide linkages have long been known to stabilize the structure of many proteins. A simple method to analyze multimeric complexes stabilized by these linkages is through non-reducing SDS-PAGE analysis. Here, this method is illustrated by analyzing the nuclear isoform of dUTPase from the human bone osteosarcoma cell line U-2 OS.
Abstract
The structures of many proteins are stabilized through covalent disulfide linkages. In recent work, this bond has also been classified as a post-translational modification. Thus, it is important to be able to study this modification in living cells. A simple method to analyze these cysteine-stabilized multimeric complexes is through a two-step method of non-reducing SDS-PAGE analysis and formaldehyde cross-linking. This two-step method is advantageous as the first step to uncovering multimeric complexes stabilized by disulfide linkages due to its technical ease and low cost of operation. Here, the human bone osteosarcoma cell line U-2 OS is used to illustrate this method by specifically analyzing the nuclear isoform of dUTPase.
Introduction
Disulfide linkages have long been known to stabilize the structure of many proteins. In recent work, this bond has also been classified as a reversible post-translational modification, acting as a cysteine-based "redox switch" allowing for the modulation of protein function, location and interaction1,2,3,4. Thus, it is important to be able to study this modification. A simple method to analyze these cysteine-stabilized multimeric complexes is through non-reducing SDS-PAGE analysis5. SDS-PAGE analysis is a technique used in many laboratories, where the results can be obtained and interpreted quickly, easily, and with minimal costs, and is advantageous over other techniques used to identify disulfide linkages such as mass spectrometry6,7 and circular dichroism8.
One important step in determining if this method is an appropriate technique to aid in a study is to thoroughly examine the primary sequence of the protein of interest to insure there are cysteine residue(s) present. Another helpful step is to research any previous crystal structures published or use a bioinformatics application to explore the three dimensional structure of the protein of interest to visualize where the cysteine residue(s) may be located. If the residue(s) is present on the outside surface it may be a better candidate to form a disulfide linkage rather than a cysteine residue buried on the inside of the structure. However, it is important to note that proteins may undergo structural changes upon substrate interactions or protein-protein interactions allowing these residues to then become exposed to the environment as well.
Identified multimeric complexes can then be verified with chemical cross-linking using formaldehyde. Formaldehyde is an ideal cross-linker for this verification technique due to the high cell permeability and short cross-linking span of ~2-3 Å, ensuring detection of specific protein-protein interactions9,10. Here, this method is illustrated by analyzing the nuclear isoform of dUTPase from the human bone osteosarcoma cell line U-2 OS11. However, this protocol can be adapted for other cell lines, tissues and organisms.
Protocol
1. Blocking Free Cysteine Residues Using Iodoacetamide
- Grow U-2 OS cells in a 6 cm2 dish to 50%–60% confluency in minimum essential medium with high glucose containing 10% fetal bovine serum and 1% sodium pyruvate at 37 °C in 5% CO2.
- Make a fresh stock of 10 mM iodoacetamide just prior to use, then discard any unused reagent.
- Add 0.1 mM final concentration iodoacetamide directly to the cell culture media. Gently rock the dish at room temperature for 2 min.
2. Harvesting the Cells
- Aspirate the media from the cells.
- Wash the cells with 5 mL of cold phosphate-buffered saline (PBS) three times. Aspirate the final PBS wash solution then add 1 mL of cold PBS.
- Scrape the cells off the bottom of the dish using a cell scraper. Using a 1 mL pipette draw up the PBS and cell suspension and dispense all the liquid into a 1.5 mL microcentrifuge tube.
- Spin the cells at 7,500 x g for three min at 4 °C. Aspirate the PBS, leaving the cell pellet behind. The cell pellet can be stored at -80 °C until processing
3. Extraction of Protein
- Prepare 100 µL of a 1x lysis buffer diluted in ddH2O (See Table of Materials). Add phenylmethylsulfonyl fluoride (PMSF) immediately before use to a 1 mM final concentration.
- Add 50 µL of the 1x lysis buffer directly to cell pellet and suspend. Incubate this on ice for 5 min.
- Sonicate for 8 s using the constant pulse mode at 40% (see Table of Materials for sonicator; adjust as necessary for different sonicators), keeping the extract on ice.
- Spin the extract at 4 °C for 5 min at 16,000 x g. The supernatant is the soluble protein fraction. Bradford analysis can be performed to determine protein concentration if desired.
4. Sample Preparation
- Take 10 µL of the soluble protein extract and add 10 µL of a 1x Laemmli SDS-sample buffer (4% SDS, 20% glycerol, 0.004% bromophenol blue, and 0.125 M Tris-HCl, pH 6.8). Do not add any reducing reagents.
- Keep samples on ice if SDS PAGE analysis will be performed that day; for long term storage, -20 °C is appropriate. Right before running the gel, heat the sample for 5 min at 85 °C.
5. In Vivo Formaldehyde Cross-linking of Endogenous Proteins in U-2 OS Cells
- Grow U-2 OS cells in a 175 cm2 flask to 70%–80% confluency (see section 1).
- Perform the formaldehyde crosslinking reaction
- In a fume hood, aliquot a 37% formaldehyde solution purchased from commercial sources. Add the formaldehyde fixative directly to the medium to a final concentration of 1% and incubate at room temperature with gentle agitation for 15 min.
- To quench the reaction, add 1.25 M glycine to a final concentration of 0.125 M and incubate at room temperature with gentle agitation on a rocker for 5 min.
- Wash the cells with 5 mL of cold PBS three times. Aspirate the final PBS wash solution and add 10 mL of PBS. Scrape the cells off the bottom of the flask using a cell scraper.
- Using a 10 mL pipette, draw up the PBS and cell suspension and dispense all of the liquid into a 15 mL conical centrifuge tube. Spin the cells at 500 x g for 2 min at 4 °C. Aspirate the PBS, leaving the cell pellet behind.
6. Fractionation of Nuclei
- Prepare 10 mL of homogenization buffer: 0.25 M sucrose, 1 mM EDTA, 10 mM HEPES, and 0.5% BSA at pH 7.4. Add PMSF immediately before use to a 1 mM final concentration and 3mL of nuclei suspension buffer (0.1% Triton X-100 in PBS).
- Add 5 mL homogenization buffer directly to the cell pellet and suspend completely. Centrifuge the suspension at 500 x g for 2 min at 4 °C then discard the supernatant.
- Suspend the pellet in 5 mL of homogenization buffer. Use a tight-fitting glass-Teflon homogenizer, dounce homogenize cells at 10 strokes/500 rpm.
- Centrifuge the suspension at 1,500 x g for 10 min at 4 °C, then discard the supernatant.
NOTE: Use the supernatant for isolation of mitochondria by centrifuging at 10,000 x g for 10 min. - Suspend the pellet in 1 mL nuclei suspension buffer and incubate on ice for 10 min.
- Centrifuge at 600 x g for 10 min. Discard the supernatant.
- Suspend the pellet in 1 mL nuclei suspension buffer and centrifuge again. Discard the supernatant. The final pellet will be isolated nuclei.
7. Extraction of Protein
- Repeat section 4, except adding 25 µL of the 1x lysis buffer directly to the cell pellet then suspending.
8. Sample Preparation
- Prepare two samples for SDS-PAGE by taking 10 µL each of the soluble protein extract and add 10 µL of 2x Laemmli SDS-sample buffer and 1 µL of 2-Mercaptoethanol (BME).
- Heat one sample for 5 min at 37 °C and the second sample for 15 min at 98 °C to reverse the formaldehyde cross link.
9. SDS-PAGE Analysis
- Prepare 1 L of 1x Tris-glycine running buffer (25 mM Tris, 192 mM Glycine, 0.1% (w/v) SDS).
- Set up the SDS-PAGE running apparatus.
NOTE: This protocol uses a 16% precast TGX SDS-Page. Of note, any percentage gel can be used.- Per the manufacturer protocol, open the package the gel is stored in and remove the cassette.
- Remove the comb that is lining the wells and the tape from the bottom of the cassette.
- Place the gel into the running apparatus.
- Fill the chamber with the 1x running buffer until the wells are submerged in liquid. Using a plastic pipet, rinse out the wells with the running buffer.
- Load the samples on the gel along with 10 µL prestained standard marker.
- Run the gel at 200 V until the dye front is approximately 1 cm from the bottom of the gel.
10. Western Blot
- Prepare 1 L of 1x transfer buffer (25 mM Tris, 192 mM Glycine, 20% methanol) and store at 4 °C until use.
- Carefully remove the gel and open the cassette. Using a razor, carefully cut and discard the stacking gel. Pick up the gel using one corner and place it into a tray with transfer buffer, allowing it to rock gently for 5 min.
- While the gel is rocking, place an ice block (stored in -20 °C) into the transfer tank and add the transfer buffer to ¾ full.
- Wet the polyvinylidene fluoride (PVDF) membrane by soaking it in 100% methanol for 30 s.
- Once the 5 min have passed, prepare to set up the transfer in a tray with transfer buffer. The transfer buffer should be enough to completely submerge the blot. Set up the blot transfer as follows: at bottom, the bottom of the cassette holder (black); then a thick sponge, extra thick blotting paper, the polyacrylamide gel, a PVDF membrane, extra thick blotting paper, and a thick sponge; then finally the top of the cassette holder (white) at top.
- Lock the cassette and place the unit into the transfer tank with the PVDF membrane towards the positive anode and the gel towards the negative. Top off the unit with transfer buffer until your transfer is completely submerged.
- Place the unit on top of a stir plate. Add a stir bar to the unit and begin stirring at 125 rpm.
- Run at 100 V for 60 min.
- While the transfer is running, prepare a solution of 5% powdered milk dissolved in Tris-buffered saline, with Tween-20, pH 7.5 (TBST).
- Dismantle the unit and remove the membrane, noting which side was in contact with the gel; ensure this side stays up in the tray for the remaining steps.
- Soak the membrane for 2 min in Tris-buffered saline (TBS) followed by blocking the membrane by incubating in 5 mL of 5% milk-TBST solution for 30 min at room temperature.
- Replace the milk with a fresh 4 mL of 5% milk-TBST solution and add the primary antibody at the proper dilution (in this case, our dilution is 1:2,000 for the dUTPase antibody) and incubate over night with gentle rocking at 4 °C.
- The following day, remove the blot from the 4 °C rocker to a room temperature rocker, discarding the primary antibody solution.
- Do three quick washes with TBST, using enough liquid to completely submerge the blot, followed by 3 washes, 5 min each, rocking slowly.
- Following the last wash, add 5 mL of the 5% milk-TBST solution and rock the blot for 15 min then discard.
- Add secondary antibody diluted in 5% milk-TBST at the desired concentration. (In this case, 1:5,000 dilution of goat anti-rabbit diluted in 5 mL of 5% milk-TBST solution)
- Incubate at room temperature, rocking for 1 h, then discard the solution.
- Do three quick washes with TBST followed by 3 washes, 5 min each, rocking slowly, then discard the final wash and add 5 mL of TBS.
- Prepare a 1:1 mixture of an ECL chemiluminescent detection solutions (1 mL of each reagent for a final volume of 2 mL) and add this directly to the blot incubating for 1 min.
- Remove the membrane using tweezers and dab the corner with a laboratory wipe, removing any excess solution. Place the membrane in shrink wrap and place the protein side up in an x-ray cassette.
- Expose the membrane to x-ray film. The time of exposure will vary.
Representative Results
Nuclear dUTPase forms an intermolecular disulfide linkage forming a stable dimer configuration through the interaction of two cysteine residues positioned at the third amino acid of each monomeric protein11. This is demonstrated in Figure 1A,B. To ensure this disulfide linkage was not a nonspecific interaction due to migration abnormalities in the non-reduced environment,t the inclusion of a proper control was essential. Of note, nuclear dUTPase is one of four isoforms present in humans. Three of the four isoforms have a unique amino-terminal domain while sharing a common catalytic core11,12. As seen on the western blot, the monomeric confirmation of dUTPase in human cells at the time of harvesting is a combination of at least three of the isoforms of dUTPase (the mitochondria isoform, the nuclear isoform, and a truncated version notated at M24), all of which are recognized by our polyclonal antibody. The nuclear isoform is the only isoform that contains a cysteine residue in its unique amino terminal domain. Due to the nuclear isoform being only a small percentage of the monomeric state of the proteins seen on the western blot, it was exposed longer to demonstrate the dimeric state of that isoform.
The mitochondrial isoform lacks the cysteine residue present in nuclear isoform and was used as a control for this intermolecular disulfide linkage. As seen in Figure 1C, isolation of mitochondria followed by a western blot analysis demonstrated that this isoform under non-reducing conditions did not form a disulfide linkage and migrated to the predicted molecular weight for the monomeric protein.
To confirm this complex can form a multimeric complex, formaldehyde cross-linking was performed. Isolated nuclei were subjected to 1% formaldehyde treatment followed by denaturing SDS-PAGE/western blot analysis under reducing conditions. As demonstrated in Figure 2, the dimerization was visualized. When the cross-link was reversed by incubating the sample at 95 °C for 15 min, the complex was destabilized and could be visualized in its monomeric state.
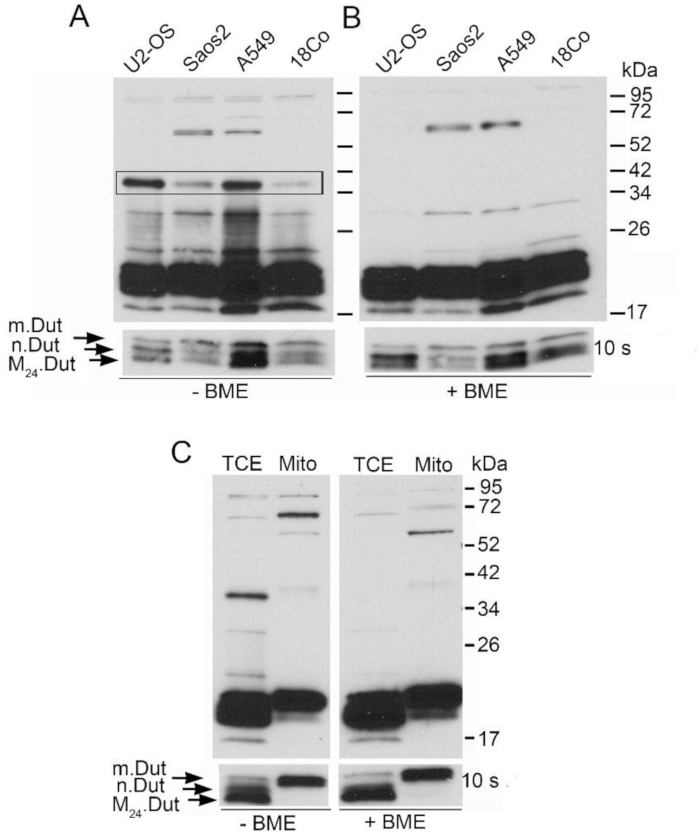
Figure 1: Demonstration of intermolecular disulfide bond formation in the nuclear dUTPase protein. (A) A western blot analysis of total cell extracts (TCE) in the absence of the reducing agent, beta-mercaptoethanol (BME), demonstrates a multimeric complex formation in asynchronous populations of U-2 OS, Saos2, A549, and 18CO as indicated by the black box. (B) This complex disappears with the addition of BME in all four cell lines examined, indicating the presence of a disulfide linkage. (C) A western blot analysis of TCE and purified mitochondrial extracts (Mito) derived from U-2 OS cells, ±BME, shows no multimeric complex formation in the -BME sample. The lower panels in panels A, B, and C demonstrate exposure to X-ray film for 10 s, showing the monomeric state of the three isoforms of dUTPase. The upper panels in A and B were exposed for 1 min, while the upper panel in C was exposed for 2 min. Equivalent amounts of protein were applied to each lane. Blots were probed with a polyclonal specific antibody against the conserved carboxyl-terminal domain of dUTPase. This figure has been modified from Rotoli et al.11 Please click here to view a larger version of this figure.
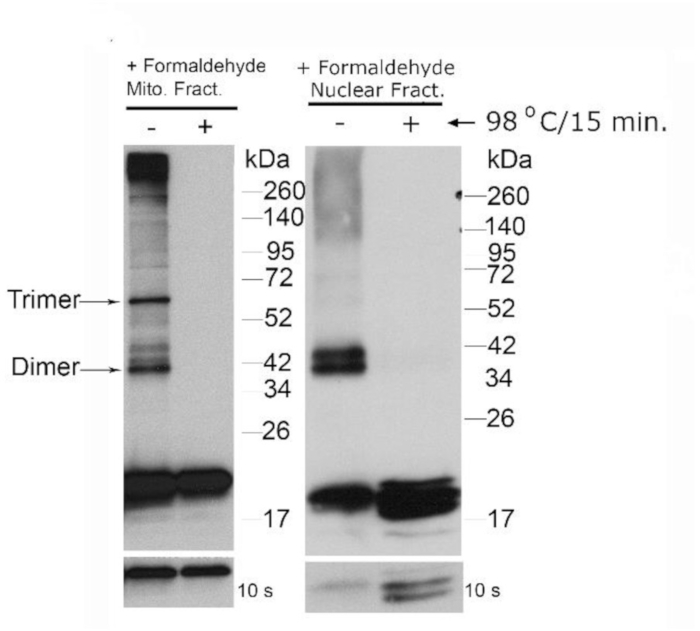
Figure 2: Formaldehyde cross-linking of nuclear dUTPase demonstrate multimeric complex formation. U-2 OS cells were incubated with 1% formaldehyde for 15 min. Nuclei (N) were isolated then analyzed by western blot using a specific polyclonal antibody against the conserved carboxyl-terminal domain of dUTPase (+ formaldehyde). To reverse the formaldehyde cross-links, extracts derived from the nuclear preparations were mixed with SDS-PAGE buffer then heated to 98 °C for 15 min in the presence of BME (+formaldehyde, 98 °C for 15 min). The observed heterogeneity (i.e. doublet bands of nDut) seen with the preparations remain to be explained, but may be due to anomalous migration due to the formaldehyde treatment. The lower panel is exposed to X-ray film for 10 s, while the upper panel is exposed to X-ray film for 2 min. This figure has been modified from Rotoli et al.11 Please click here to view a larger version of this figure.
Discussion
The method outlined here gives a straight-forward protocol for the analysis of multimeric complexes stabilized through disulfide linkages. This protocol can easily be adapted to other cell culture lines, tissues and organisms allowing for a broad range of applications.
An important step in this procedure is to ensure the disulfide linkages are not a consequence of the extraction procedure. Any free cysteine residues can be blocked using iodoacetamide13. This alkylating agent will bind covalently to cysteine residues though their thiol group, blocking the formation of new disulfide bonds. However, if the disulfide bond is present at the time of treatment this agent will not disrupt it. Optimization of the iodoacetamide can be done using a variation of concentration and time. However, over exposure to this reagent will cause cell death.
An additional step to this protocol that may be helpful is the optimization of the formaldehyde cross-linking. A variation of the percent of formaldehyde can be used as well as the time of cross-linking9,14. It is important to note that as the percent of formaldehyde as well as time increases, so do the chances of forming non-specific interactions. Downstream confirmation of the multimeric complex is also necessary. A useful technique to determine the molecular weight and identity, if the complex is a heteromultimeric, is mass spectrometry. Also, site directed mutagenesis can be a suitable technique to determine the cysteine residues responsible for the disulfide linkage.
Lastly, this two-step method of non-reduced SDS-PAGE analysis and formaldehyde cross-linking verification is beneficial due to its technical ease and low cost. It can be a first step in uncovering multimeric complexes stabilized by disulfide linkages.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
We gratefully appreciate the efforts put forth by Dr. Jennifer Fischer for the purification of the dUTPase polyclonal antibody and Kerri Ciccaglione for all her efforts to help edit this manuscript. This research was partially supported by a grant from the New Jersey Health Foundation (Grant #PC 11-18).
Materials
| 16% precast TGX gels | ThermoFisher | Xp00160 | |
| 175 cm2 Flask | Cell star | 658175 | |
| 18CO | ATCC | CRL-1459 | |
| 6 cm2 dish | VWR | 10861-588 | |
| A549 | ATCC | CCL-185 | |
| Amersham ECL detection kit | GE | 16817200 | |
| Blot transfer apparatus | Biorad | 153BR76789 | |
| BME | Sigma Aldrich | M3148 | |
| Bradford protein reagent | Biorad | 5000006 | |
| Bromophenol Blue | |||
| BSA | Cell signaling | 99985 | |
| Cell lysis buffer | Cell signaling | 9803 | |
| Centrifuge | Eppendor | 5415D | |
| DMEM | Gibco | 11330-032 | |
| Drill | |||
| EDTA | Sigma Aldrich | M101 | |
| Electrophoresis apparatus | Invitrogen | A25977 | |
| Extra thick western blotting paper | ThermoFisher | 88610 | |
| Fetal bovine serum | Gibco | 1932693 | |
| Formaldehyde | ThermoFisher | 28908 | |
| Glass-teflon homogenizer | |||
| Glycerol | Sigma Aldrich | 65516 | |
| Glycine | RPI | 636050 | |
| Heat block | Denville | 10285-D | |
| Hepes | Sigma Aldrich | H0527 | |
| Hydrochloric acid | VWR | 2018010431 | |
| Iodoacetamide | ThermoFisher | 90034 | |
| Kimwipe | Kimtech | 34155 | |
| Methanol | Pharmco | 339000000 | |
| Non-fat dry milk | Cell signaling | 99995 | |
| PBS | Sigma Aldrich | P3813 | |
| PMSF | Sigma Aldrich | 329-98-6 | |
| Posi-click tube | Denville | C2170 | |
| Power supply | Biorad | 200120 | |
| Prestained marker | ThermoFisher | 26619 | |
| PVDF membrane | Biorad | 162-0177 | |
| Rocker | Reliable Scientific | 55 | |
| Saos2 | ATCC | HTB-85 | |
| SDS | Biorad | 161-0302 | |
| Secondary antibody | Cell signaling | 70748 | |
| Small cell scraper | Tygon | S-50HL class VI | |
| Sodium chloride | RPI | S23020 | |
| Sodium pyruvate | Gibco | ||
| Sonicator | Branson | 450 | |
| Sponge pad for blotting | Invitrogen | E19051 | |
| Stir plate | Corning | PC353 | |
| Sucrose | Sigma Aldrich | S-1888 | |
| Tris Base | RPI | T60040 | |
| Tris Buffered Saline, with Tween 20, pH 7.5 | Sigma Aldrich | SRE0031 | |
| Tris-Glycine running buffer | VWR | J61006 | |
| Triton X-100 | Sigma Aldrich | T8787 | |
| Tween 20 | Sigma Aldrich | P9416 | |
| U-2 OS | ATCC | HTB-96 | |
| X-ray film | ThermoFisher | 34090 |
Referenzen
- Klomsiri, C., Karplus, P. A., Poole, L. B. Cysteine-based redox switches in enzymes. Antioxidants and Redox Signaling. 14 (6), 1065-1077 (2011).
- Antelmann, H., Helmann, J. D. Thiol-based redox switches and gene regulation. Antioxidants and Redox Signaling. 14 (6), 1049-1063 (2011).
- Brandes, N., Schmitt, S., Jakob, U. Thiol-based redox switches in eukaryotic proteins. Antioxidants and Redox Signaling. 11 (5), 997-1014 (2009).
- Groitl, B., Jakob, U. Thiol-based redox switches. Biochimica et Biophysica Acta. 1844 (8), 1335-1343 (2014).
- Stern, B. D., Wilson, M., Jagus, R. Use of nonreducing SDS-PAGE for monitoring renaturation of recombinant protein synthesis initiation factor, eIF-4 alpha. Protein Expression and Purification. 4 (4), 320-327 (1993).
- Gorman, J. J., Wallis, T. P., Pitt, J. J. Protein disulfide bond determination by mass spectrometry. Mass Spectrometry Reviews. 21 (3), 183-216 (2002).
- Li, X., Yang, X., Hoang, V., Liu, Y. H. Characterization of Protein Disulfide Linkages by MS In-Source Dissociation Comparing to CID and ETD Tandem MS. Journal of the American Society for Mass Spectrometry. , (2018).
- Kelly, S. M., Price, N. C. The use of circular dichroism in the investigation of protein structure and function. Current Protein and Peptide Science. 1 (4), 349-384 (2000).
- Klockenbusch, C., Kast, J. Optimization of formaldehyde cross-linking for protein interaction analysis of non-tagged integrin beta1. Journal of Biomedicine and Biotechnology. 2010, 927585 (2010).
- Gavrilov, A., Razin, S. V., Cavalli, G. In vivo formaldehyde cross-linking: it is time for black box analysis. Briefings in Functional Genomics. 14 (2), 163-165 (2015).
- Rotoli, S. M., Jones, J. L., Caradonna, S. J. Cysteine residues contribute to the dimerization and enzymatic activity of human nuclear dUTP nucleotidohydrolase (nDut). Protein Science. , (2018).
- Caradonna, S., Muller-Weeks, S. The nature of enzymes involved in uracil-DNA repair: isoform characteristics of proteins responsible for nuclear and mitochondrial genomic integrity. Current Protein and Peptide Science. 2 (4), 335-347 (2001).
- Macpherson, L. J., et al. Noxious compounds activate TRPA1 ion channels through covalent modification of cysteines. Nature. 445 (7127), 541-545 (2007).
- Carey, M. F., Peterson, C. L., Smale, S. T. Chromatin immunoprecipitation (ChIP). Cold Spring Harbor Protocols. 2009 (9), (2009).

