Use of Atomic Force Microscopy to Measure Mechanical Properties and Turgor Pressure of Plant Cells and Plant Tissues
Summary
Here, we present atomic force microscopy (AFM), operated as a nano- and micro-indentation tool on cells and tissues. The instrument allows the simultaneous acquisition of 3D surface topography of the sample and its mechanical properties, including cell wall Young’s modulus as well as turgor pressure.
Abstract
We present here the use of atomic force microscopy to indent plant tissues and recover its mechanical properties. Using two different microscopes in indentation mode, we show how to measure an elastic modulus and use it to evaluate cell wall mechanical properties. In addition, we also explain how to evaluate turgor pressure. The main advantages of atomic force microscopy are that it is non-invasive, relatively rapid (5~20 min), and that virtually any type of living plant tissue that is superficially flat can be analyzed without the need for treatment. The resolution can be very good, depending on the tip size and on the number of measurements per unit area. One limitation of this method is that it only gives direct access to the superficial cell layer.
Introduction
Atomic force microscopy (AFM) belongs to the scanning probe microscopy (SPM) family, where a tip with a radius of usually a few nanometers scans the surface of a sample. The detection of a surface is not achieved via optical or electron-based methods, but via the interaction forces between the tip and the sample surface. Thus, this technique is not limited to topographic characterization of a sample surface (3D resolution which can go down to a few nanometers), but also allows the measurement of any type of interaction forces such as electrostatic, van der Waals or contact forces. Furthermore, the tip can be used to apply forces at the surface of a biological sample and measure the resulting deformation, the so-called "indentation", in order to determine its mechanical properties (e.g., Young's modulus, viscoelastic properties).
Mechanical properties of plant cell walls are essential to be taken into account when trying to understand mechanisms underlying developmental processes1,2,3. Indeed, these properties are tightly controlled during development, in particular since cell wall softening is required to allow cells to grow. AFM can be used to measure these properties and study the way they change between organs, tissues or developmental stages.
In this paper, we describe how we use AFM to measure both cell wall mechanical properties and turgor pressure. These two applications are demonstrated on two different AFM microscopes and are detailed here after.
Protocol
1.Measure of Cell Wall Mechanical Properties
NOTE: Example of the developing gynoecium of Arabidopsis is presented.
- Preparation of the biological samples
- Collect a closed flower bud at stage 9 to 10 (about 0.5 mm long) according to published stages determination for Arabidopsis4. Under a binocular, using fine tweezers, carefully open the bud to check the stage of development and collect the gynoecium located at the center of the flower.
- Put the gynoecium on double side tape placed at the center of the cover of a small Petri dish (diameter of 5 cm).
NOTE: As an alternative, biocompatible glue can also be used for more efficient immobilization of the sample upon indentation. - Rapidly add water until the sample is completely covered. This avoids dehydration and reduces adhesion of the tip to the sample. Alternatively, submerge the sample in liquid medium, such as Arabidopsis apex culture medium5.
- AFM calibration
- Set the cantilever spring constant k high enough to allow the deformation of the sample surface up to the desired indentation, but not too high to avoid loss of sensitivity.
NOTE: As a rough rule of thumb, if Young's modulus of the sample is known, the order of magnitude of the spring constant can be chosen as k≈E * δ, where δ is the desired indentation. - Use an R = 400 nm spherical-ended tip with 15 µm tip-cantilever distance.
NOTE: The tip radius is directly related to lateral resolution. Generally, for indentation on biological materials, choose rounded tips (R greater than 10-20 nm) or colloidal probes. Small colloidal probes may be tricky to use due to the small distance between tip end and cantilever that can touch sample surface. - Switch on the software and place the head horizontally at least 2-3 h before the experiment: this will allow the head to thermalize and will avoid thermally-induced cantilever-laser relative movements. If the microscope is equipped with a CellHesion module (extending the available Z piezo range to 100 µm instead of 15 µm), switch on its controller first and then select CellHesion mode when starting the software.
- Mount the cantilever on the glass block and mount the block on the head. Place a droplet of a few microliters of ultrapure water on the tip to avoid the formation of air bubbles when the tip is dipped into water.
- Place a hard sample (cleaned glass slide or sapphire) and add 30-50 µL of ultrapure water.
NOTE: The calibration procedure described here is the sometimes-called contact calibration. First, a force curve is made on a stiff and flat surface and then the oscillation spectrum of the thermally excited cantilever is recorded in order to calculate the spring constant. Other calibration protocols exist and will briefly be described in the discussion paragraph. - Place the head on the stage (be careful to lift the Z motors high enough). Use the optical image to roughly place the laser on the cantilever.
- Move the laser along the main axis of the cantilever monitoring the sum signal on the photodiode.
NOTE: When standard cantilevers are used, a sum greater than 0.5 V should be obtained. - Move the laser along the other direction and maximize the sum signal in order to position the laser at the middle of the cantilever. This will minimize the cross-talk between lateral and vertical deflection.
- Move the laser along the main axis of the cantilever monitoring the sum signal on the photodiode.
- Measurement of the deflection sensitivity
NOTE: The photodiode reads laser displacement and provides a signal in volts. In order to be able to measure deflection in metric unit, deflection sensitivity must be measured.- Set the instrument in Contact → Force spectroscopy. Set a Relative setpoint to 2 V, Z length to 0.5 µm, and Extend speed to 2 µm/s (Sample rate to 10000 Hz) and select Z Closed loop.
- Open the Calibration manager and select the contact part of the force curve (that should be linear) to make a linear fit: the inverse of the slope gives the deflection sensitivity. The photodiode reading is now calibrated in metric unit.
- Determination of spring constant. In Calibration manager, select Spring constant to run a thermal spectrum acquisition. To average the signal for a longer time, select the ∞ symbol. The power spectrum of the thermally excited cantilever may show several peaks; draw a selection around the one placed at lowest frequency to fit it.
NOTE: If thermal tune is performed in liquid, the resonance peak will be broader and its frequency lowered compared to nominal one.
- Set the cantilever spring constant k high enough to allow the deformation of the sample surface up to the desired indentation, but not too high to avoid loss of sensitivity.
- Force spectroscopy experiment set-up and acquisition
- Place the sample on the AFM stage and place the head over the sample.
NOTE: Be sure the head has been retracted enough to avoid a hard contact between the tip and the sample surface. - Let the cantilever thermalize for a few minutes.
- In QI mode, approach with a Setpoint force of 50 nN.
- Set a Z length of 4 µm and a scan area to 80 x 80 µm2 with a number of pixels of 40 x 40. In the Advanced imaging settings panel, set the mode to constant speed. Set extend and retract speed to 200 µm/s and Sample rates to 25 kHz.
- Start scanning and use this rapid low-force scanning for checking if the sample moves. Verify that the scanned area is free from debris or deflected cells and locate a region of interest as flat as possible to perform the measurements.
NOTE: In order to appreciate the real sample tilt, the line levelling should be set to OFF or to Constant. An excessive tilt angle between the indentation axis and the surface will have an effect on the measured Young's modulus5. - Once an area of interest has been located, select a region of 40 x 40 to 60 x 60 µm2 around it and increase the pixel number to reach 2 pixels/µm. Increase the Setpoint to 500 nm to obtain 100-200 nm of indentation. Adjust this value, if necessary, at the beginning of the experiment. Decrease the Z length to 2 µm. Decrease extend and retract speed to 100 µm/s and increase the Sample rate to 50 kHz.
- Start scanning and save the output (generally composed by an Image and a Data file).
- At the end of the measurement day, remove the tip holder and rinse it gently with ultrapure water and 70% EtOH.
- Dry and remove the cantilever. For further experiments with the same tip, consider cleaning it with a wet-cleaning protocol and, if possible, a follow-up plasma O2 treatment. Do not let the water dry on the cantilever and/or tip holder to avoid salt crystallization.
- Place the sample on the AFM stage and place the head over the sample.
- Data Analysis (for 6.x Data Processing software version)
- Open Data Processing software and load the Data file.
- Click on Use this map for batch processing button in order to use the same analysis parameters on all the curves of the map.
- In Load pre-defined process, select Hertz fit.
- Use the first tab to verify or change the calibration parameters.
- In the second tab, remove an offset (or an offset plus a tilt) from the baseline to set its average value to 0.
- In the third tab, estimate the point of contact (POC) position by considering it as the first point crossing the 0 force when coming down from the setpoint value along the extend curve.
- The tab Vertical tip position calculates tip movement by subtracting the cantilever deflection from Height Measured. At this step, use Unsmoothed height (raw data) for the following fit, by checking the corresponding checkbox.
- In Elasticity fit tab, select the appropriate fit model. If no or weak adhesion is visible on retract curves (corresponding to less than 10% of the setpoint force or to the maximum average force at the selected indentation depth), the Model type should be set to Hertz/Sneddon and extend curve should be used. In case of stronger adhesion, DMT model, which stands for the Derjaguin-Muller-Toporov model, should be preferred and fit should be performed on retract curves (refer to manual for details on the available contact models and related formulae).
- Set tip geometrical parameters based on nominal tip shape. Here, Tip Shape is sphere and Tip Radius is 400 nm.
- Set the Poisson's Ratio to 0.5 as it is conventionally done for biological material (corresponding to incompressible material).
- Fit up to specific indentation. By default, the fit is performed over the whole curve. If the fit has to be done up to a specific indentation, first check the Shift Curve checkbox; this will shift the origin of the curve based on newly determined baseline and POC values.
- Add a second Elasticity fit routine by clicking on the icon on the main window.
- Set again all the fit parameters and specify in X min the desired indentation. Add as many steps as necessary (2 to 3 steps should be enough) in order to refine the determination of the POC position and subsequently the calculated indentation depth. The process can be saved at this point.
- Click on Keep and apply to all to iterate the previous steps on all the curves of the map.
- Save the results. An image and a .tsv files will be obtained.
2. Measure of Turgor Pressure
NOTE: An example of the oryzalin-treated inflorescence meristem of Arabidopsis is presented.
- Preparation of biological sample
- Collect Arabidopsis inflorescence meristem (IM) treated with the microtubule depolymerizing drug oryzalin from in vitro plantlet grown on medium containing the polar auxin transport inhibitor 1-N-Naphthylphthalamic acid (NPA) following published method6.
- Mounting IM sample following one of the two options
- For long-term monitoring: mount the sample in a Petri-dish containing Arabidopsis apex culture medium (ACM)7,8 and 0.1% plant preservation mixture (PPM), to prevent contamination. Suspend the IM tip above ACM surface and support the base of IM with a drop of 2% agarose.
- For measurement with rapid solution changes:mount the sample in a Petri-dish holding a small piece of adhesive mastic, and quickly seal the gap between the mastic and the sample base with bio-compatible glue. Wait for the glue to solidify (less than 2 min), then submerge the sample in liquid ACM containing 0.1% PPM.
NOTE: Make sure the sample surface is not coated with agarose or glue when fixing the sample base. AFM measurement of turgor pressure requires the sample to be stably mounted, and the previous mounting methods provide acceptable stability. Depending on the sample, other mounting methods might be used, like double-sided tape, poly-lysine, etc.
- AFM calibration
- Turn on the AFM acquisition software and choose the PeakForce QNM (large amplitude) measurement mode.
- Perform calibration following the same principle described in steps 2.1 to 2.6.
- In the Check parameters window, set Scan size to 0. In the Ramp window, set Ramp size between 200 nm and 500 nm, Trig threshold between 2 V and 5 V and Number of samples to 2048 or higher.
- Align the cantilever tip with the calibration sample and click Approach.
- Upon contact, go to Ramp window and click the button Continuous ramp. On the curve's linear regime, determine the slope by clicking on the Update Sensitivity button. Repeat the deflection sensitivity measurement several times and manually update the calibration with the measurement average by opening the Tab Detector from the menu Calibration.
- Retract the AFM head and remove the calibration sample.
NOTE: It is suggested to completely retract AFM head by choosing tab Stage → Initialize to prevent accidental hard contact upon sample change.
- Force spectroscopy experiment set-up and acquisition
- If the sample is not submerged yet, submerge it with liquid ACM containing PPM.
- In the acquisition software, specify measurement parameters as follows:
- In Check parameter window, set Spring constant to the cantilever's manufactured spring constant or the determined spring constant as in step 2.8. In this example, it is set at 42 N/m.
- Set Tip radius to 400 nm in this example.
- Set Sample Poisson's ratio to 0.5, since water contributes mainly to turgor pressure.
- Set Sample/Line to 128 to ensure rapid acquisition.
- Set Scan rate to 0.2 Hz.
- Set Scan size to 1 µm.
NOTE: Setting scan rate and scan size small may effectively prevent AFM scan-triggered sample damage. It is recommended reducing these two parameters upon any sample change. - In Ramp window, set Ramp size to 5 µm.
NOTE: It is better setting ramp size bigger than intended indentation depth for better baseline acquisition. - Set Trig threshold to maximum.
- Set Number of samples to 4608.
- Optionally, in Microscope → Engagement parameters tab, reduce the following parameters to prevent strong contact-induced sample damage. Set Peak Force Engage Setpoint to 0.3 V (default 0.5 V). Set Engage int. gain to 0.5 (default 0.75). Set SPM engage step to 4 µm (default 15 µm).
- Place and align the sample under the AFM head, and approach until cantilever is submerged but not in contact with the sample surface.
NOTE: While approaching, lightly blow on the surface of liquid ACM until a liquid bridge is formed between the cantilever and the liquid surface. This usually prevents hard contact. - With care, manually approach towards the sample. When the probe is relatively close to sample surface, click Approach.
- Upon contact, gradually increase Scan size and/or Scan rate until a desired balance without damaging the sample and/or the cantilever.
NOTE: The scan size is limited by the surface curvature and roughness of the sample. In the case of oryzalin-treated IM, 50 x 50 µm2 scan area can be achieved with scan rate of 0.3 Hz. - While scanning, determine if the measurement region is as desired. Relocate if needed. When satisfied, click the button Point and Shoot to initiate the point and shoot window.
- Before recording the scan, choose an appropriate image channel that may facilitate clear identification of cell contours. Often, Peak force error, DMT Modulus, LogDMT Modulus or Dissipation is suitable. Specify save directory and file name. Then click Ramp on next scan to initiate recording.
NOTE: A string of a dot and three numbers will be automatically added after your designated file name (like .000). This number automatically increases by 1 upon each saved scan repetition. - When scan is complete, the software interface will be automatically redirected to Ramp window. Click on the scanned image to specify the positions to indent.
NOTE: Before recording indentations, it is better choosing several landmarks to perform test indentations by clicking Ramp only, in case parameter alternation is required (for Ramp size, Piezo position, etc.). Indentation depth needs to be bigger than the cell wall thickness (ideally determined separately by transmission electron microscopy). - Choose at least three indentation sites per cell near its barycenter, and repeat indentation three times per site. This would yield at least nine force curves per cell for further analysis. When satisfied with indentation site placement, click Ramp and capture. The force-indentation curves are automatically saved to the designated directory.
- When the ramps are completed, relocate to a different position for tile measurement, or retract scan head and change sample.
- When finished, clean the cantilever as in steps 1.3.8 and 1.3.9.
- Data analysis
- In the analysis software, open the *.mca file. This shows the position of each force curve on the scanned image. If desired, pre-select force curves for analysis.
- Open one force curve to be analyzed, usually in the format of x0000y.00z, where x is the specified file name in the save directory while y and z are automatically registered numbers denoting indentation sequence and scan number.
- Click the Baseline correction button and drag the blue dash lines on the force curve until Extend Source Baseline Start and Extend Source Baseline Stop are at 0% and 80%, respectively. Click Execute.
NOTE: Alternatively, Extend Source Baseline Stop may be set at different values, as long as it is still within the baseline and not beyond the contact point. - Click the Boxcar Filter button and click Execute to smooth force curve.
- Click the Indentation button.
- In the Input window, set the Active Curve auf Extend.
- Set the Fit Method auf Linearized Model and Include Adhesion Force auf Yes.
- Set Max Force Fit Boundary to 99% and Min Force Fit Boundary to 75%.
- Set Fit Model auf Stiffness (Linear).
NOTE: This setup will compute the apparent stiffness k for turgor pressure deduction. In this example, the fit boundaries reflect a stiffness fit of around 1.5 µm indentation depth.
- Force curves can be analyzed in batch. Click the Run History button, specify report directory and add all other force curves that require the same treatment. When satisfied, click Run. By default, the fit will be stored as a *.txt file.
- When k is batch fitted, click History → 5 Indentation to return to the indentation window.
- Change Max Force Fit Boundary to 10% and Min Force Fit Boundary to 0%.
- Set Fit Model auf Hertzian (Spherical).
NOTE: This will compute the cell wall Young's modulus E for turgor pressure deduction. In this example, the fit boundaries reflect a Hertzian fit (using spherical probe) of around 0.4 µm indentation depth.
- Repeat step 2.4.6 for batch fit for E.
- Open the *.00z file (z is the automatically registered scan number) to display the different scan channels. In Höhe channel window, click Section button. This will allow the measurement of sample's surface curvature that is required for turgor pressure deduction.
- Draw a line across the long axis of one cell, move the dash line boundaries to the cell edges, and record the Radius value r1. Repeat this for the short axis to retrieve radius r2. Calculate cell's surface mean curvature κM and Gaussian curvature κG using the two radii measurement as follows:
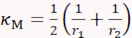 and
and 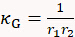
- Draw a line across the long axis of one cell, move the dash line boundaries to the cell edges, and record the Radius value r1. Repeat this for the short axis to retrieve radius r2. Calculate cell's surface mean curvature κM and Gaussian curvature κG using the two radii measurement as follows:
- Deduce turgor pressure P using the thin-shell model published9 as follows:
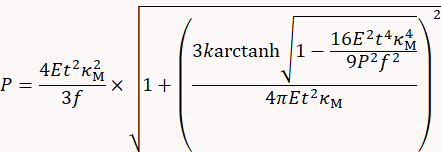
with
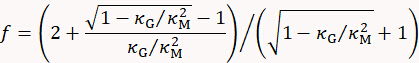
where t is cell wall thickness determined by e.g. electron microscopy.
NOTE: The deduction of turgor pressure is a fitting process, where iterations are required. Four iterations are generally able to reproduce stable product, however more iterations can be done (for example 100 times). - Calculate mean E, k and P per cell. Also, register the intracellular variability (e.g., standard deviation) for documentation.
Representative Results
Figure 1A and Figure 1B show a screenshot illustrating the result of the steps 1.3.4 to 1.3.6 of the protocol, used to locate a region of interest where to acquire the QI map. It is worth mentioning that the region of interest has been chosen in order not to be on a tilted surface (i.e., as flat as possible). Actually, as noticed by Routier et al.5, if the indentation axis is not perpendicular to the surface, the Young's modulus measured can be underestimated. This effect is visible on the upper-right corner of C or D panels of Figure 1, where part of the cells' border looks softer than the rest of the cell. The artefacts induced by this effect will be far stronger in the case of meristems, where the local tilt angle can easily overcome 40° over 50 x 50 µm2 scan areas. In our laboratory, we are currently developing an algorithm, based on the approach described by Routier et al.5, to correct those artefacts (paper in preparation). Figure 1C and Figure 1D show Young's modulus maps obtained after the analysis detailed in the fourth paragraph of the protocol. In particular, panel C represents the Young's modulus obtained analyzing the whole indentation, up to the user-defined force setpoint, while panel D shows the result of the analysis of the first 100 nm of indentation (as described in step 1.4.9). Here, the 2 maps look highly similar, because during the experiment set-up, the setpoint has been chosen in order to obtain average indentations around 100 nm. The variation of the analyzed indentation can sometimes lead to better highlight sample heterogeneities, that can be helpful for identifying the location or for providing information on the behavior of internal structures (e.g., Costa et al.10)
The force curve in Figure 2 shows two effects that are worth mentioning. First of all, it can be noticed that if the approach part of the force curve actually ends at the setpoint force of 500 nN, the downward tip movement keeps on going, meaning the final force applied by the tip is higher than expected (here 1,000 nN). This is due to the fact that the feedback loop monitoring the cantilever deflection does not act instantaneously, so its finite reaction time introduces a lag between deflection threshold detection and piezo movement stop. Furthermore, especially when using CellHesion which moves the sample holder, inertia of the moving parts comes into play, making this time lag longer. This issue can be limited by using the head piezo, in which case the user must be really careful in the case of measurements on very rough or tilted samples, or by reducing the ramp velocity, which anyway limits the resolution and/or the number of samples scanned (furthermore for too slow maps, the sample growing can become an issue). Generally, if the force curves are far enough from one another (based on the indentation model of choice, the radius of the indented area can be calculated given the indentation depth and used as minimum separation distance), the measurements performed in different positions along the sample should not be correlated and if the analysis is performed on the approach curve, overcoming the force threshold should not be an issue. Anyway, if the sample is delicate or if repeated scans are done on the same area, the scientist may want to try to minimize this overshoot. Unfortunately, there is not a rule of thumb to determine the amount of this overshoot, since it depends on the piezo used, on the ramp velocities, but also on material properties: the stiffer the material, the faster the variation of the deflection signal in time and, given a finite feedback response time, the higher the overshoot.
The second thing to notice is the waving on retract curve highlighted by the ellipse. Such waving can be an indicator of sample moving/vibrating under the action of the tip. In this case, the user can try to change the scanned area (sometimes other part of the microscope but the tip can touch the sample, or the sample can be insufficiently well fixed to its support) or the sample if several of them have been prepared on the same support. If the feature is always there, it is better to change the fixation method, in this case switching from the double-sided tape fixation to biocompatible glues.
Figure 3 depicts the determination of key parameters for turgor pressure deduction. Every parameter, except for cell wall thickness11 which was separately determined by electron microscopy to be ~740 nm, can be retrieved from AFM scans and indentations. Following the fitting process in step 2.4.10, the turgor pressure is deduced to be 1.50 MPa for this force curve. Pooling all eighteen force curves in this cell yields average values of E = 6.77 ± 1.02 MPa, k = 14.18 ± 2.45 N/m and P = 1.45 ± 0.29 MPa (mean ± standard deviation).
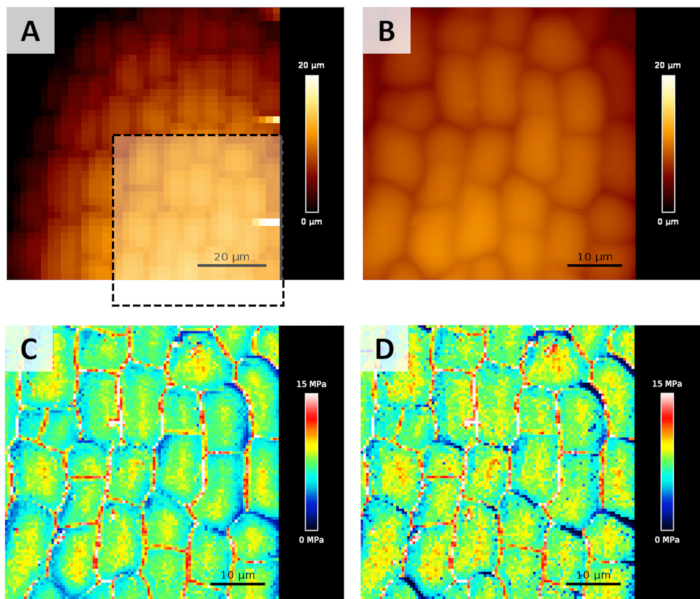
Figure 1: Topography maps (Height Measured) typically acquired during an experiment. (A) A first low resolution/low force map is recorded to find a suitable region to scan. (B) A smaller high resolution/higher force map is acquired for the calculation of Young's modulus (on the area highlighted by the black dotted square). (C) Young's modulus maps calculated from B map, as detailed in step 1.4. Here, the Young's modulus map has been obtained analyzing the whole indentation on each force curve. (D) Young's modulus map obtained analyzing only the first 100 nm of indentation. Please click here to view a larger version of this figure.
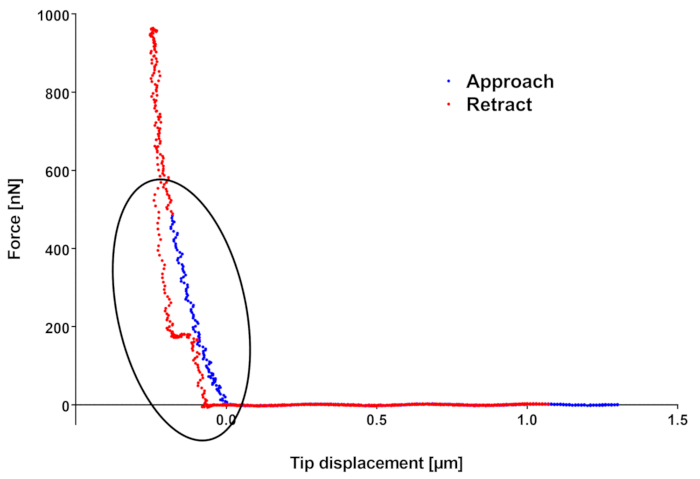
Figure 2: Example of force curve. In blue the approach, in red the retract segment. Force maximum (located on retract curve) is different from the force setpoint because of finite feedback loop reaction time and because of inertia. The waving in retract curve can be representative of a deficiency in sample fixation to its support. Please click here to view a larger version of this figure.
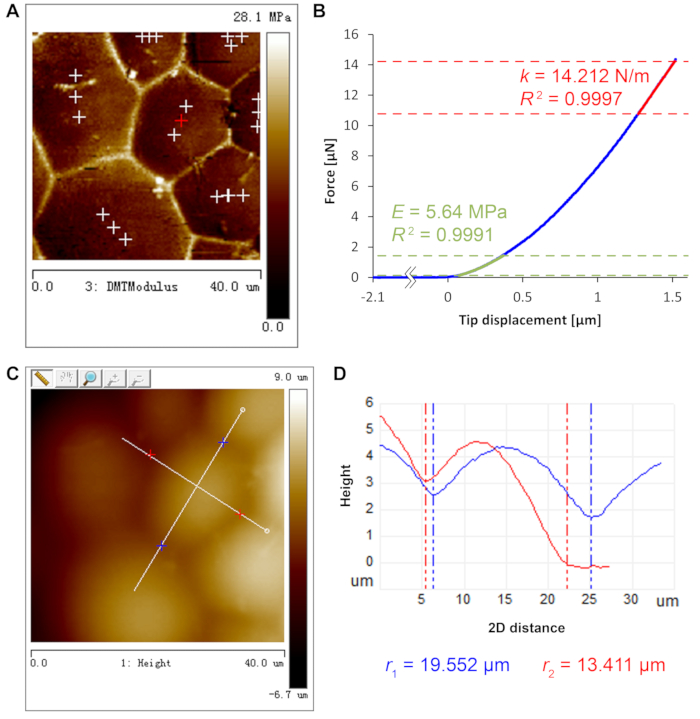
Figure 3: AFM measurements for turgor pressure deduction. (A) DMT modulus map shows clear cell contour to supervise the positioning of deep-indentation sites near cell barycenter. Indentation sites are marked by crosses. (B) Force curve of deep indentation at the red cross position in A. Cell wall Young's modulus E (green) and sample apparent stiffness k (red) are fitted at different regimes of the curve as detailed in step 2.4. R2 denotes coefficient of determination of the fits. (C) Height map with manually determined long and short axes of a, depicted by white lines, of the same cell analyzed in panels A and B. (D) Height profile of the long and short axes of the cell in Panel C (blue, long axis red, short axis) and the fitted radii of curvature (r1 and r2). Please click here to view a larger version of this figure.
Discussion
The emergence of shapes in plants is mainly determined by the coordinated rate and direction of growth during time and space. Plant cells are encased in a rigid cell wall made of a polysaccharidic matrix, which glues them together. As a result, cell expansion is controlled by the equilibrium between turgor pressure pulling on the cell wall, and stiffness of the cell wall resisting to this pressure. In order to understand the mechanisms underlying development, it is important to be able to measure both cell wall mechanical properties as well as turgor pressure in different tissues or cells of a given organ. As demonstrated in this paper, AFM is the method of choice in this context.
There are several critical steps in the protocol. The first one is the tissue preparation which should be fast enough to avoid dehydration. This can be critical in particular if we want to measure turgor pressure. Very important is also a correct fixation of the sample to its substrate: an unstable sample can lead to very obvious effects like a macroscopic movement that can be easily detected optically, or a strong deformation of force curves. Anyway, sample vibration or bending can be subtle to detect, introducing artefacts into the results. Finally, another critical step concerns the choice of the indentation parameters. This is determinant for the quality of the results.
When measuring mechanical properties on large areas (over 40-50 µm scan sizes), height differences can be high, making it difficult to properly track the surface and realizing force curves without reaching the limits of Z piezo range. In order to overcome this issue, a CellHesion module has been used. This module adds an additional Z piezo, located into the sample stage, having a 100 µm range, which allows large areas acquisition without approaching dangerously Z piezo limits.
The first limitation of the method is that we can only measure superficial cell layers. However, recently authors have used AFM on tissue sections12,13,14. Although this must be done on fixed material, it can give access to mechanical properties of deep cell layers. Alternatively, deeper indentations have also been performed to infer mechanical properties of internal tissues on living samples, albeit with bigger sacrifice on spatial resolution15. A second limitation can be linked to sample geometry, as a surface with a steep slope can be problematic since the tip is no more perpendicular to the sample. We need to restrict the range of angles within which measures are reliable and we are currently developing an algorithm to correct potential artefacts linked to this phenomenon. Also, some tissues are covered with trichomes which can block the measurements. Finally, indentation measures mechanical properties in a perpendicular direction regarding the cell surface and we can wonder if this can be easily linked to cell wall extensibility associated to growth capacity. Several studies suggest it is the case15,16.
It is worth doing here a more general observation about the measurement/application of forces by AFM. As described in the protocol section, in order to be able to transform laser displacements on the photodiode to forces, a calibration has to be performed. In this calibration one parameter, the deflection sensitivity, allows to convert laser displacements to actual cantilever deflection (generally measured in nm), so this factor calibrates photodiode output voltages (along the vertical axes) to displacements in nm. The second factor is the spring constant K, measured in N/m, which converts vertical cantilever deflections in force units (generally nN), since the flexible cantilevers behave like springs. Even if the procedure appears simple, a robust and reproducible calibration of K for AFM force spectroscopy measurements is still an issue, because of different error sources that can affect the different steps of the procedure. For example, the measurement of the deflection sensitivity realizing a force curve on a stiff surface, can lead to incorrect values if the tip slips on the surface, or if the surface is not perfectly cleaned or also if surface charge induce electrostatic repulsion. In all of the previous cases, the contact part of the force curve will be deformed (further tilted or curved instead of linear).
The contact procedure described in the protocol section is only one of several existing calibration procedures. There are at least 2 more methods that can be used in a relatively easy and inexpensive way: the Sader method (also called non-contact when implemented in some AFM software) or the reference cantilever method. Sader method17 has been originally developed by John Sader for V-shaped cantilevers and then for rectangular ones18 and does not need the acquisition of a force curve on a stiff substrate. Instead, the user needs to specify (out of the temperature as in the contact method), the length and width of the cantilever and the density and viscosity of the fluid where the cantilever is (generally air, water or a water-like fluids). Then a thermal spectrum is acquired and both deflection sensitivity and spring constant are measured. This method is advantageous when using very sharp or functionalized tips that may be damaged doing a force curve on a stiff substrate.
Both the previous methods relate to the acquisition of the oscillation spectrum of a thermally-excited cantilever. When stiff cantilevers are used at room temperature, the mean oscillation amplitude can be lower than 0.1 nm, making the resonance peak smaller and more difficult to detect. Anyway, at least for both instruments used in this paper, the resonance peak can actually be detected in air and in water and fitted for measuring the spring constant (considering that when the measurement is done in liquid, the resonance frequency shifts toward lower values and the peak broadens).
A third method doesn't use a thermal tune, but instead a reference cantilever or a reference elastic structure, with a known spring constant (generally with an uncertainty around or below 5%), in order to determine the cantilever K. In this case a first force curve needs to be acquired on a stiff substrate in order to measure the deflection sensitivity (which comes again along with all possible artefacts affecting the shape of curve). Then the stiff substrate is replaced by the reference cantilever (generally a tip-less cantilever with a calibrated spring constant) and another (or few others) force curve is performed, recording again the value of the deflection sensitivity. The cantilever spring constant is then calculated as:
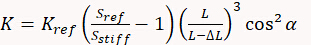
where Kref is the spring constant of the reference cantilever, Sref the deflection sensitivity measured on it, L is its length and Sstiff is the deflection sensitivity measured on the stiff substrate. ΔL is the offset between the tip and the end of the reference cantilever and depends on the alignment done by the user and finally α is the tilt angle of the cantilever, specified by the AFM manufacturer. The same method can be used on elastic structures specifically designed for cantilever or indenter calibration. The advantage of those structures (that are generally circular) is that K at their center is constant and doesn't depend on any geometrical parameter or on the precise positioning of the AFM tip on them, so removing L and ΔL from the previous formula.
The AFM user has to keep in mind that all those calibration methods are affected by different error sources (deflection sensitivity determination in first and third, the use of thermal tune in the case of stiff cantilevers for first and second and geometrical parameters and alignment for the third as well as calibration accuracy of Kref in the case reference cantilever is used) and that contact methods are potentially harmful for the tip (especially the third, where calibration imposes the use of two different samples). This implies that the spring constant will always be determined up to a certain precision and so will be the measured/applied forces (see Sikora19 for a comprehensive review of different calibration methods and their precision. This means that Young's moduli and turgor pressures will also be affected by cantilever calibration. Anyway, other even more important error sources are involved when calculating those quantities (like the use of simplified models for Young's modulus determination) so it will always be a challenge to obtain absolute values out of those kinds of measurements. What is important is to obtain values that are of the right order of magnitude and that are coherent with each other, meaning that if the experiment is repeated by the same user on the same sample, the values should be compatible. In order to obtain it, the calibration must be done carefully, and error sources must be minimized (for example limiting as much as possibly acoustic noise/vibrations). One possible strategy focused to increasing the coherence of repeated calibrations has been recently proposed in a paper by Schillers et al.20, where thanks to the collaboration of 11 European labs, a protocol (called SNAP) to compensate for errors in deflection sensitivity determination has been developed. In this paper the authors use directly calibrated cantilevers for their measurements, anyway the same protocol can be applied to non-calibrated cantilevers, simply calibrating them the first time with the method of choice and then considering the first spring constant as the reference ("calibrated") value.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
We would like to thank the PLATIM team for their technical support, as well as Arezki Boudaoud and members of the Biophysic team at the RDP lab for helpful discussions.
Materials
| Growth medium | |||
| 1000x vimatin stock solution | used to make ACM, composition see Stanislas et al., 2017. Add to ACM after autoclaving, before pouring. | ||
| 1-N-Naphthylphthalamic acid (NPA) | Sigma-Aldrich/Merck | 132-66-1 | add to Arabidopsis medium, 10 μM. Add after autoclaving, before pouring. |
| agar-agar | Sigma-Aldrich/Merck | 9002-18-0 | add to Arabidopsis medium, 1% w/v. |
| agarose | Merck Millipore | 9012-36-6 | used to make solid ACM, 0.8% w/v. |
| Arabidopsis medium | Duchefa Biochimie | DU0742.0025 | For in vitro arabidopsis culture, 11.82g/L. |
| Calcium nitrate tetrahydrate | Sigma-Aldrich/Merck | 13477-34-4 | add to Arabidopsis medium, 2mM. |
| MURASHIGE & SKOOG MEDIUM | Duchefa Biochimie | M0221.0025 | Basal salt mixture, used to make ACM, 2.2g/L. |
| N6-benzyladenine (BAP) | Sigma-Aldrich/Merck | 1214-39-7 | used to make ACM, 555 nM. Add to ACM after autoclaving, before pouring. |
| oryzalin | Sigma-Aldrich/Merck | 19044-88-3 | for oryzalin treatement, 10 μg/mL. |
| plant preservation mixture (PPM) | Plant Cell Technology | used to make ACM, 0.1% v/v. Add to ACM after autoclaving, before pouring. | |
| Potassium hydroxide | Duchefa Biochimie | 1310-58-3 | used to make Arabidopsis medium and ACM, both pH 5.8. |
| sucrose | Duchefa Biochimie | 57-50-1 | used to make ACM, 1% w/v. |
| Tools for AFM | |||
| BioScope Catalyst BioAFM | Bruker | The AFM used for turgor pressure measurement in this protocol. | |
| Nanowizard III + CellHesion | JPK (Bruker) | The AFM used for measuring mechanical properties. | |
| Patafix | UHU | D1620 | |
| Reference elasitic structure | NanoIdea | 2Z00026 | |
| Reprorubber-Thin Pour | Flexbar | 16135 | biocompatible glue. |
| Spherical AFM tips | Nanoandmore | SD-SPHERE-NCH-S-10 | Tips used for measuring mechanical properties. |
Referenzen
- Du, F., Guan, C., Jiao, Y. Molecular mechanisms of leaf morphogenesis. Molecular Plant. 11, 1117-1134 (2018).
- Cosgrove, D. J. Growth of the plant cell wall. Nature Reviews Molecular Cell Biology. 6, 850-861 (2005).
- Dumais, J. Can mechanics control pattern formation in plants?. Current Opinion in Plant Biology. 10, 58-62 (2007).
- Smyth, D. R., Bowman, J. L., Meyerowitz, E. M. Early flower development in Arabidopsis. The Plant Cell. 2, 755-767 (1990).
- Routier-Kierzkowska, A. L., et al. Cellular force microscopy for in vivo measurements of plant tissue mechanics. Plant Physiology. 158 (4), 1514-1522 (2012).
- Corson, F., et al. Turning a plant tissue into a living cell froth through isotropic growth. Proceedings of the National Academy of Sciences of the United States of America. 106, 8453-8458 (2009).
- Hervieux, N., et al. A mechanical feedback restricts sepal growth and shape in Arabidopsis. Current Biology. 26, 1019-1028 (2016).
- Stanislas, T., Hamant, O., Traas, J., Lecuit, T. Chapter 11 – In-vivo analysis of morphogenesis in plants. Methods in Cell. 139, 203-223 (2017).
- Beauzamy, L., Derr, J., Boudaoud, A. Quantifying hydrostatic pressure in plant cells using indentation with an atomic force microscope. Biophysical Journal. 108 (10), 2448-2456 (2015).
- Costa, K. D., Sim, A. J., Yin, F. C. P. Non-Hertzian Approach to Analyzing Mechanical Properties of Endothelial Cells Probed by Atomic Force Microscopy. Journal of Biomechanical Engineering. 128 (2), 176-184 (2006).
- Beauzamy, L., Louveaux, M., Hamant, O., Boudaoud, A. Mechanically, the shoot apical meristem of Arabidopsis behaves like a shell inflated by a pressure of about 1MPa. Frontiers in Plant science. 6 (1038), 1-10 (2015).
- Majda, M., et al. Mechanochemical polarization of contiguous cell walls shapes plant pavement cells. Developmental Cell. 43 (3), 290-304 (2017).
- Torode, T. A., et al. Branched pectic galactan in phloem-sieve-element cell walls: implications for cell mechanics. Plant Physiology. 176, 1547-1558 (2018).
- Farahi, R. H., et al. Plasticity, elasticity, and adhesion energy of plant cell walls: nanometrology of lignin loss using atomic force microscopy. Scientific Reports. 7, 152 (2017).
- Peaucelle, A., et al. Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Current Biology. 21, 1720-1726 (2011).
- Cosgrove, D. J. Diffuse growth of plant cell walls. Plant Physiology. 176, 16-27 (2018).
- Sader, J. E., Larson, I., Mulvaney, P., White, L. R. Method for the calibration of atomic force microscope cantilevers. Review of Scientific Instruments. 66 (7), 3789-3798 (1995).
- Sader, J. E., Chon, J. W. M., Mulvaney, P. Calibration of rectangular atomic force microscope cantilevers. Review of Scientific Instruments. 70 (10), 3967-3969 (1999).
- Sikora, A. Quantitative Normal Force Measurements by Means of Atomic Force Microscopy Towards the Accurate and Easy Spring Constant Determination. Nanoscience and Nanometrology. 2 (1), 8-29 (2016).
- Schillers, H., et al. Standardized Nanomechanical Atomic Force Microscopy Procedure (SNAP) for Measuring Soft and Biological Samples. Scientific Reports. 7 (1), (2017).

